Abstract
As a transcription factor, the critical tumor suppressor p53 directly regulates the transcription of hundreds of genes, leading to cell cycle arrest, apoptosis, cellular senescence and differentiation. While it has been assumed that p53 transcription activity is critical for tumor suppression, this assumption has been increasingly contested by recent findings of transcription-independent roles of p53 in apoptosis as well as the findings that none of the mutant mice lacking important p53 transcription targets are cancer prone. Based on previous findings that p53 transcription activity is abolished in p53QS (Leu25Trp26 to Gln25Ser26) knock-in mouse cells after DNA damage, to determine the importance of transcription activity of p53 in tumor suppression, we generated a knock-in mice that can conditionally express p53QS protein in a Cre-dependent manner. By breeding the knock-in mice with Lck-Cre transgenic mice that specifically express Cre in thymocytes, we demonstrate that p53-dependent suppression of thymic lymphomas is abolished in thymocytes expressing high levels of p53QS protein. In addition, p53QS protein is accumulated in some of the thymic tumors. Therefore, p53 transcription activity induced by DNA damage is required for tumor suppression. Together with the findings that disruption of various p53-dependent functions individually fails to promote cancer, our findings indicate that various transcription-dependent functions of p53 must collaborate to efficiently suppress tumorigenesis.
Keywords: p53, transcription activity, tumor suppression, metastasis
As the guardian of the genome, tumor suppressor p53 plays multiple roles in both somatic and stem cells to maintain genetic stability, including cell cycle arrest that allows time for the repair of DNA damage, apoptosis and senescence that prevent the cells with damaged genome from replicating, cellular differentiation that eliminates the stem cells with damaged DNA from the self-renewing pool (Ko and Prives, 1996; Michael and Oren, 2002; Xu, 2005). Structural and functional analyses of p53 indicate that it is a transcription factor with a sequence-specific DNA binding domain in the central region, transcriptional activation domains at the N-terminus, and a tetramerization domain at the C-terminus (Ko and Prives, 1996). p53 directly regulates the expression of hundreds of genes that play important roles in p53-dependent functions (Wei et al., 2006). For example, p21 and 14-3-3σ are required for mediating p53-dependent cell cycle G1/S and G2/M cell cycle checkpoints respectively (Oren, 2003). Puma is required for p53-dependent apoptosis after DNA damage (Oren, 2003). Plasminogen activator inhibitor-1 is required for p53-dependent replicative senescence (Kortlever et al., 2006). p53 can induce the differentiation of embryonic stem (ES) cells after DNA damage by directly suppressing the expression of Nanog, which is required for the self-renewal of ES cells (Lin et al., 2005).
While it is clear that the transcription activity is important for p53-dependent functions in response to various stresses, the importance of p53 transcription activity in tumor suppression remains to be established. In this context, while p53-dependent cell cycle G1/S arrest after DNA damage is abolished in p21−/− mice, these mice are genetically stable and are not cancer prone (Brugarolas et al., 1995; Deng et al., 1995). Despite the abolishment of p53-dependent apoptosis, Puma-deficient mice are not cancer prone (Jeffers et al., 2003; Villunger et al., 2003). In addition, the importance of p53 transcription activity in tumor suppression is contested by recent studies that have identified transcription-independent roles of p53 in apoptosis and tumor suppression (Schuler and Green, 2005).
To determine the importance of p53-dependent transcription in apoptosis, others and us took the advantage of two missense mutations (Leu22Trp23 to Gln22Ser23) at the N-terminus of human p53 that abolish the transcriptional activities of p53 to independently establish the p53QS (Leu25Ser26 of mouse p53 to Gln25Ser26) knock-in ES cells and mice (Chao et al., 2000; Johnson et al., 2005; Lin et al., 1994). While transcription-independent role of p53 in apoptosis remains intact in p53QS knock-in cells (Chipuk et al., 2004), p53-dependent transcription and apoptosis are abolished in p53QS knock-in cells after DNA damage (Chao et al. 2000; Johnson et al. 2005). Therefore, p53-dependent transcription is critical for p53-dependent apoptosis after DNA damage.
To determine the physiological importance of p53-dependent transcription in tumor suppression, we introduced the p53QS-Neo allele into mouse germline (Fig. 1a). The detail of the targeting construct and strategy to generate p53QS-Neo allele in ES cells was described by us previously (Chao et al., 2000). It is known that the PGK-Neo cassette in the targeted allele suppresses the expression of the targeted gene (Chao et al., 2000; Inlay et al., 2002), the p53QS-Neo mice were bred with CMV-Cre mice that express Cre at the zygote stage and have been routinely used to excise the LoxP-flanked PGK-Neo gene from various p53 knock-in mice (Chao et al., 2003; Chao et al., 2006a; Chao et al., 2006b; Song et al., 2007). Similarly to previous findings in an independently established p53QS mice that normal expression of p53QS allele leads to embryonic lethality (Johnson et al., 2005), no offsprings harboring the PGK-Neo-deleted allele could be found from the breeding of p53QS-Neo and CMV-Cre mice.
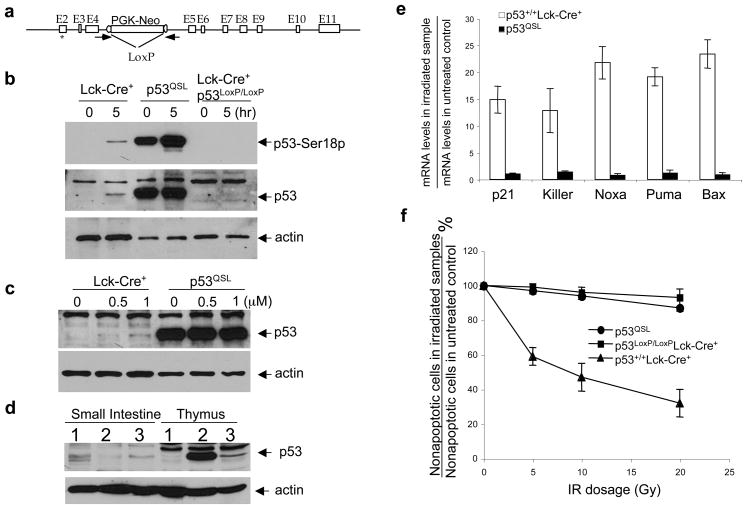
Figure 1.
Expression and function of p53 in the thymocytes of p53QSL mice. (a) The p53QS-Neo knock-in allele. The exons are represented by open boxes. PCR primers to screen for the LoxP/Cre-mediated deletion are indicated by arrowheads. The QS mutations are indicated by an asterisk. The generation of the knock-in ES cells were described previously (Chao et al., 2000). (b) Expression of p53 protein in the thymocytes of Lck-Cre+, p53QSL and Lck-Cre+p53LoxP/LoxP mice before and 5 hours after IR (10Gy). Lck-Cre+ mice express the Cre enzyme specifically in the thymocytes but not in other cell types (Lee et al., 2001). In p53LoxP/LoxP mice, the exons 2–10 of the p53 gene are flanked by LoxP sites and can be deleted from the genome in a Cre-dependent manner (Jonkers et al., 2001). Protein extracts from 2×106 thymocytes were resolved on 10% SDS PAGE gel and transferred to nitrocellulose membrane, which was probed with a polyclonal phosphor-specific antibody against p53 phosphorylated at Ser18 (Cell Signaling Technology, Danvers, MA) or ploclonal antibody against p53 or β-actin (Santa Cruz Biotechnology). The membrane was subsequently probed with a horseradish peroxidase-conjugated secondary antibody and developed with Supersignal Pico reagents (Thermo Scientific, Rockford, IL). (c) Induction of p53 protein levels in Lck-Cre+ and p53QSL thymocytes 8 hours after treatment with increasing concentration of doxorubicin. (d) Expression of p53 protein in the small intestine and thymus of Lck-Cre+, p53QSL and Lck-Cre+p53LoxP/LoxP mice 1 hour after whole body IR (10Gy). No p53 protein can be detected in the brain and liver of these mice of all genotypes by Western blotting (data not shown). (e) Induction of p53 target genes in Lck-Cre+ and p53QSL thymocytes 5 hours after 10Gy of IR. Total RNA from thymocytes was isolated using Trizol (Invitrogen, Carlsbad, CA) and RNAeasy Mini Kit (Qiagen, Valencia, CA), reverse-transcribed using Superscript II RT (Invitrogen). Real-time PCR was performed with an AjBI Prism 7000 (Applied Biosystems, Foster City, CA) with Power SyberGreen PCR Master Mix (Applied Biosystems, Foster City, CA). The PCR conditions were: 10 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C. The average Ct value for each gene was determined from triplicate reactions and normalized with the levels of GAPDH as previously described (Chao et al., 2006a). The primers were described previously (Chao et al., 2006a; Lin et al., 2005). Mean value from three independent experiments are presented with standard derivation. (f) p53-dependent apoptosis of Lck-Cre+, p53QSL and Lck-Cre+p53LoxP/LoxP thymocytes 10 hours after increasing dosages of IR. Single cell suspension of thymocytes derived from 4-week-old mice was cultured in DMEM supplemented with 5% FBS and 25 mM HEPES at pH 7 at a density of 106 cells/ml. The cells were irradiated and the percentage of apoptotic cells was analyzed 10 hr later by staining with Annexin V-FITC as described (Chao et al., 2006a).
p53−/− mice mostly develop thymic lymphomas, indicating the critical roles of p53 in tumor suppression in mouse thymocytes (Donehower et al., 1992; Jacks et al., 1994). Therefore, to determine the physiological importance of p53 transcription activity in tumor suppression, p53QS-Neo mice were bred with Lck-Cre transgenic mice that express Cre specifically in the thymocytes. p53QS-NeoLck-Cre+ mice, denoted p53QSL mice, were born with expected mendelian ratio and could mature into adulthood. When assayed by PCR, the PGK-Neo gene was deleted from the targeted alleles in the thymocytes of all homozygous p53QSL mice analyzed (>20 mice), indicating that Lck-Cre transgene expresses sufficient Cre enzyme in thymocytes of p53QSL mice for efficient LoxP/Cre-mediated deletion of the PGK-neo gene. Consistent with previous findings that QS mutation disrupts the interaction between p53 and Mdm2 leading to p53 stabilization (Chao et al., 2000; Lin et al., 1994), significantly higher p53 protein levels were detected specifically in the thymocytes derived from p53QSL mice than those from Lck-Cre+ control mice (Fig. 1b, c, d). In addition, the expression of p53 is specifically silenced in the thymocytes of Lck-Cre+p53LoxP/LoxP mice (Figure 1b, c, d). Similarly to what was observed in other cell types (Chao et al., 2000; Johnson et al., 2005), while DNA damage agents such as ionizing radiation (IR) and doxorubicin significantly induce the protein levels of p53 in the Lck-Cre+ thymocytes, they did not increase the p53 protein levels in p53QSL thymocytes (Fig. 1b, c). In addition, p53 was constitutively phosphorylated at Ser18 in p53QSL thymocytes, suggesting the chronic activation of DNA damage responses responsible for p53 phosphorylation in these thymocytes (Fig. 1b). In summary, p53 is constitutively stable in p53QSL thymocytes. Despite the greatly increased protein levels of p53, p53-dependent induction of target genes was abolished in p53QSL thymocytes after IR, confirming that p53-dependent transcription is essentially abolished in p53QSL thymocytes after DNA damage (Fig. 1e). Consistent with this finding, similar to that in p53−/− mice, p53-dependent apoptosis after IR is abolished in p53QSL thymocytes (Fig. 1f).
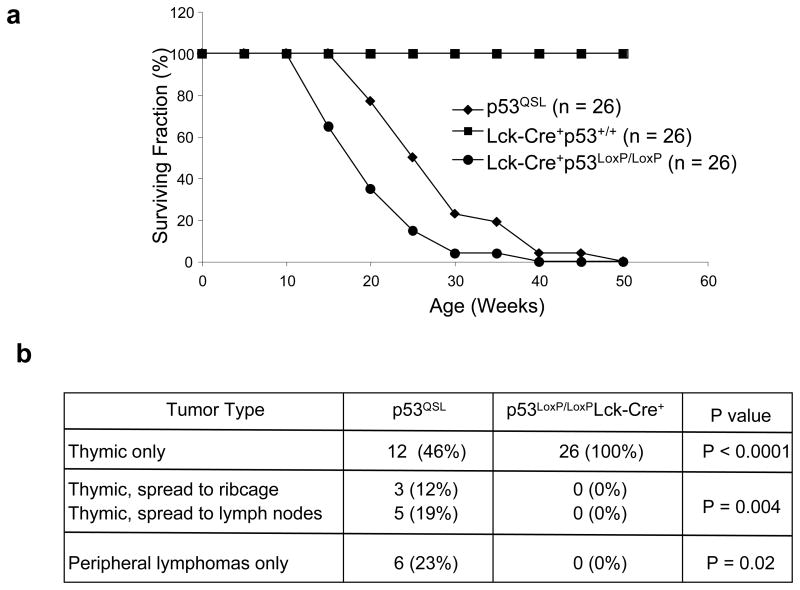
To determine the contribution of p53 transcription activity to tumor suppression, the tumorigenesis in p53QSL mice were monitored. As controls, conditional p53 knockout mice (p53LoxP/LoxP) were bred with Lck-Cre+ mice to specifically eliminate p53 in the thymocytes (Jonkers et al., 2001). As expected, none of the Lck-Cre+p53+/+ control mice died of tumors within 50 weeks of age (Fig. 2a). The p53LoxP/LoxPLck-Cre+ mice were all succumbed to thymic lymphomas by 50 weeks of age, further supporting the critical roles of p53 in suppressing thymic lymphomas (Fig. 2a, b). p53QSL mice uniformly died of tumors by 50 weeks of age, and the tumors are predominantly thymic and peripheral lymphomas (Fig. 2a, b). Since there is no statistically significant difference in the survival between p53LoxP/LoxPLck-Cre+ and p53QSL mice, we concluded that p53-dependent tumor suppression is abolished in p53QSL thymocytes.
Figure 2.
Tumorigenesis of p53QSL mice. (a) Survival curve of Lck-Cre+, p53LoxP/LoxPLck-Cre+ and p53QSL mice. N shows the number of mice monitored. The P value for the difference of survival between p53LoxP/LoxPLck-Cre+ and p53QSL mice is 0.46. (b) Tumor spectrum of p53LoxP/LoxPLck-Cre+ and p53QSL mice. The p values are shown.
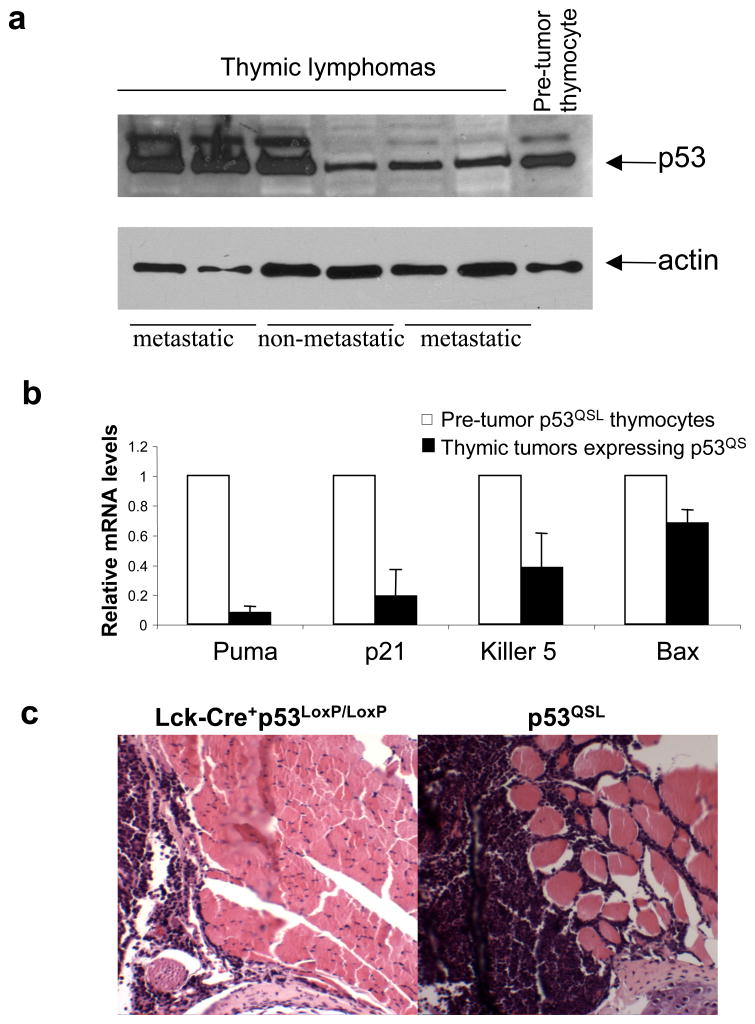
Previous studies have suggested that p53QS protein retains some transcription activity in response to non-genotoxic stresses (Johnson et al., 2005). If p53QS protein still retains significant tumor suppression activities, the expression of p53QS protein should be selected against in the tumors developed in p53QSL mice. However, p53 protein was accumulated to high levels in some of the thymic lymphomas of p53QSL mice (Fig. 3a). Sequencing analysis of the p53 gene in two of these tumors indicate that only QS mutations but no other mutations are present in the p53 gene. The basal levels of p53-dependent transcription of target genes were reduced in the thymic tumor cells when compared with that in pre-tumor p53QSL thymocytes, indicating that the remnant transcription activity of p53QS is selected against in p53QSL thymic tumor cells as seen in most human cancer cells that harbor wild type p53 (Fig. 3b). Similarly to thymic tumors observed in p53−/− mice (Donehower et al., 1992; Jacks et al., 1994), none of thymic tumors in p53LoxP/LoxPLck-Cre+ mice exhibited metastatic phenotypes to invade other tissues. However, about 40% of thymic lymphomas detected in p53QSL mice were metastatic and invaded other tissues (Fig. 2b, 3c). In addition, peripheral lymphomas were frequently detected in p53QSL mice (Fig. 2b). Therefore, similarly to what has been observed in p53 gain of function mutant knock-in mice, the constitutively stable p53QS protein could behave like a gain-of-function p53 cancer mutant to promote metastasis by disrupting the functions of other proteins (Lang et al., 2004; Olive et al., 2004; Song et al., 2007).
Figure 3.
Metastatic phenotypes of thymic tumors in p53QSL mice. (a) p53 protein is accumulated to high levels in some of the thymic tumors derived from p53QSL mice. The metastatic and non-metastatic tumors are indicated. (b) The relative expression of p53 target genes in the tumors expressing p53QS versus in pre-tumor p53QSL thymocytes. The mRNA levels of each gene were determined by quantitative real time PCR and standardized by the mRNA levels of GAPDH. Mean value from three tumors are presented with standard derivation. (c) Representative histological image of one metastatic thymic tumor in p53QSL mice that invade the skeleton muscles of the rib cage as well as the one from p53LoxP/LoxPLck-Cre+ mice. Tumor samples were fixed in 10% buffered formalin, embedded in paraffin and sliced. All sections were stained with hematoxylin and eosin for histological assessment as previous described (Chao et al., 2006a).
With recent discovery of the transcription-independent roles of p53 in apoptosis and tumor suppression (Moll et al., 2005), the requirement of p53-dependent transcription in tumor suppression has been challenged. Since transcription-independent roles of p53 are retained by p53QS in both transfected cells and knock-in cells (Chipuk et al., 2004), our findings provide definitive evidence that p53 transcription activity induced by DNA damage is required for p53-dependent tumor suppression. Together with the findings that the disruption of a single p53-dependent function does not lead to increased tumorigenesis, our findings support the conclusion that these p53-dependent functions cooperate to suppress tumorigenesis in vivo.
Acknowledgments
We thank T. Lin and C. Chao for their help in constructing the p53QS mice. This work was supported by a NIH grant (R01 CA94254) to Y.X.
References
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem. 2003;278:41028–41033. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. Embo J. 2006a;25:2615–2622. doi: 10.1038/sj.emboj.7601167. Epub 2006 Jun 2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. Embo J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, et al. Acetylation of Mouse p53 at Lysine 317 Negatively Regulates p53 Apoptotic Activities after DNA Damage. Mol Cell Biol. 2006b;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct Activation of Bax by p53 Mediates Mitochondrial Membrane Permeabilization and Apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the [kappa] light chain intronic enhancer and 3[prime] enhancer in [kappa] rearrangement and demethylation. Nat Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and-independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A Critical Role for Dnmt1 and DNA Methylation in T Cell Development, Function, and Survival. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53 and Mdm2 families in cancer. Curr Opin Genet Dev. 2002;12:53–59. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Current Opinion in Cell Biology. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Schuler M, Green DR. Transcription, apoptosis and p53: catch-22. Trends Genet. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Xu Y. A new role for p53 in maintaining genetic stability in embryonic stem cells. Cell Cycle. 2005;4:363–364. doi: 10.4161/cc.4.3.1529. [DOI] [PubMed] [Google Scholar]