Abstract
The changeabilities of individual modules of aminoacyl-tRNAs are poorly understood, despite the relevance for evolution, translational accuracy and incorporation of unnatural amino acids. Here, we dissect the effect of successive changes in four domains of Ala-tRNA3Ala on translation in a purified system. Incorporating five amino acids, not one, was necessary to reveal major effects on yields of peptide products. Omitting tRNA modifications had little affect, but anticodon mutations were very inhibitory. Surprisingly, changing the terminal CCA to CdCA was sometimes inhibitory and non-cognate amino acids were sometimes compensatory. Results have implications for translational fidelity and engineering.
Keywords: tRNAAla, purified translation, protein synthesis, peptide, unnatural amino acid, Escherichia coli
1. Introduction
Aminoacyl-tRNAs (AA-tRNAs) contain four modular domains with very precise boundaries: an AA, an invariant 3'-terminal CCA, a three-base anticodon and a tRNA body. Domain swaps occurred extensively in evolution [1,2] and are also important for translational fidelity and engineering. Most studies of anticodon swaps have focused on nonsense suppressor tRNAs, which usually function less efficiently in translation than other tRNAs [3]. The misacylated tRNAs characterized best in translation are the natural precursors Asp-tRNAAsn and Glu-tRNAGln, where low affinities for elongation factor Tu (EF-Tu) inhibit delivery to the ribosome, thus preserving translational accuracy [4]. Many unnatural AAs have been incorporated with moderate efficiencies in translation using nonsense suppressor [5] and sense suppressor [6-10] tRNAs.
The limited understanding of the interchangeability of individual domains of AA-tRNAs in translation is mostly due to experimental challenges. One challenge is making single domain changes in a tRNA. For example, an anticodon change frequently also changes the AA charged because anticodons are major positive determinants for AA-tRNA synthetases [11]. Another challenge is interpreting results in in vivo and crude in vitro translation systems: effects may occur at the level of transcription, pre-tRNA processing, tRNA modification, AA charging and/or translation. In order to circumvent these challenges, we combined two technologies: a simplified, purified, E. coli translation system lacking AA-tRNA synthetases [12] and chemoenzymatic synthesis of AA-tRNAs [5,13]. The latter technology has the advantage of enabling independent switching of AA and anticodon, but it also generally introduces three additional unnatural changes: a penultimate deoxyribose linkage for ease of chemical synthesis, and the omission of tRNA modifications (Fig. 1) combined with small changes in 5’- and 3’-terminal sequences for ease of tRNA preparation by in vitro transcription. In model cases, these three additional types of changes had little effect on incorporation yields of single AAs [5,14,15]. However, significantly lower product yields were obtained when incorporating three to five unnatural L-AAs in a row using tRNAAsn- and tRNAPhe-based synthetic adaptors (tRNAAsnB and tRNAPheB; [6,16]).
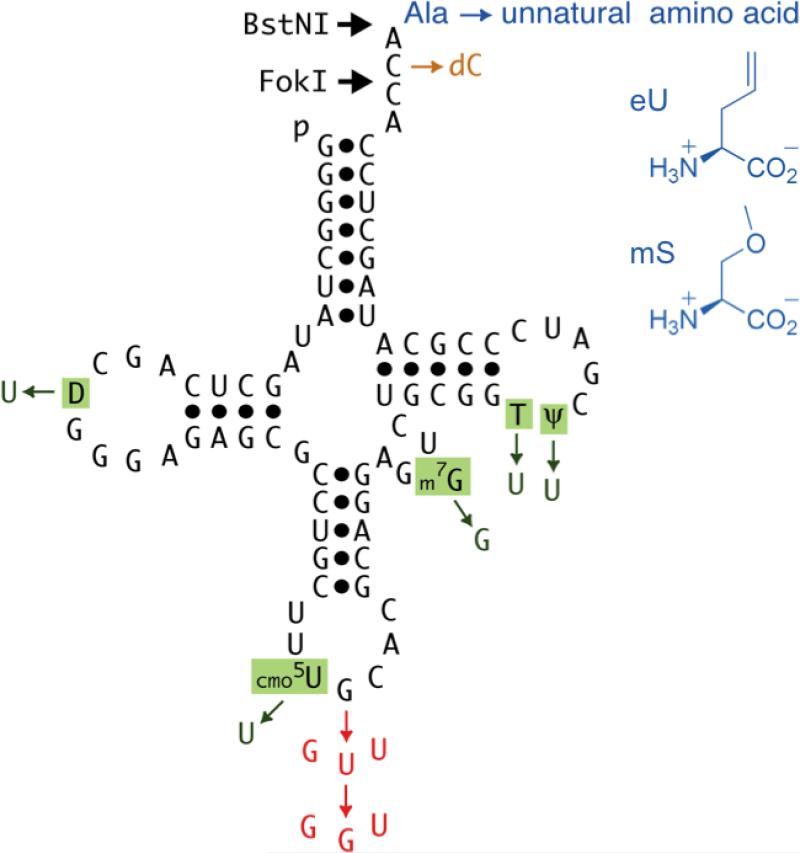
Fig. 1.
Wild-type E. coli tRNA3Ala isoacceptor (black; [32]) and synthetic tRNAAlaB species which contain from one to four different types of changes (green, red, orange and blue). Run off transcripts of plasmids cut with BstNI or FokI terminate at the positions shown with arrows. Note that tRNA2Ala is closely related to tRNA3Ala and that these isoacceptors have identical anticodons that read the same Ala codons (see tRNA database website at http://www.trna.uni-bayreuth.de/).
In principle, the effects of individual domain changes on multiple AA incorporations could be determined simply by changing one domain at a time from the wild-type AA-tRNA in a purified system. Unfortunately, this was not practical with tRNAAsnB (for synthetic reasons discussed in ref. [16]) or with tRNAPheB (because the polyPhe product is insoluble). Here, we overcome these experimental limitations by synthesizing polyAla using a tRNAAlaB adaptor (Fig. 1; [17,18]). This tRNA also has the unusual benefits of having charging determinants independent of the anticodon ([11]; thus allowing charging of anticodon mutants with Ala [17,18]) and having a 5’-terminal sequence of pGGG that coincides with the optimal T7 RNA polymerase promoter (thus avoiding the need for potentially confounding mutations in 5’- and 3’-terminal sequences [16]).
2. Materials and methods
New materials were prepared by standard methods as described [6,12,16,19]. Purified translations were also performed as described [16], except the final concentration of tRNATotal was adjusted to 160 uM taking into account any tRNATotal added as a component of wild-type tRNATotal charged with Ala, Asn or Thr. Translations also contained 0.5 uM each of initiation factors 1-3 and elongation factors Ts and G, 2.5 uM elongation factor Tu, 0.5 uM purified ribosomes, 1 uM appropriate mRNA, 0.2 uM (limiting) fMet-tRNAifMet, 0.5 uM C-terminal, 3H-labeled Val-tRNAVal, and upstream-encoded, unlabeled elongator AA-tRNAs at the following estimated concentrations: 0.5 uM for single incorporation or 2.5 uM for five straight incorporations. Additional details are given in the supplementary material.
3. Results
Effect of an unnatural AA-tRNAAlaB substrate on ribosomal peptide synthesis
Incorporation of unnatural AAs into peptides by the translation apparatus can be inefficient, so we chose a radioactive pure translation assay for sensitivity and quantitation of full-length peptide products. Another advantage of this assay is that it encompasses a number of controls, being specific for products initiated by fMet and terminated by Val (the only 3H-labeled amino acid provided), with measured yields being dependent on both added mRNA and test elongator AA-tRNA prepared from pure components. To test the suitability of the tRNAAlaB body for assaying the effects of individual domain changes, we first used a wild-type tRNAAlaBUGC sequence (Fig. 1 left) for the ribosomal polymerization of unnatural AA eU (Fig. 1 top right) using mRNAs encoding MTAV, MTA2V and MTA5V (Fig. S1A). In comparison with maximal product yields in translations incorporating the wild-type Ala-tRNAAla substrates (prepared from tRNATotal, pure Ala and pure AlaRS), saturation to give the same peptide yields occurred when one or two unnatural eU incorporations were templated (Fig. S1B). However, a significant drop in yield was observed when templating five straight incorporations of eU-tRNAAlaBUGC compared with five of wild-type Ala-tRNAAla, despite using excess unnatural substrate (Fig. S1B). Though Ala and eU incorporation into the peptide product were not directly demonstrated, the fidelity of our in vitro translation system regarding incorporation of several other natural AAs and also eU and mS has been rigorously established by product comigration on HPLC with authentic synthetic marker peptides [6,16,20].
These results support the conclusion from our prior studies using tRNAAsnB and tRNAPheB [6,16] that differences in product yields from different substrates often only become apparent when templating several, not single, incorporations of those substrates. Measuring multiple, as opposed to single, AA incorporations also proved superior for differentiating activities of various substrates in other purified [9,10], partially purified [7], crude [21] and in vivo translation systems [22,23].
Effect of nucleoside modifications
Next, we prepared substrates with smaller changes between them (Figs. 1 and 2). As predicted from Fig. S1 and discussed below, all substrates gave maximal peptide product yields upon single (Fig. 2B), not five, incorporations (Fig. 2C). Ala-tRNAAlaB-BstNIUGC differs from wild-type Ala-tRNA3Ala in lacking all five post-transcriptional nucleoside modifications (Fig. 1). These changes had a minimal effect on incorporation of five Ala's (- mods; Fig. 2C). This is consistent with single Ala incorporation yield from unmodified tRNAAla transcripts in a crude translation system ([17]; with the proviso that this system probably contained modification enzymes), with minimal effects on kinetics of single Phe insertions upon removing all 10 modifications from E. coli tRNAPhe [14,15] and with knowledge of only minor functions of modifications in translation [24].
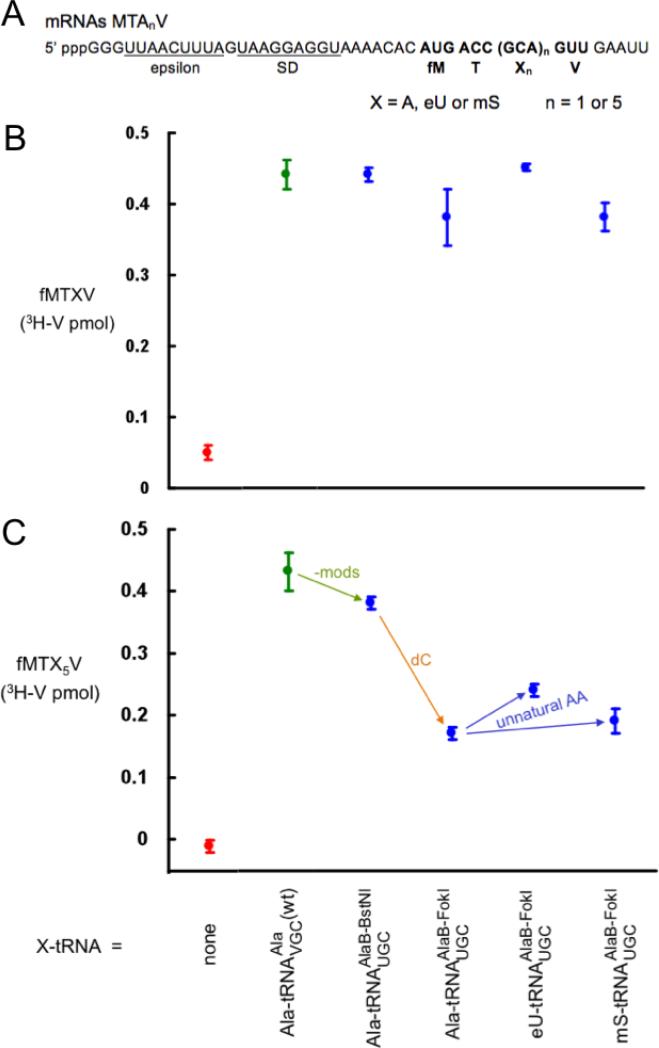
Fig. 2.
Effect of AA-tRNA domain changes on product yields using a wild-type anticodon. (A) mRNA sequences. (B) Yields from single incorporations of the test substrates. (C) Yields from five straight incorporations of the test substrates. Background d.p.m. obtained in translations without mRNA were subtracted (consistently about 25% of the positive control signal), and standard deviations from three independent determinations are shown.
Effect of the penultimate 2’OH group
The only difference between Ala-tRNAAlaB-BstNIUGC (prepared by charging the full-length transcript with Ala) and Ala-tRNAAlaB-FokIUGC (prepared from the truncated transcript by ligating to NVOC-amino protected pdCA-Ala followed by photo-deprotection of the amino group) is the removal of one oxygen atom (dC; Fig. 1). Surprisingly, this change caused a major (55%) decrease in yield (Fig. 2C). Control experiments argued against putative technical causes such as incomplete photo-deprotection (supplementary material and refs. [15,16,18]). This major effect on the incorporation of five Ala's, not one, contrasts with no effect within experimental error on single incorporations in a crude translation system [5]. However, the implicated importance of the penultimate 2’OH is not unreasonable given that it resides on a C that forms a base pair with E. coli G2553 of 23S ribosomal RNA [25] and that there is only one nucleotide between this C and the amino acid. Note that it is formally possible that the lack of modifications, which had a minimal effect on incorporation (Fig. 2C), somehow potentiated the effect of the dC. Ruling out this possibility is not experimentally tractable in our system.
Effect of non-cognate L-AAs
Next, we programmed synthesis of fM(mS)5V and fM(eU)5V (Fig. 2C) by charging with unnatural AAs mS or eU instead of Ala (Fig. 1). These unnatural changes made no difference or even increased the yield of peptide product, respectively, in comparison with synthesis of fMA5V from the same tRNAAlaB-FokIUGC but charged with cognate Ala (Fig. 2C). This is consistent with Fig. 6 of ref. [16] which showed that tRNAAsnB and tRNAPheB each charged with mS and eU gave similar or increased yields, respectively, compared with wild-type AA-tRNAs. These results were surprising in light of the broadened “thermodynamic compensation” hypothesis which proposes that evolution has optimized the pairing of each AA with its tRNA body for EF-Tu and ribosomal binding and performance in translation [26,27].
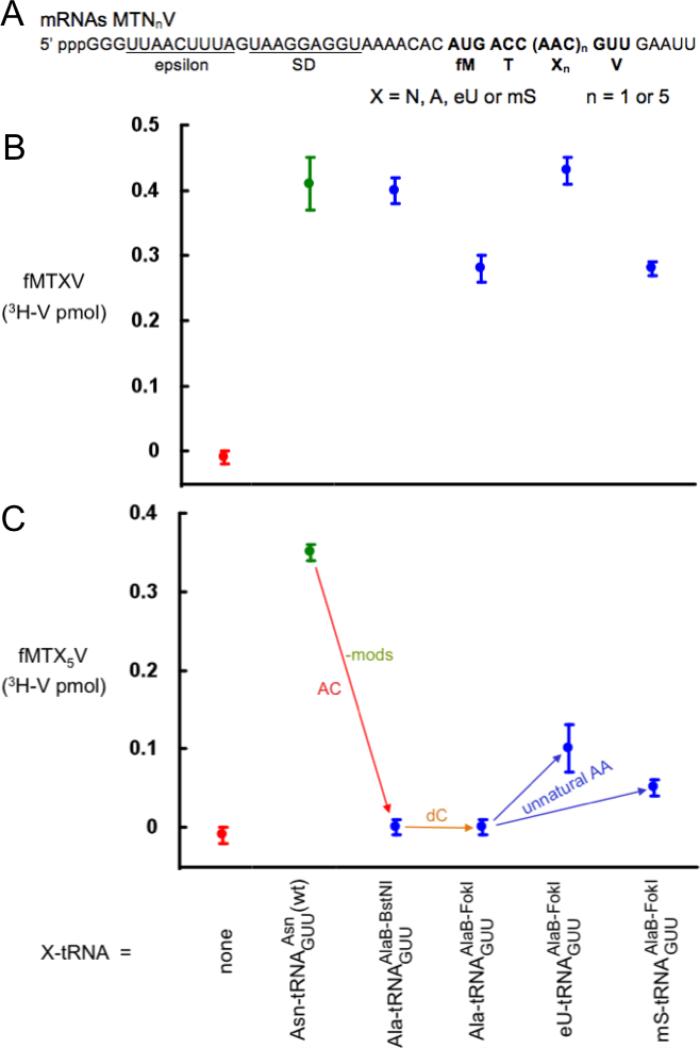
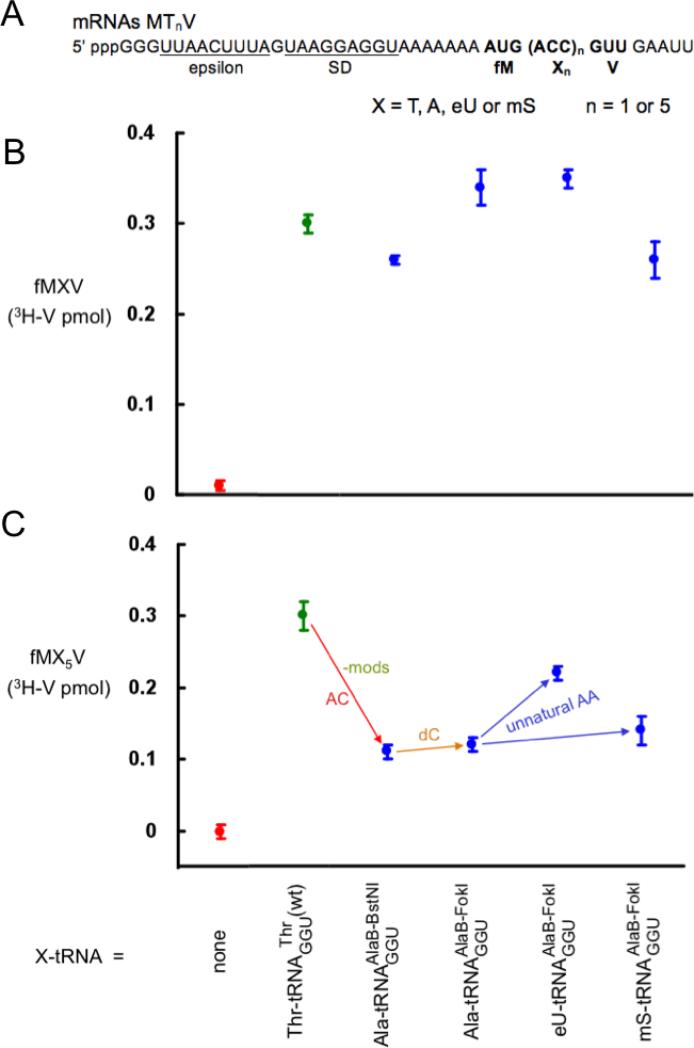
Effect of anticodon mutations
Both anticodon transplants (Fig. 1) had major inhibitory effects on five, not, single incorporations (- mods/AC; Figs. 3 and 4). This conclusion is based on the fact that the unmodified substrates shown by the left-most blue symbol in Figs. 3 and 4 only differed from the efficient unmodified substrate shown by the left-most blue symbol in Fig. 2 by the anticodon swaps alone. Again, it is formally possible that the lack of modifications, which had a minimal effect on incorporation (Fig. 2), somehow potentiated the effects of anticodon swaps. Ruling out this possibility is not experimentally tractable in our system.
Fig. 3.
Effect of AA-tRNA domain changes on product yields using the GUU mutant anticodon. (A) mRNA sequences. (B) Yields from single incorporations of the test substrates. (C) Yields from five straight incorporations of the test substrates. Note that because the anticodon change in the test substrates (blue) required codon changes in the mRNAs, this in turn required using a very different wild-type substrate (green) as a positive control for these mRNAs. The Ala-tRNA3Ala wild-type substrate also gave similar yields (the green points in Figs. S1 and 2); this positive control is more informative because it only differs from the left-hand test substrate by the lack of modifications and the anticodon change. For simplicity, the Ala-tRNA3Ala positive control is not replotted here but the “-mods” and “AC” differences are written adjacent to the left arrow.
Fig. 4.
Effect of AA-tRNA domain changes on product yields using the GGU mutant anticodon. (A) mRNA sequences. (B) Yields from single incorporations of the test substrates. (C) Yields from five straight incorporations of the test substrates (see note in Fig. 3 legend).
It is well known that mutating the anticodon can decrease function in translation [3,28], as nucleotides adjacent to the anticodon likely affect the efficiency of codon recognition [29]. However, we cannot conclude from our studies that wild-type anticodons might always be more efficient: ranking our five different tRNA body-anticodon combinations used for incorporating five straight eU's versus control incorporations with all-wild-type tRNAs gives tRNAAlaBUGC(wt) (ca. 60% peptide product yield) > tRNAAlaBGGU (30%) = tRNAAsnBGGU (30%; [6]) > tRNAAsnBGUU(wt) (5%; [16]) > tRNAAlaBGUU (0%). Unexpectedly, the variable that best correlates with yield is the anticodon sequence, irrespective of its tRNA body. Consistent with this was the finding that the least efficient of the three tRNAAsnB substrates and the least efficient of the three tRNAPheB substrates in Fig. 6 of ref. [16] also both had the GUU anticodon. It is doubtful that the low activity of GUU is due to absence of the queuosine (Q) modification because this modification apparently slightly decreases the stability of pairing with C [24]. Note that there is a rough correlation between yields of products with five straight eU's and the theoretical stability of anticodon-codon base pairing [24] (a perfect correlation would have had the yield with anticodon UGC = anticodon GGU because all anticodon bases form Watson-Crick pairings with the codons chosen for our mRNAs). But this may be an over-simplification because the most efficient tRNAs in Fig. 6 of ref. [16] formed the anticodon-codon interactions of lowest theoretical stability.
The two anticodon swaps also provided opportunities to evaluate the effects of the dC and unnatural AA changes in the setting of an unnatural tRNA sequence. The only surprising result in comparison with analogous data with the wild-type tRNA sequence (Fig. 2) was that the dC change did not further decrease the already decreased incorporation efficiency of tRNAAsnBGGU (Fig. 4C). Thus, the inhibitory effects of the GGU anticodon change and dC on five incorporations were not additive. Additive inhibitory effects of the dC change could not be assayed for the GUU anticodon with five incorporations because the rC version was inactive in this assay (left-most blue symbol in Fig. 3C), but there was evidence of a small additive effect with this severe anticodon mutant upon single incorporation (Fig. 3B). The effects of unnatural AA changes were consistent with analogous data with the wild-type tRNA sequence (Fig. 2): mS had little effect and eU even increased the yields (Figs. 3 and 4).
4. Discussion
Evolution of AA-tRNAs required translational incorporation of anticodon mutants and mischarged tRNA bodies. Based on our results, it seems probable that evolution was (and still is) restricted by low incorporation efficiencies of anticodon mutants, but it might have been aided by the apparently minimal or even stimulatory effect of mischarging. The high incorporation yield of our completely unmodified Ala-tRNAAla is consistent with the logical notion that primordial tRNAs were simpler.
Our stimulatory effect of mischarging on translation incorporation suggests that the descrimination by EF-Tu against misacylated tRNAs by weak binding, as occurs with Asp-tRNAAsn and Glu-tRNAGln [4], is unlikely to be general. Indeed, some mischarged AA-tRNAs bind similarly or more tightly than wild-type AA-tRNAs to EF-Tu [26]. Proofreading by AA-tRNA synthetases is presumably more important for minimizing incorporation of mischarged tRNAs [4].
Our results immediately suggest how to improve efficiencies of incorporation of unnatural AAs for protein engineering [5] and for creation of de novo genetic codes towards evolution of peptidomimetic ligands [6]. The use of unnatural L-AAs and unmodified tRNAs were apparently not the causes of inefficient incorporations here. Rather, use of the pdCA-AA charging method and anticodon mutants were problematic. The incorporation of the dC could be avoided by ligating on pCpA-AAs [30] or by using the flexizyme ribozyme to charge full-length tRNA transcripts [31]. Inhibitory effects of anticodon mutants might be overcome by testing many different tRNA body-anticodon combinations, transplantating extended anticodons [29], or using native tRNA body sequences.
Supplementary Material
Acknowledgements
We are grateful to Drs. Zhongping Tan, Virginia Cornish and Måns Ehrenberg for materials and Dr. R. Edward Watts for comments on the manuscript. This work was supported by the National Institutes of Health and the American Cancer Society.
List of abbreviations
- AA or X
amino acid
- U
unnatural AA
- x-tRNAyz
x = charged AA, y = AA specificity of either the wild-type isoacceptor or the wild-type isoacceptor upon which the chemoenzymatic sequence is based, z = either the wild-type isoacceptor designation or the anticodon sequence (5’ to 3’) of the chemoenzymatic tRNA sequence
- V
cmo5U
- fM
Formylmethionine
- mS
O-methylserine
- eU
2-amino-4-pentenoic acid, (also known as allylglycine; structure shown in Fig. 1)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary material
More detailed materials and methods, additional references and Fig. S1.
References
- 1.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–81. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 2.Saks ME, Sampson JR, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–70. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 3.Raftery LA, Yarus M. Systematic alterations in the anticodon arm make tRNAGlu-Suoc a more efficient suppressor. EMBO J. 1987;6:1499–1506. doi: 10.1002/j.1460-2075.1987.tb02392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibba M, Soll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–8. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 5.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. The use of 5′-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res. 1989;17:9649–60. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forster AC, Tan Z, Nalam MNL, Lin H, Qu H, Cornish VW, Blacklow SC. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci U S A. 2003;100:6353–7. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel A, Millward SW, Roberts RW. Encodamers: unnatural peptide oligomers encoded in RNA. Chem Biol. 2003;10:1043–50. doi: 10.1016/j.chembiol.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Merryman C, Green R. Transformation of aminoacyl tRNAs for the in vitro selection of “drug-like” molecules. Chem Biol. 2004;11:575–82. doi: 10.1016/j.chembiol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Josephson K, Hartman MC, Szostak JW. Ribosomal synthesis of unnatural peptides. J Am Chem Soc. 2005;127:11727–35. doi: 10.1021/ja0515809. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami T, Murakami H, Suga H. Ribosomal synthesis of polypeptoids and peptoid-peptide hybrids. J. Am. Chem. Soc. 2008;130:16861–16863. doi: 10.1021/ja806998v. [DOI] [PubMed] [Google Scholar]
- 11.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forster AC, Weissbach H, Blacklow SC. A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal Biochem. 2001;297:60–70. doi: 10.1006/abio.2001.5329. [DOI] [PubMed] [Google Scholar]
- 13.Hecht SM, Alford BL, Kuroda Y, Kitano S. “Chemical aminoacylation” of tRNA's. J Biol Chem. 1978;253:4517–20. [PubMed] [Google Scholar]
- 14.Harrington KM, Nazarenko IA, Dix DB, Thompson RC, Uhlenbeck OC. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993;32:7617–22. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- 15.Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci U S A. 2009;106:50–4. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster AC. Low modularity of aminoacyl-tRNA substrates in polymerization by the ribosome. Nucleic Acids Res. 2009;37:3747–55. doi: 10.1093/nar/gkp240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C, Kudlicki W, Odom OW, Kramer G, Hardesty B. In vitro protein engineering using synthetic tRNA(Ala) with different anticodons. Biochemistry. 1993;32:7939–45. doi: 10.1021/bi00082a015. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Tan Z, Dickson LG, Nalam MNL, Cornish VW, Forster AC. Specificity of translation for N-alkyl amino acids. J Am Chem Soc. 2007;129:11316–7. doi: 10.1021/ja0734871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Z, Blacklow SC, Cornish VW, Forster AC. De novo genetic codes and pure translation display. Methods. 2005;36:279–90. doi: 10.1016/j.ymeth.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Tan Z, Forster AC, Blacklow SC, Cornish VW. Amino acid backbone specificity of the Escherichia coli translation machinery. J Am Chem Soc. 2004;126:12752–3. doi: 10.1021/ja0472174. [DOI] [PubMed] [Google Scholar]
- 21.Hohsaka T, Ashizuka Y, Sasaki H, Murakami H, Sisido M. Incorporation of two different nonnatural amino acids independently into a single protein through extension of the genetic code. J. Am. Chem. Soc. 1999;121:12194–12195. [Google Scholar]
- 22.Bonekamp F, Dalboge H, Christensen T, Jensen KF. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J. Bacteriol. 1989;171:5812–5816. doi: 10.1128/jb.171.11.5812-5816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–80. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 24.Curran JF. Modified nucleosides in translation. In: Grosjean H, Benne R, editors. Modification and editing of RNA. ASM Press; Washington, D.C.: 1998. pp. 493–516. [Google Scholar]
- 25.Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A-site. Mol. Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 26.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–8. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 27.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–23. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 29.Yarus M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science. 1982;218:646–52. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- 30.Baldini G, Martoglio B, Schachenmann A, Zugliani C, Brunner J. Mischarging Escherichia coli tRNAPhe with L-4′-[3-(trifluoromethyl)-3H- diazirin-3-yl]phenylalanine, a photoactivatable analogue of phenylalanine. Biochemistry. 1988;27:7951–9. doi: 10.1021/bi00420a054. [DOI] [PubMed] [Google Scholar]
- 31.Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–9. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 32.Lund E, Dahlberg JE. Spacer transfer RNAs in ribosomal RNA transcripts of E. coli: processing of 30S ribosomal RNA in vitro. Cell. 1977;11:247–62. doi: 10.1016/0092-8674(77)90042-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.