Abstract
Understanding of the cell cycle control logic in Caulobacter has progressed to the point where we now have an integrated view of the operation of an entire bacterial cell cycle system functioning as a state machine. Oscillating levels of a few temporally-controlled master regulator proteins in a cyclical circuit drive cell cycle progression. To a striking degree, the cell cycle regulation is a whole cell phenomenon. Phospho-signaling proteins and proteases dynamically deployed to specific locations on the cell wall are vital. An essential phospho-signaling system integral to the cell cycle circuitry is central to accomplishing asymmetric cell division.
Keywords: Caulobacter, cell cycle, cell regulation, systems biology, robustness, cell biology
Biological systems are often characterized as “complex”, but this apparent complexity decreases with study and analysis of the system [1]. The reduction of perceived complexity with increased understanding is particularly apparent in the case of bacterial cell cycle regulation. One important insight has been the recognition that regulatory circuits involve far more than transcriptional networks. Rather, we now recognize essential roles for regulatory protein localization, the specific location of genes on the chromosome, and the evolving topology of the cell. Modeling of the Caulobacter cell cycle regulatory circuit has progressed so that the functioning of the cellular system can be analyzed from an engineering perspective [2]. Indeed, engineering simulations of the operation of Caulobacter’s cell cycle control system show that the regulatory circuit design mirrors design approaches that human engineers use to achieve reliable asynchronous electrical circuits [2].
In following sections, we address the operation of the cell cycle from a systems-level perspective with special consideration for how the changing spatial organization of the cell affects regulation of cell cycle progression. We describe dynamic phospho-signaling systems that are integral to the cell cycle control system. We show how the cell links the progression of chromosome replication to the ordered appearance of global transcriptional regulators. Finally, we consider how the cell cycle control system is designed for robust operation.
Architecture of the Caulobacter cell cycle control system
Caulobacter crescentus divides asymmetrically to produce two different progeny, a swarmer cell and a stalked cell, each with distinct morphological features and regulatory programs (Fig. 1A). The swarmer cell is motile for a short interval before differentiating into a stalked cell identical to its sibling. The cell cycle system is comprised of multiple modular subsystems that implement cellular growth and reproduction. An integral control system constructed using biochemical and genetic logic circuitry organizes the timing of initiation of each of these modular functions.
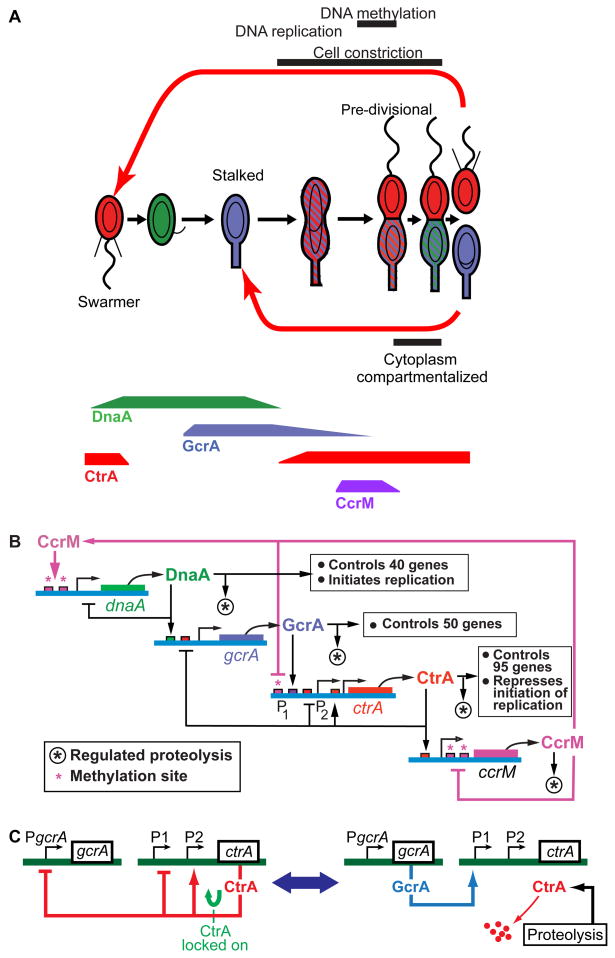
Figure 1. Caulobacter cell cycle control systems.
(A) Caulobacter cell cycle. The stalked daughter cell always re-enters the cell cycle as a stalked cell. In contrast, the swarmer daughter cell has an interval of motility before differentiating into a stalked cell equivalent to its sibling and entering the stalked cell cycle. Shading shows temporal and spatial localization patterns of the DnaA, CtrA, and GcrA regulatory proteins. Varying protein concentrations over the cell cycle are indicated below for four master regulators. The circles and theta structures inside the cell depict progression of chromosome replication. (B) Four proteins (DnaA, GcrA, CtrA, and CcrM) create a cyclical genetic circuit, the “core engine” that drives the Caulobacter cell cycle [3,8]. DnaA, GcrA, and CtrA are transcriptional regulators that control activation of modular subsystems, and CcrM is a DNA methyltransferase. (C) Simplified illustration of the bistable switch that causes alternate synthesis and destruction of CtrA. A tightly coupled phospho-signaling pathway (see Fig. 2) controls both the activation of proteolysis and the phosphorylation state of CtrA.
The cell cycles of both Caulobacter daughter cell types have a cyclical genetic circuit – a cell cycle engine --comprised of the DnaA, GcrA, CtrA and CcrM master regulatory proteins (Fig. 1B) that directly control the temporal transcription of over 200 genes [3–4]. These proteins are synthesized and cleared from the cell one after the other over the course of the cell cycle (Fig. 1A). The cyclical variation of these four master regulatory proteins controls activation of modular subsystems in the appropriate sequence and timing relative to each other. In both the swarmer and stalked cell cycles, DNA replication can only be initiated after CtrA is cleared from the cell and active DnaA accumulates. Several signaling pathways described later operate together to ensure timely and reliable elimination of CtrA from the cell.
The subcircuit involving CtrA and GcrA is centrally involved in the cyclical variation of CtrA concentration over the cell cycle. Figure 1C shows on the left the situation when CtrA is locked into a high concentration state by positive autoregulation. At key points in the cell cycle this feedback loop is interrupted by accelerated CtrA proteolysis (Fig. 1C, right), and CtrA is cleared from the cell (Fig. 2A). Accelerated CtrA proteolysis is triggered by different events in the swarmer and stalked cell cycle, but activated over the same phospho-signaling pathway (Figs. 2B). In the case of the stalked daughter cell, clearance of CtrA is triggered by compartmentalization of the cytoplasm well before cell separation (Fig. 2A) [5–6]. In the case of the swarmer daughter cell, the trigger for CtrA clearance at the swarmer to stalked cell transition is not known. Later we describe the remarkable configuration of interlocked, spatially-distributed phospho-signaling pathways that trigger elimination of CtrA from the cell by a localized protease complex. This phospho-signaling system monitors the topology of the cell and is central to establishing daughter cell asymmetry.
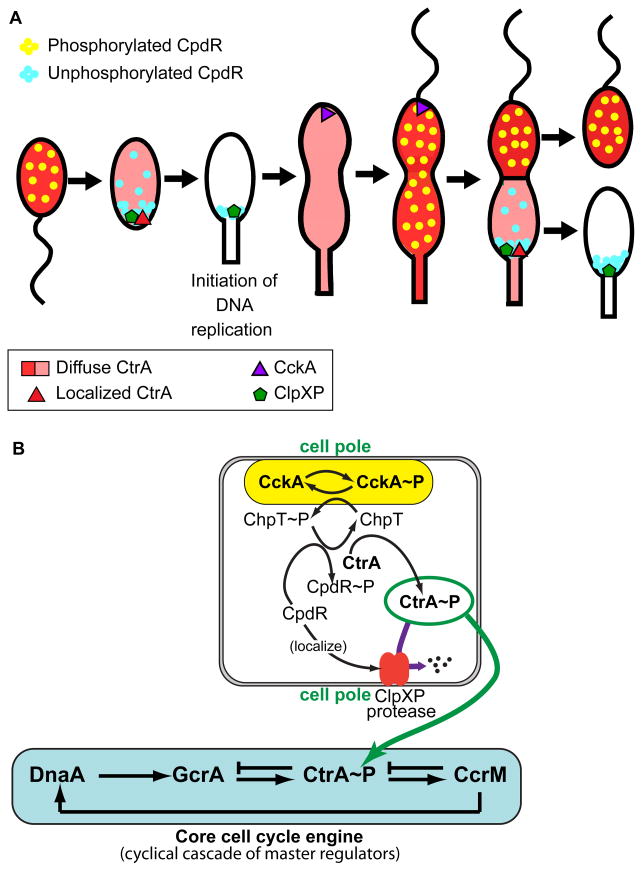
Figure 2. Dynamic protein localization controls CtrA stability and phosphorylation.
(A) Dynamic changes in the complement of phospho-signaling proteins localized at the swarmer cell pole and stalked pole drive polar organelle development and switching between accumulation and destruction of the key master regulator CtrA~P. Localization and activation of the ClpXP protease occurs in the newly differentiated stalked cell and in the daughter stalked cell compartment prior to the completion of cell division. (B) A phospho-signaling pathway originating at the polar-localized CckA histidine kinase controls both localization/activation of the ClpXP protease and the phosphorylation state of CtrA.
DnaA and CtrA have opposing roles in controlling the initiation of chromosome replication [7]. The DnaA protein binds to and opens the origin of replication facilitating replication initiation, whereas the CtrA protein binds to the origin and blocks replication initiation. In addition to opening the replication origin, DnaA activates the transcription of at least 40 genes, including the gene encoding the GcrA global transcription factor (Fig. 1B). GcrA, in turn, controls the transcription of 50 genes, many of which are involved in chromosome replication and segregation, while activating the transcription of ctrA [4,7]. CtrA, which accumulates following the initiation of replication, prevents the re-initiation of chromosome replication, while directly activating the transcription of 95 cell cycle-regulated genes. CtrA turns off the transcription of gcrA and activates transcription of the ccrM gene encoding an essential DNA methyltransferase that is required to facilitate the transcription of dnaA, thereby completing the cyclical operation of the core cell cycle engine.
A DNA methylation ratchet affects timing of cell cycle regulatory gene transcription
The level of expression of two of the four cell cycle master regulator proteins, CtrA and DnaA, is coupled to the progression of DNA replication by the DNA methylation-state change that occurs upon passage of a replication fork through their respective genes [8–10]. Caulobacter exploits the ordered replication of its circular chromosome initiated from a single origin to synchronize the time of transcription of these regulatory genes with the ordered replication of the chromosome [8]. At the start of DNA replication, the chromosome is in the fully methylated state, and the dnaA gene is transcribed preferentially from a fully methylated promoter. However, the dnaA gene is near the chromosomal origin of replication (Cori), and upon passage of the replication fork it becomes hemimethylated, and thus down-regulated [8]. This methylation state control of dnaA transcription is particularly important due to DnaA’s central role in the initiation of chromosome replication. The enzyme that remethylates DNA, CcrM, only accumulates near the completion of DNA replication. After remethylation of the DNA, CcrM is rapidly both deactivated and cleared from the cell [11–12]. Remethylation of the chromosome by CcrM during a short time window near the end of replication enables dnaA transcription in preparation for the next cell cycle. DnaA proteolysis is also activated shortly after initiation of DNA replication. This DnaA regulatory strategy for preventing re-initiation of chromosome replication and assures that Caulobacter cells under one, and only one, round of DNA replication per cell cycle [13].
Transcription of the ctrA gene is also modulated by the methylation state of its promoter. ctrA is positioned much further from the origin than DnaA, and one of its two promoters can be activated only when in the hemimethylated state [10]. This regulatory configuration prevents re-synthesis of CtrA too early in the chromosome replication process and provides further protection against reinitiation of DNA replication. We have shown through modeling studies that this circuit design enhances Caulobacter fitness [2]. Thus, this methylation-based regulatory mechanism provides a feedback signal that helps synchronize progression of the core cell cycle engine with the progress of chromosome replication and, as a result, to enhance reliability of cell cycle control by assuring that there is one and only one round of replication per cell cycle.
Transiently polar-localized protease complexes and phospho-signaling proteins control CtrA degradation
Prior to the completion of cell division, fission of the inner membrane produces two cellular compartments inside a contiguous outer membrane, each containing one of the duplicated chromosomes (Fig. 1A) [5–6]. This cytoplasmic compartmentalization event triggers the divergent genetic programs in the nascent stalked and swarmer cell compartments [5,14–15]. The distinct transcriptional regulatory program followed by each sibling upon compartmentalization is determined by phospho-signaling proteins that are differentially localized to the two cell poles prior to compartmentalization. These asymmetrically positioned signaling proteins initiate localized proteolysis of the CtrA master regulator in only the daughter stalked cell compartment at the time of cell compartmentalization (Fig. 2A) [14–18]. Later, after the completion of cell division and a period of swarmer cell motility, an unknown cue signals the onset of the swarmer to stalked cell transition and CtrA proteolysis by the polar ClpXP protease.
To clear CtrA from the nascent stalked cell, the ClpXP protease complex is localized to the stalk-bearing cell pole (Fig. 2A) [19]. The transient polar localization of ClpXP requires the function of the polar CpdR protein (Fig. 2B) [20]. Simultaneously, the CtrA substrate is brought to the activated polar protease by the combined action of the RcdA localization factor and the PopA cyclic di-GMP effector protein [19,21]. The control of CpdR polar localization is linked to the phospho-signaling cascade [20], while the control of PopA polar localization is mediated by the cyclic di-GMP second messenger system [21]. Thus, two different signaling systems, each intimately connected to the three dimensional deployment of their regulatory components, direct cell cycle progression by controlling the time and place of CtrA proteolysis.
The phospho-signaling network controls localized proteolysis in the stalked cell through the CpdR protein. CpdR localizes to the cell pole only in its unphosphorylated state, and thereby localizes and activates the ClpXP protease complex (Fig. 2A) [20]. The pathway that phosphorylates CpdR initiates with the CckA histidine kinase, the same kinase pathway that phosphorylates and activates CtrA (Fig. 2B). This double-barreled approach to control of the level of CtrA~P, the activated form of CtrA, facilitates rapid changes in its cellular concentration. The CckA histidine kinase is active when positioned at the cell pole. In the new stalked cell, CckA is not at the cell pole and is thus inactive; consequently, CpdR remains in the unphosphorylated state so that it facilitates the polar localization of the ClpXP protease and thus CtrA degradation (Fig. 2A). This remarkably elegant circuit created using networked phospho-signaling pathways is another factor leading to robust regulation of the CtrA master regulator.
Asymmetric cell division
The spatially distributed CckA-related phospho-signaling pathways play an essential role in the operation of the overlapping, but not identical, cell cycles of the Caulobacter swarmer and stalked cell types. As described earlier, this signaling system controls both the initiation of differentiated development in the incipient swarmer and stalked cell compartments and in the elimination of CtrA~P that is essential to initiation of chromosome replication in early stalked cells (Fig. 2A). Bacillus subtilis also undergoes a division that is asymmetric at the morphological and molecular levels during sporulation [22]. Like Caulobacter, several of the alpha-proteobacteria (Brucella abortus, Sinorhizobium meliloti, and Agrobacterium tumefaciens) divide asymmetrically into daughter cells of slightly different sizes, and they also incorporate many conserved elements of the CtrA-related Caulobacter cell cycle regulatory machinery [23]. Molecular level asymmetry has been examined in very few species. B. abortus is one of these cases, and it was shown to have two asymmetrically polar-localized histidine kinases, PleC and PdhS. The B. abortus PdhS protein is homologous to Caulobacter’s PleC and DivJ histidine kinases which are also asymmetrically polar-localized [24]. There is growing evidence that asymmetric bacterial division is quite widespread. Certainly in the alpha-proteobacteria, this is the case, and many components of the Caulobacter cell cycle control system are conserved in many of these species.
There are parallels between the cellular strategy for asymmetric cell division in eukaryotic cells and Caulobacter cells (Fig. 3). Diversification of eukaryotic cell types occurs in two ways: (1) two initially identical daughter cells can become different because they encounter different environments that influence their subsequent development, or (2) the factors determining cell fates are differentially inherited by the two daughter cells so that their development diverges [25]. Caulobacter cells implement two distinct methods of cellular differentiation. Asymmetric partitioning of polar-localized regulatory proteins into the daughter cells during cytokinesis, as described earlier, produces the distinctive swarmer and stalked cells. Subsequently, in the swarmer daughter cell at the swarmer-to-stalked cell transition, the swarmer cell differentiates into a stalked cell equivalent to its sibling. It remains to be determined whether the swarmer-to-stalked cell differentiation event is triggered by internal signals, e.g., nutrient status or cell size, or if is it a preprogrammed event that occurs after a programmed delay.
Figure 3. Parallels between the strategy for asymmetric cell division in some stem cells and in Caulobacter cells.
Key requirements are to establish the cell’s axis of polarity, orient the chromosome, distributed critical proteins spatially along the polarity axis, and divide the cell. The asymmetrically sequestered proteins direct the ongoing differential development of the distinctive daughter cells.
The Caulobacter stalked cell is similar to a stem cell in that it divides recurrently to produce a differentiated “offspring” -- the swarmer cell -- while retaining its own identity (Fig. 3). In stem cells, asymmetric cell division involves four steps [25]: (1) establish an axis of polarity, (2) set up the mitotic spindle oriented along the axis of polarity, (3) distribute cell fate determinants asymmetrically along the axis, and (4) divide and pursue distinctive cell fates as dictated by the cell fate determinants in each cell. While the molecular details are completely different, Caulobacter has adopted a similar strategy for accomplishing asymmetric cell division.
Caulobacter cells are intrinsically polarized, with polar markers positioned in the cell membrane at the point of cell division that establish the polarity of the daughter cells [26–27]. In addition, before initiation of DNA replication, organization of the chromosome is polarized, with the origin of replication located at the stalked pole and the terminus at the opposite pole [28], and genetic loci are positioned along the length of the cell in a manner linearly correlated with their position on the chromosome [29]. Since chromosome replication and segregation occur simultaneously in bacteria, there is no step equivalent to the orientation of the mitotic spindle seen in eukaryotic cells. Rather, the orientation of the Caulobacter chromosome prior to initiation of chromosome replication is already determined with regard to cell polarity, and the chromosome is ordered in a specific direction just as in a stem cell. Differential location of histidine kinase signaling proteins (the Caulobacter cell fate determinants) at the poles of the nascent daughter cells then leads to the distinct cell fates of the swarmer and stalked daughter cells.
Feedback signals pace progress of the cell cycle engine
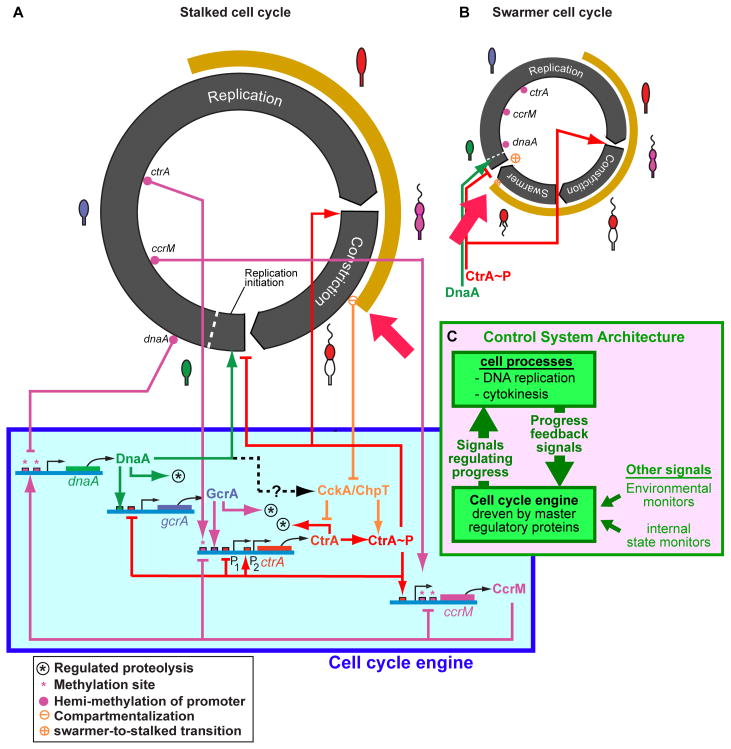
Cyclically varying concentrations of the four master regulators comprising the cell cycle engine (Fig. 1A, 1B) control the transcription of more than 200 other proteins to effect timely implementation of the many subsystems that accomplish cell growth, polar organelle biogenesis, chromosome replication, and cell division. Figure 4 shows key signals from the cell cycle engine that couple into initiation of chromosome replication and cell constriction. The differing pattern of CtrA presence in the swarmer and stalk cell cycles is indicated by the tan arc exterior to the black bands denoting cell cycle progression (Figs. 4A, 4B). The red arrows point to the great differences in timing of clearance of CtrA from the cell within each of these cell cycles. These timing differences result from the differential activation of the CckA-related phospho-signaling pathway within the two cell cycles as shown in Figure 2B.
Figure 4. Whole cell view of the cell cycle control system.
The cell cycle engine drives both the stalked (A) and swarmer (B) cell cycles by activating numerous subsystems in a precisely controlled order. The duration of DNA replication and FtsZ-ring constriction in the cell cycle are approximately to scale. The cell cycle engine shown below the stalked cell cycle controls activation of the processive reactions that implement DNA replication and cell constriction. Feedback signals (C) activated by events in the progression of the cell cycle synchronize the system. The circuit design assures that timing errors do not occur, by halting or slowing the cell cycle engine so that subsystems are not activated until necessary precursor events have occurred [2]. Cell stage is indicated by the cell-type icons on the perimeter. The distinct difference in timing of the presence of CtrA (tan arcs and red arrows) in the swarmer and stalked cell cycles is controlled by the phospho-signaling mechanisms shown in Figure 2.
Each process activated by the proteins of the cell cycle engine involves a cascade of many reactions. The longest subsystem cascade is DNA replication, which involves about 2 million DNA synthesis reactions for each arm of the chromosome over about 40 minutes. While the average time for each individual synthesis reaction can be computed from the observed total time, the actual reaction time for each reaction is stochastic, not deterministic. There is significant inevitable cell-to-cell variation around the average rate of progress of DNA synthesis and in the overall time to complete replication of the chromosome. As described above, feedback signals, including the effects of the methylation ratchet, pace progression of the cell cycle engine to match progress of events in each particular cell. This control system organization, with a controller (the cell cycle engine) driving a complex system, with modulation by feedback signals from the controlled system comprises a closed loop control system (Fig. 4C).
There are additional signals arising from environmental monitors (e.g., nutrient levels, oxygen monitors) or internal state monitors (e.g., DNA damage). Figure 4 shows only the DNA replication and cytokinesis processes (that dominate timing of the stalked cell cycle) and the interpolated motile phase in the swarmer cell cycle. But, there are a myriad of other subsystems that have to be activated in parallel. For example, the flagellum is constructed by a cascade of reactions involving about 40 genes that is itself paced by internal feedback signals and checkpoints.
Reliability of the Caulobacter cell cycle control system
A critical design consideration for the cell cycle system is reliable (or robust) operation in spite of inevitable stochastic variability of reaction rates within the cell and equally inevitable unpredictable variability in the environment. The rates of progress of the parallel independent reaction cascades comprising cellular subsystems are inherently unpredictable due to both internal genetic noise and wide variations in environmental conditions. Yet, some pathways must be completed in a particular order for success in cell duplication and division. Using simulation and formal analysis of the design of the Caulobacter cell cycle control circuit, we have shown that the system is remarkably well-designed to prevent timing-related failures and that the overall circuit design is an essential factor in achieving robust, highly-reliable cell cycle execution [2]. This analysis of the Caulobacter system provides further evidence that biological regulatory systems conform to principles that engineers use to design regulatory systems in other engineering domains [30]. It also suggests that engineering analysis of alternative candidate regulatory circuit hypotheses to identify the subset of the designs that are robust to timing errors can be a useful method for focusing experimental investigations.
The cell cycle control circuit includes numerous features to assure that the cell cycle completes successfully under all contingencies. The methylation ratchet discussed earlier is one such feature. Another, even more subtle mechanism, arises from the continued low-level basal expression from the dnaA promoter when in the repressive hemimethylated state. Previously we would have dismissed this experimental observation as “imperfect” repression, but analysis now shows it is a regulatory feature that prevents cell cycle failure in the small fraction of cells where the downstream GcrA regulatory protein is completely proteolyzed by chance before the CtrA promoter region is hemimethylated. Without the basal expression from dnaA, cell cycle progression would be blocked, and the cell would die when this condition occurred, but the dnaA basal expression will produce enough DnaA to “reboot” the cell cycle and rescue this cell [2].
These examples demonstrate that there are regulatory mechanisms, including epigenetic mechanisms, whose role is specific to countering low-probability stochastic contingencies. These are not mechanisms where alternative subsystems are activated in the face of environmental challenges, such as the heat shock response or activation of alternative metabolic pathways. Rather, they are permanently present features of the cell cycle control circuit whose purpose is to forestall potential fatal failures whenever a predictable, but possibly low probability, stochastic variation in reaction rates would otherwise lead to a dangerous timing glitch. Identifying these features of the regulatory circuitry experimentally is inherently quite difficult. Some of these contingency design elements will be important under conditions encountered in the wild, but not usually investigated in the lab. Caulobacter is adapted to live in clear lakes and streams where there are low and highly variable nutrient levels. Growth under such restricted nutrient conditions where the rate of progression of reaction cascades is slowed might increase sensitivity to stochastic variations in reaction rates.
Another problem is that the phenotypes of mutations affecting the efficacy of these prophylactic regulatory mechanisms will have low penetrance since the failure of the mechanism will only be relevant to the small subset of cells in the population at any time that are experiencing a low probability stochastic excursion of the relevant reaction rates. Mutations in a contingency mechanism designed to deal with an occasional critical situation would be highly relevant to the fitness of the mutant strain in the wild, but unlikely to be noticed in the laboratory. Eukaryotic regulatory circuitry surely includes such mechanisms for forestalling contingent risks. Mutations in these mechanisms will also produce low penetrance phenotypes. Such mutations will manifest as rare abnormalities occurring with no particular pattern. Identifying the genetic cause of such conditions will be particularly difficult since it requires identifying the cause of an intermittent failure which is always difficult.
Summary
Perhaps the most striking aspect of Caulobacter cellular control is the degree to which bacterial cell cycle regulation is a whole cell phenomenon. It is not just changing cellular concentration of transcriptional regulators driving the cell cycle. Rather, there are many additional cell cycle regulatory proteins, particularly phospho-signaling proteins and protease complexes, whose correct functioning depends on where they are positioned in the cell and the 3-dimensional topology of the cell at that time [31]. During the Caulobacter cell cycle, the spatial structure of the cell changes in a tightly prescribed manner. Landmarks are implanted in the new cell poles at the time of cell division by the preceding cell generation where they contribute to establishment of cell polarity by determining the positioning of polar organelles, e.g., the stalk, flagellum, and pili [26–27]. The chromosome is also positioned in a prescribed linear order with respect to polar-positioned regulatory proteins, and this positioning is an integral feature of regulation of initiation and execution of chromosome replication. The physical organization of the cell -- its topology and the specific positioning of regulatory proteins -- is tightly integrated into the genetic and biochemical signaling pathways of cell cycle control. The physical orientation of the chromosome within the cell is a predetermined element of cellular polarity, carefully maintained from one cell generation to the next, and an integral element of asymmetric division. All these observations and others not addressed here (see [32]) support the observation that “it takes a cell to make a cell.”
There is rapid and accelerating progress in understanding the integrated operations of the bacterial cell, particularly of the cell cycle regulatory system and the coupling of the cell cycle controls with the mechanisms that implement the cell cycle, and the sensor/response mechanisms that interface with the environment. There are realistic prospects that a whole cell regulatory model of the Caulobacter cell will be possible within the next decade and surely models of other bacteria will follow in short order thereafter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rouse WB. Engineering complex systems: Implications for research in systems engineering. Transactions on Systems, Man, and Cybernetics--Part C. 2003;33:154–156. [Google Scholar]
- 2.Shen X, Collier J, Dill D, Shapiro L, Horowitz M, McAdams HH. Architecture and inherent robustness of a bacterial cell-cycle control system. Proc Natl Acad Sci U S A. 2008;105:11340–5. doi: 10.1073/pnas.0805258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collier J, Murray SR, Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. Embo J. 2006;25:346–56. doi: 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–7. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- 5.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–82. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci U S A. 2003;100:8235–40. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–53. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 8.Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A. 2007;104:17111–6. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens CM, Zweiger G, Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–9. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. Embo J. 2002;21:4969–77. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–77. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shier VK, Hancey CJ, Benkovic SJ. Identification of the active oligomeric state of an essential adenine DNA methyltransferase from Caulobacter crescentus. J Biol Chem. 2001;276:14744–51. doi: 10.1074/jbc.M010688200. [DOI] [PubMed] [Google Scholar]
- 13.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–45. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 14.McGrath PT, Viollier P, McAdams HH. Setting the Pace: Mechanisms tying Caulobacter cell-cycle progression to macroscopic cellular events. Current Opinion in Microbiology. 2004;7:192–197. doi: 10.1016/j.mib.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–90. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 16.McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–7. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 17.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–25. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 19.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–47. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 20.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barak I, Wilkinson AJ. Where asymmetry in gene expression originates. Mol Microbiol. 2005;57:611–20. doi: 10.1111/j.1365-2958.2005.04687.x. [DOI] [PubMed] [Google Scholar]
- 23.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12:361–5. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, Letesson JJ, Jacobs-Wagner C, De Bolle X. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. Embo J. 2007;26:1444–55. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–85. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 26.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–23. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 27.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–37. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 28.Thanbichler M, Shapiro L. Chromosome organization and segregation in bacteria. J Struct Biol. 2006;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–62. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Samad H, Kurata H, Doyle JC, Gross CA, Khammash M. Surviving heat shock: control strategies for robustness and performance. Proc Natl Acad Sci U S A. 2005;102:2736–41. doi: 10.1073/pnas.0403510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009 doi: 10.1126/science.1175685. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harold FM. Molecules into cells: specifying spatial architecture. Microbiol Mol Biol Rev. 2005;69:544–64. doi: 10.1128/MMBR.69.4.544-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]