Abstract
Phenylalanine ammonia lyase (PAL) has long been recognized as a potential enzyme replacement therapeutic for treatment of Phenylketonuria. However, various strategies for the oral delivery of PAL have been complicated by the low intestinal pH, aggressive proteolytic digestion and circulation time in the GI tract. In this work, we report 3 strategies to address these challenges. First, we used site-directed mutagenesis of a chymotrypsin cleavage site to modestly improve protease resistance; second, we used silica sol-gel material as a matrix to demonstrate that a silica matrix can provide protection to entrapped PAL proteins against intestinal proteases, as well as a low pH of 3.5; finally, we demonstrated that PEGylation of AvPAL surface lysines can reduce the inactivation of the enzyme by trypsin.
Introduction
Phenylketonuria (PKU) is an autosomal recessive metabolic disorder leading to the impairment of phenylalanine hydroxylase (PAH). The consequent accumulation of phenylalanine can in turn result in severe irreversible neurological complications [1]. Despite the fact that PKU is one of the best studied metabolic disorders, a specific and effective therapeutic for the treatment of classical PKU is unavailable. Noting the fact that the defect or deficiency of PAH is the underlying cause of PKU, recombinant forms of PAH from various origins were previously examined as potential candidates for use as an enzyme replacement therapeutic. However, due to the obligatory requirement of a tetrahydrobiopterin cofactor for the activity of PAH, as well as the complex regulatory properties of the enzyme, further development of PAH as a potential therapeutic appeared to be challenging [2–3]. In contrast, phenylalanine ammonia lyase (PAL) enzymes derived from bacterial or yeast origins proved to be comparatively robust enzymes that are capable of degrading phenylalanine to the metabolically harmless trans-cinnamic acid and small quantities of ammonia [3]. For this reason, PAL has been intensively examined over the past few decades as a potential lead enzyme for the development of an enzyme substitution therapy for PKU [4–7]. We have recently shown that injection of a PEGylated form of PAL is capable of significantly reducing plasma phenylalanine levels while exhibiting favorable pharmacokinetic and pharmacodynamic properties [8–9]. This PEG-PAL molecule has since entered phase 2 clinical trials for the treatment of PKU.
As protein therapeutics become more common, and as PKU patients require lifelong enzyme replacement, a non-invasive or oral formulation of PAL would be highly desirable. Taking advantage of the enterorecirculation of phenylalanine in the intestine, it is appropriate to focus on developing oral formulations of entrapped PAL that would allow phenylalanine to diffuse from the blood stream into the GI tract, and be metabolized before being reabsorbed [4, 7, 10]. Measures to inhibit the degradation of PAL protein by digestive enzymes have resulted in activity retention, permitting a metabolic response of phenylalanine pools to the enzyme, in vivo. Such efforts revealed the potential for intestinal PAL to act as a therapeutic in PKU. Herein, we report an engineered mutant of Anabaena variabilis PAL (AvPAL) aimed at improving the enzyme’s resistance to chymotrypsin, as well as two chemical modification strategies to confer protease resistance to AvPAL for possible oral delivery.
Materials and Methods
Mutagenesis, expression and purification of an AvPAL triple mutant
AvPAL variants (triple mutant; TM-AvPAL) were derived through point mutations on the AvPAL double mutant (DM-AvPAL) clone reported previously [8]. The mutations were introduced using a QuikChange site-directed mutagenesis kit (Strategene, CA, USA). Variants of the AvPAL gene were cloned into a pET28a vector and expressed in E. coli BL21 (DE3) host cells (Novagen, NJ, USA). The cell culture was grown in LB broth with 30 μg/ml Kanamycin at 37 °C, agitated at 250 rpm to achieve a turbidity of 0.8 OD600 before being induced with 1 mM IPTG at 18 °C for 16 h. The cell pellet was then harvested by centrifuging at 5000 g for 10 min at 4 °C. The pellet was then lysed by suspending in 150 mM NaCl, 25 mM Tris-Cl, and EDTA-acid-free protease inhibitor cocktail tablets (Roche Diagnostics GMbH, Mannheim, Germany) pH 8.0 before passing through a microfluidizer set at 18 000 psi process pressure. After cell-disruption, the lysate was ultracentrifuged at 45 000 rpm for 45 min, and the supernatant was applied to a Ni-NTA agarose resin for affinity purification of the His-tagged protein using 200 mM Imidazole, 150 mM NaCl, 25 mM Tris-Cl, pH 8.0. Ammonium sulfate was then added to the eluent to a final concentration of 2 M and stirred for 30 min at 4 °C. The precipitated protein was isolated by ultracentrifugation before solubilization in 150 mM NaCl, 25 mM Tris-Cl, pH 8.0 and solvent exchang with the same buffer.
Activity Assay
The PAL activity assay was performed by adding 50 μl of protein solution of appropriate concentration into 950 μl of 22.5 mM phenylalanine in 100 mM Tris-Cl pH 8.5. Activity of the PAL was monitored through the observation of trans-cinnamic acid formation at absorption of 290 nm at room temperature.
Protease digestion
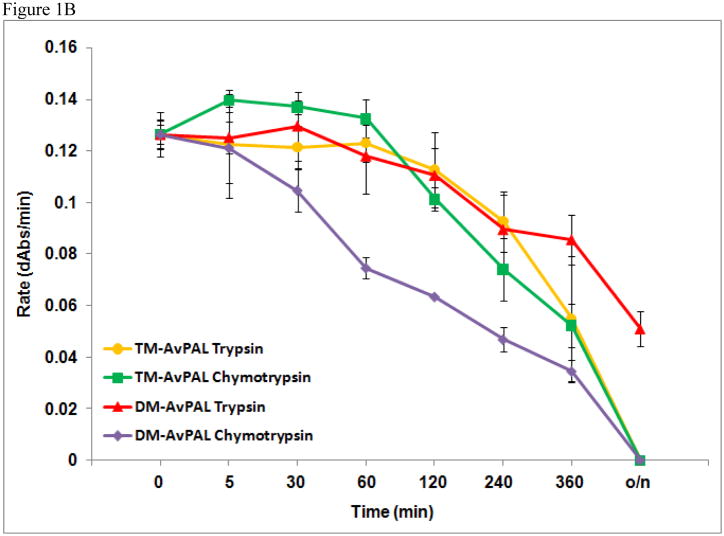
Resistance to proteolytic digestion of TM-AvPAL was demonstrated as per the limited proteolysis digestion protocol previously reported [8], using 40 μg/ml of chymotrypsin or trypsin in 0.1 M Tris and 150 mM NaCl pH 8.5 at 37 °C. The concentration of TM-AvPAL used for these protease resistance studies was 0.10 mg/ml. The data points present are the average of triplicate sets done concurrently (Figure 1B).
Figure 1.
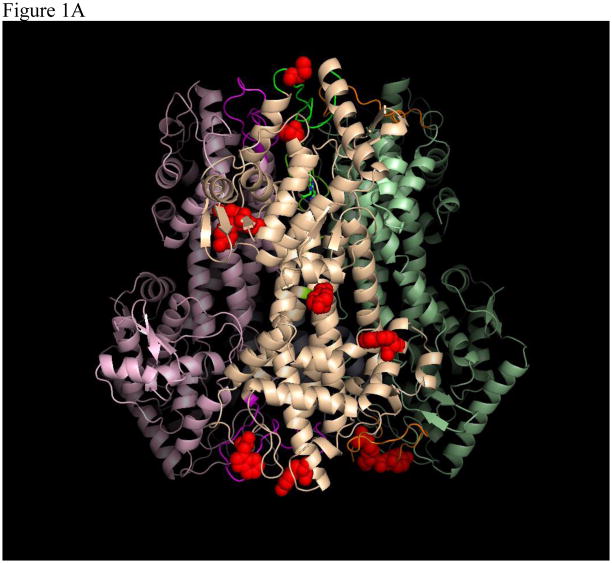
(A) Tetrameric structure of AvPAL (PDB 2NYN). Surface-exposed potential trypsin and chymotrypsin cleavage sites on one out of the four monomers are represented by red spheres. (B) Comparison of protease resistance between TM-AvPAL and DMAvPAL incubated with chymotrypsin or trypsin (40 μg/ml) solutions at pH 8.0. The various plots were normalized at time zero to facilitate comparison.
PEGylated TM-AvPAL and TM-AvPAL-silica particles were examined for protease resistance by incubating with a significantly higher concentration of chymotrypsin and trypsin in USP pH 6.8 solutions. The 4 sets of assay conditions include: (1) Control conditions involving USP pH 6.8, (2) 5 mg/ml Trypsin in USP pH 6.8 solution, (3) 5 mg/ml Chymotrypsin in USP pH 6.8 solution, and (4) Acidic conditions involving USP pH 3.5 solution. . The deliberate increase in protease concentration is to demonstrate the protection conferred in the elevated protease concentrations of 5 mg/ml, as well as to facilitate possible diffusion of the protease into the silica matrix. Protease resistance studies for PEG-TM-AvPAL were conducted at TM-AvPAL concentrations of 0.10 mg/ml. Protease digestion was performed at 37 °C for 4 h and PAL activity was assayed at regular intervals.
Parallel to incubation with proteases, TM-AvPAL-silica particles were also incubated with USP pH 3.5 or USP pH 6.8 buffers to verify for possible protection against low pH [11]. TM-AvPAL-silica particles prepared in accordance to the protocol described in the next section were resuspended in appropriate volumes of protease or buffer solutions to give a final concentration of 0.5 mg/ml encapsulated TM-AvPAL and incubated at 37 °C, agitated at 250 rpm. At appropriate time intervals, the TM-AvPAL-silica particles were removed from the protease or buffer solutions by centrifugation at 5000 rpm, and washed with USP pH 6.8 buffer solution before resuspending in 22.5 mM phenylalanine in 100 mM Tris-Cl pH 8.5 to make up a final encapsulated TM-AvPAL concentration of 0.2 mg/ml. The reactions were allowed to incubate for an additional 15 min at 37 °C before terminating by the addition of 5 % v/v glacial acetic acid. PAL activity of the encapsulated TM-AvPAL was determined by assaying for amounts of trans-cinnamic acid at 280 ± 10 nm. All assays were performed in triplicates.
Thermal Stability Studies
The thermal stability of TM-AvPAL was examined using the method reported earlier by Wang et al. [8]. Briefly, 5 μl of 1:250 diluted SYPRO Orange was added to 2 μg of protein and made up to volume with 50 mM Tris, 150 mM NaCl, pH 8.0. Thermal stability studies were then performed using a Bio-Rad MyIQ iCylcer real-time polymerase chain reaction instrument over a temperature range of 20 – 95 °C at a scan rate of 0.5 °C/min. Samples containing additive ligands (Phenylalanine or trans-cinnamic acid) were prepared by incubating the protein samples with 5 mM concentrations of the ligands for 10 min prior to initiation of the stability studies.
Formulation of TM-AvPAL-Silica particles
Silica sol was freshly prepared from sodium silicate solutions using a protocol modified from that previously described by Bhatia et al [12]. Briefly, 2.875 g of sodium silicate solution was mixed with 7 ml of deionized water. Eight grams of Dowex 50WX8-100 ion-exchange resin was then added to the resulting solution and stirred for 5 min. The resin was removed from the solution by vacuum filtration, and the final pH of the filtrate was adjusted to pH 1.8. From the resulting sol, 765 μl was combined with 73 μl of 0.2 M NaOH to bring the pH of the sol above pH 4, and immediately followed by addition of the protein solution of the desired concentration. The final mixture was then rapidly transferred into a stirring solution of liquid paraffin with 1 % w/w Span 80 to disperse the protein-sol mixture before gelation occurred. The emulsion was allowed to stir for 30 min at room temperature before storing at 4 °C overnight without stirring. The protein-silica particles were harvested by centrifugation at 5000 rpm for 10 min, and washed twice with deionized water. Loading efficiency of the gelation process was shown to be close to 100 % due to the transition from sol to gel, and was evaluated by assaying for protein content of wash using Bradford assay.
Images of the protein-silica particles were captured through a QImaging Retiga 2000R camera mounted on a Nikon eclipse 50i laboratory light microscope at 10 × magnification.
PEGylation of TM-AvPAL
PEGylation of TM-AvPAL was performed using Sunbright ME-050HS N-hydroxysuccinimide activated 5 kDa polyethylene glycol (PEG; NOF America Corporation, NY, USA). The amount of PEG to be used was added at a 3:1 ratio to the total number of lysine molecules present on each subunit of TM-AvPAL. The protein solution was added dropwise to a stirring solution of PEG dissolved in 200 mM potassium phosphate buffer pH 8.0, with the final protein concentration of the reaction set at 8 mg/ml. The PEGylation reaction was then allowed to proceed for 4.5 h before stopping by transferring the reaction mixtures to ice. The solution was then dialyzed against 0.5 M Tris-Cl solution pH 8.5 overnight to remove excess PEG molecules. Completion of the PEGylation reaction was verified by analyzing the samples on SDS-PAGE using 10% Bis-Tris gels and 4–12% Tris-Glycine native gels (Invitrogen, CA, USA). The final PEGylated protein was concentrated to appropriate concentrations using 100 kDa centrifugal filters, and assayed for PAL activity, before storing at −20 °C until ready to use.
Results and Discussion
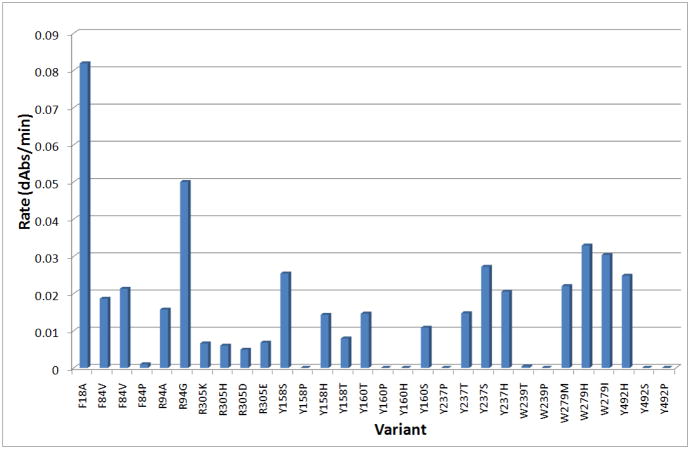
In our previous work, we established that C503S and C506S mutations to AvPAL (henceforth termed DM-AvPAL) are necessary to reduce aggregation while maintaining the catalytic efficiency of the enzyme. In an attempt to improve the protease resistance of AvPAL, we also reported the limited proteolysis of DM-AvPAL with chymotrypsin and trypsin [8]. Candidate sites identified to be amenable for additional protein engineering include those with susceptible protease cleavage sites, such as residues F18, R94, and R305, as well as surface exposed residues Y158, Y160, Y237, W279, Y304, Y492, and W531 (Figure 1A). Through conservative substitution of these residues, it was found that only the F18A variant (henceforth termed TM-AvPAL) exhibited activity comparable to the native enzyme (Figure 2). Thermal stability of TM-AvPAL was examined as reported in our earlier work on DM-AvPAL [8]. TM-AvPAL was shown to exhibit a slightly lower thermal stability as compared to DM-AvPAL, but had a stability that is still significantly higher than RtPAL (Table 1). Compared to the DM-AvPAL reported in our earlier work, TM-AvPAL exhibits modest improvements in resistance against chymotrypsin digestion (Figure 1B). As noted previously, it is important not to focus on any one specific enzyme characteristic when developing a therapeutic and a broader view must be taken incorporating the fact that half life, stability, activity, and pharmacokinetic/pharmacodynamic factors are all important.
Figure 2.
Summary of activity for DM-AvPAL variants. Protein concentration used was 0.07mg/ml enzyme. Unmodified DM-AvPAL had an activity rate of ~0.1 dAbs/min (data not shown).
Table 1.
Comparison of Tm (°C) shifts of PAL variants.
| PAL species | Tm |
|---|---|
| DM-AvPAL[8] | 78.4 ± 0.5 |
| + Phe | 85.0 ± 0.0 |
| + tCA | 84.6 ± 0.5 |
| TM-AvPAL | 75.7 ± 0.7 |
| + Phe | 78.7 ± 0.1 |
| + tCA | 79.3 ± 0.2 |
| RtPAL | 66.4 ± 0.2 |
| + Phe | 67.1 ± 0.1 |
| + tCA | 66.8 ± 0.3 |
The first chemical modification strategy explored to further improve the protease resistance of TM-AvPAL utilized the silica sol-gel matrix for entrapping the enzyme. Silica matrix was originally created to assay a variety of enzymes by entrapping the proteins in optically transparent matrices while preserving their biochemical properties [13]. This has since been applied to solid-phase assays for a variety of protein targets, as well as drug delivery systems of chemicals and, to some extent, protein therapeutics [14–15]. Additional work also suggested that silica matrices may preserve the activity of entrapped enzymes against extremes of pH [11]. Corollary to these observations, the silica matrix appears to be useful as a potential delivery system for entrapping and delivering TM-AvPAL to the intestines.
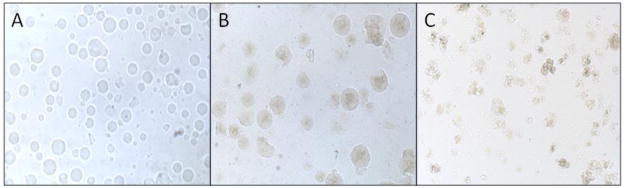
TM-AvPAL-silica particles were prepared based on the strategy by Bhatia et al. [12] and further dispersed into liquid paraffin to form discrete microspheres (Figure 3). Formulation of the TM-AvPAL-silica particles, using sol with an initial pH of 1.8, achieved a final formulation pH of ~6 as estimated with pH indicator strips. The rapid gelation rate of the silica matrix upon raising the sol pH precluded accurate determination of formulation pH. These TM-AvPAL-silica particles were then viewed using a light microscope under 10 × magnification. Unlike BSA-silica particles that formed transparent and discrete particles (Figure 3A), TM-AvPAL-silica particles were translucent, and maintained discrete microspheres up to a protein load of approximately 13.8 mg/ml (Figure 3B and 3C). Any further increase in final formulation pH or protein load resulted in particles of poor morphology and protease protection (data not shown).
Figure 3.
BSA-silica and TM-AvPAL-silica particles. 10× magnification of (A) 13.8 mg/ml BSA, (B) 7 mg/ml TM-AvPAL, and (C) 13.8 mg/ml TM-AvPAL in silica matrix.
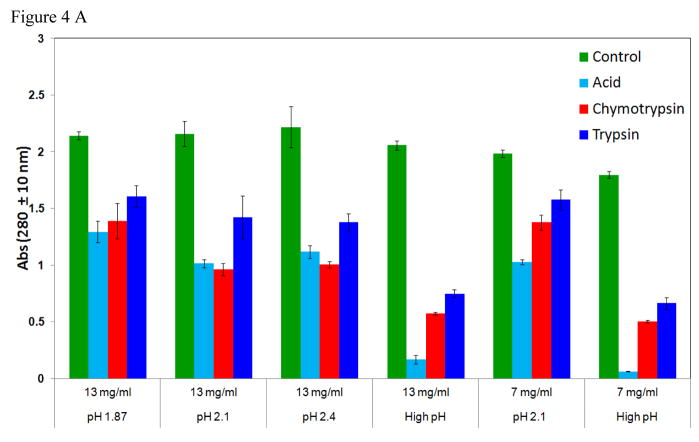
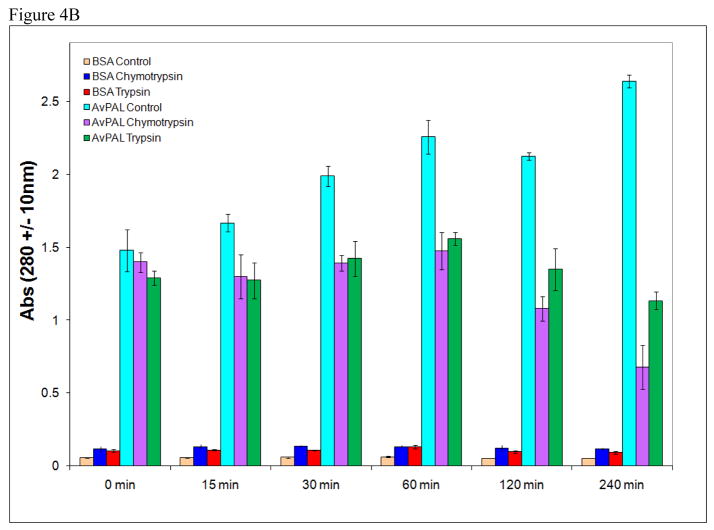
In order to demonstrate the protective effect of the silica matrix, TM-AvPAL-silica particles were deliberately incubated with a higher protease concentration of 5 mg/ml chymotrypsin or trypsin solution pH 6.8 for up to 4 h. Silica particles containing protein load of up to 13.8 mg/ml TM-AvPAL exhibited resistance to inactivation upon incubation with low pH of 3.5 for up to 1 h, or incubation in 5 mg/ml chymotrypsin or trypsin solution pH 6.8 for up to 2 h (Figure 4A). In contrast to free TM-AvPAL which was inactivated within 15 min of incubation at these raised protease concentrations, TM-AvPAL-silica particles retained over 50 % of their activity even after 120 min (Figure 4B). While silica matrix had been previously examined for use as a matrix for sustained release of therapeutics, our findings suggest that silica matrix may also be utilized for providing protection for encapsulated proteins against intestinal proteases.
Figure 4.
(A) Formulation parameters of TM-AvPAL silica particles. Different concentrations of TM-AvPAL were loaded into silica matrix at various pH formulations before incubation with pH 6.8 buffer (control), acidic buffer (pH 3.5) for 1 h, or solutions containing chymotrypsin or trypsin (5 mg/ml), pH 6.8 for 2 h. “High pH” conditions referred to the final formulation pH of approximately 9 as estimated by pH indicator. (B) Incubation of TM-AvPAL-silica particles (13 mg/ml) with 5 mg/ml chymotrypsin or trypsin solutions pH 6.8.
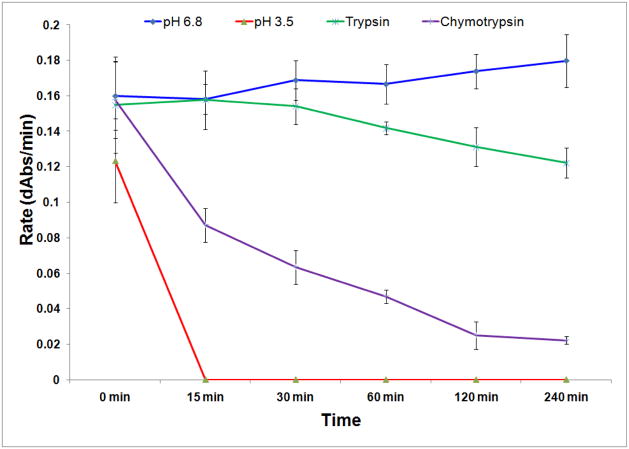
PEGylation with 5-kDA monofunctional linear polyethylene glycol (PEG) was examined as an alternative provider of protease resistance to TM-AvPAL. PEGylation is a strategy that has been established for reducing immunogenicity of proteins formulated for parenteral administration, as well as for extending circulating half-life of injectable protein therapeutics [16–17]. In the current context of developing orally administered protein therapeutics, immunogenicity of the protein is typically not relevant. Instead, the bulky N-hydroxysuccinimide activated PEG molecules that were reacted and cross-linked with TM-AvPAL through the ε-amino group of lysine’s side chain were utilized to effectively block potential trypsin cleavage sites and the consequent inactivation of the protein (Figure 5).
Figure 5.
Assay of 5 kDa PEG-TM-AvPAL. PEGylated TM-AvPAL was incubated with USP pH 3.5 buffer for 1h, or USP pH 6.8 buffer, 5 mg/ml chymotrypsin solution, or 5 mg/ml trypsin solution for 2h before assaying for catalytic activity.
Substrate affinity of TM-AvPAL and PEG-TM-AvPAL were shown to be largely unaffected by the point mutation and PEGylation, and exhibited Km comparable to the wild-type AvPAL and DM-AvPAL. However, incorporation of TM-AvPAL in silica resulted in a lower specific activity of the TM-AvPAL-silica formulation as compared to the other species (Table 2) and further optimization may be needed.
Table 2.
Comparison of activity and kinetic constants of AvPAL variants and formulations.
| Wild type AvPAL[20] | DM-AvPAL[8] | TM-AvPAL | PEG TM-AvPAL | TM-AvPAL-Silica | |
|---|---|---|---|---|---|
| Km (mM) | 0.06 | 0.05 | 0.07 | 0.07 | ND |
| Specific Activity (IU/mg) | 1.7 | 2.2 | 1.13 | 1.08 | 0.35 |
In conclusion, we have shown that point substitution of TM-AvPAL resulted in a modest improvement in the enzyme’s in vitro resistance against two prevalent intestinal enzymes. Such improvement in resistance may be further augmented through the innovative utilization of two established modalities of protein derivatization to overcome one of the classic challenges in protein drug delivery. The distinct advantages of using PEGylation and silica matrix preparations for protein delivery are the established safety and extensive applications of these reagents, as well as absence of organic solvents in the formulation process. PEGylation has been widely utilized for numerous FDA-approved protein therapeutics since the 1980s [9, 17–18], and the biocompatibility of silica matrix is being increasingly proven as a suitable delivery system [19].
Acknowledgments
This research project was supported by the NIH grant U01 NS051353. The authors would like to thank Dr. Ellen Chien for advice on thermal stability studies, and Angela Walker for assistance in manuscript preparation. T.S. Kang is supported by the National University of Singapore Overseas Postdoctoral Fellowship.
References
- 1.Donlon K, Levy H, Scriver CR. The metabolic & molecular bases of inherited disease. Chapter 77. McGraw-Hill/Medical Publishing Division; New York: 2007. Hyperphenylalaninemia: Phenylalanine Hydroxylase Deficiency. ( http://genetics.accessmedicine.com/) [Google Scholar]
- 2.Gamez A, Wang L, Straub M, Patch MG, Stevens RC. Toward PKU enzyme replacement therapy: PEGylation with activity retention for three forms of recombinant phenylalanine hydroxylase. Mol Ther. 2004;9:124–129. doi: 10.1016/j.ymthe.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Sarkissian CN, et al. A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2339–2344. doi: 10.1073/pnas.96.5.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourget L, Chang TM. Artificial cell-microencapsulated phenylalanine ammonia-lyase. Applied biochemistry and biotechnology. 1984;10:57–59. doi: 10.1007/BF02783735. [DOI] [PubMed] [Google Scholar]
- 5.Sarkissian CN, Gamez A. Phenylalanine ammonia lyase, enzyme substitution therapy for phenylketonuria, where are we now? Molecular genetics and metabolism. 2005;86(Suppl 1):S22–26. doi: 10.1016/j.ymgme.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Hoskins JA, Gray J. Phenylalanine ammonia lyase in the management of phenylketonuria: the relationship between ingested cinnamate and urinary hippurate in humans. Research communications in chemical pathology and pharmacology. 1982;35:275–282. [PubMed] [Google Scholar]
- 7.Ambrus CM, et al. Phenylalanine depletion for the management of phenylketonuria: use of enzyme reactors with immobilized enzymes. Science (New York, NY) 1978;201:837–839. doi: 10.1126/science.567372. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, et al. Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase. Journal of molecular biology. 2008;380:623–635. doi: 10.1016/j.jmb.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkissian CN, et al. Preclinical evaluation of multiple species of PEGylated recombinant phenylalanine ammonia lyase for the treatment of phenylketonuria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20894–20899. doi: 10.1073/pnas.0808421105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah RM, D’Mello AP. Strategies to maximize the encapsulation efficiency of phenylalanine ammonia lyase in microcapsules. International journal of pharmaceutics. 2008;356:61–68. doi: 10.1016/j.ijpharm.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 11.Frenkel-Mullerad H, Avnir D. Sol-gel materials as efficient enzyme protectors: preserving the activity of phosphatases under extreme ph conditions. Journal of the American Chemical Society. 2005;127:8077–8081. doi: 10.1021/ja0507719. [DOI] [PubMed] [Google Scholar]
- 12.Bhatia RB, Brinker CJ, Gupta AK, Singh AK. Aqueous Sol-Gel Process for Protein Encapsulation. Chemistry of Materials. 2000;12:2434–2441. [Google Scholar]
- 13.Ellerby LM, et al. Encapsulation of proteins in transparent porous silicate glasses prepared by the sol-gel method. Science (New York, NY) 1992;255:1113–1115. doi: 10.1126/science.1312257. [DOI] [PubMed] [Google Scholar]
- 14.Barbe C, et al. Silica Particles: A Novel Drug-Delivery System. Advanced Material. 2004;16:1959–1965. [Google Scholar]
- 15.Lebert JM, Forsberg EM, Brennan JD. Solid-phase assays for small molecule screening using sol-gel entrapped proteins. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2008;86:100–110. doi: 10.1139/O08-010. [DOI] [PubMed] [Google Scholar]
- 16.Delgado C, Francis GE, Fisher D. The uses and properties of PEG-linked proteins. Critical reviews in therapeutic drug carrier systems. 1992;9:249–304. [PubMed] [Google Scholar]
- 17.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nature reviews. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 18.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature reviews. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 19.Rigby SP, Fairhead M, van der Walle CF. Engineering silica particles as oral drug delivery vehicles. Current pharmaceutical design. 2008;14:1821–1831. doi: 10.2174/138161208784746671. [DOI] [PubMed] [Google Scholar]