Abstract
Here we report the characterization of a knock-in mouse model for the autosomal recessive disorder mucopolysaccharidosis type I-Hurler (MPS I-H), also known as Hurler syndrome. MPS I-H is the most severe form of α-L-iduronidase deficiency. α-L-iduronidase (encoded by the IDUA gene) is a lysosomal enzyme that participates in the degradation of dermatan sulfate and heparan sulfate. Using gene replacement methodology, a nucleotide change was introduced into the mouse Idua locus that resulted in a nonsense mutation at codon W392. The Idua-W392X mutation is analogous to the human IDUA-W402X mutation commonly found in MPS I-H patients. We found that the phenotype in homozygous Idua-W392X mice closely correlated with the human MPS I-H disease. Homozygous W392X mice showed no detectable α-L-iduronidase activity. We observed a defect in GAG degradation as evidenced by an increase in sulfated GAGs excreted in the urine and stored in multiple tissues. Histology and electron microscopy also revealed evidence of GAG storage in all tissues examined. Additional assessment revealed bone abnormalities and altered metabolism within the Idua-W392X mouse. This new mouse will provide an important tool to investigate therapeutic approaches for MPS I-H that cannot be addressed using current MPS I-H animal models.
Keywords: MPS I, Hurler syndrome, mouse, W402X mutation, α-L-iduronidase
INTRODUCTION
The mucopolysaccharidosis (MPS) diseases are a group of lysosomal storage disorders caused by a deficiency in one of the lysosomal enzymes that catalyzes the degradation of glycosaminoglycans (GAGs). Lysosomal accumulation of GAGs results in cellular dysfunctions, organ abnormalities, and metabolic defects through mechanisms that are not entirely understood [1-4]. Currently, there are eleven known lysosomal enzyme deficiencies that lead to seven distinct MPS diseases. MPS I is caused by a deficiency of α-L-iduronidase (EC 3.2.1.76, encoded by the IDUA gene) that leads to the lysosomal accumulation of dermatan sulfate and heparan sulfate. Depending upon the amount of residual α-L-iduronidase activity, the severity of the MPS I phenotype can vary widely. Consequently, MPS I has been categorized as having three distinct phenotypic subtypes: MPS I-Hurler (MPS I-H), the severe form; MPS I-Scheie (MPS I-S), the mild form; and MPS I-Hurler/Scheie (MPS IHS), an intermediate form. MPS I-H is a progressive disorder with multiple organ and tissue involvement that includes skeletal deformities, hearing loss, corneal clouding, heart failure and mental retardation. Patients usually die within their first decade as a result of obstructive airway disease, respiratory infection, or cardiac complications [2].
Two naturally occurring MPS I-H animal models have previously been characterized. The first was a feline model with a three-nucleotide deletion in the Idua gene that results in the loss of a single amino acid in the α-L-iduronidase protein [5, 6]. A canine model was later identified with a splice site mutation that causes retention of an intron in the Idua mRNA and leads to premature termination of α-L-iduronidase protein synthesis [7-9]. In addition, two MPS I-H mouse models have been generated using knockout strategies that disrupt the Idua gene using an insertion cassette [10, 11]. The MPS I-H animal models have phenotypes that are generally consistent with the disease manifestations of MPS I-H patients, including deficiency of α-L-iduronidase activity [11, 12], accumulation of GAGs in most tissues [6, 10, 11, 13, 14], increased urine GAG excretion [11, 15], accumulation of GM2 and GM3 gangliosides in the brain [16, 17], abnormal facial appearance [16, 18], bone deformities [11, 19], neuropathology [20, 21], and cardiac manifestations [22, 23].
These existing animal models have proven to be valuable tools to investigate the pathogenesis of MPS I-H, and to evaluate several therapeutic strategies such as stem cell transplantation [12], enzyme replacement therapy [24], and gene therapy [15, 19]. However, the current animal models are not useful in evaluating the effectiveness of other promising therapeutic approaches. In particular, the lack of MPS I-H mouse models that carry mutations identified in MPS I-H patients limits many prospective investigations since some therapeutic approaches target a specific mutation or mutation type. In order to obtain an MPS I-H animal model that can be used to investigate a wider range of therapeutic approaches, we generated an Idua-W392X knock-in mouse model that carries a nonsense mutation corresponding to the IDUA-W402X mutation, the most common mutation found in MPS I-H patients. Here we report the characterization of the Idua-W392X mouse. We evaluated the phenotype of Idua-W392X mice at three ages and found that mutant mice developed a quantifiable disease progression. We found evidence of biochemical, metabolic, and morphological abnormalities that correlate closely with the phenotype described for other MPS I-H animal models [10, 11, 14] as well with the human MPS I-H disease [2]. Thus, the Idua-W392X mouse will allow us to evaluate the efficacy of therapeutic approaches that previously have been limited by the availability of a suitable MPS I-H animal model.
MATERIALS AND METHODS
Generation of the Idua-W392X Knock-in Mouse
The Idua-W392X targeting construct was made using a 129/Sv mouse genomic DNA fragment containing Idua exons 3-14 [11]. The W392X mutation (TGG→TAG) was introduced into exon 9 of the Idua gene by site-directed mutagenesis and was verified by sequencing. A viral thymidine kinase negative selectable marker was placed directly upstream of Idua exon 3, and a loxP-flanked neomycin-resistance positive selectable marker was cloned into the BstZ17I restriction site within the intron between exons 8 and 9. The targeting construct was electroporated into 129/Sv mouse ES cells that were subsequently cultured in 250 μg/ml G418 and 2 μM gancyclovir to select for recombinants. The ES cells bearing the integrated Idua-W392X construct as determined by Southern blot were introduced into C57BL/6J mouse blastocysts and then transferred to a pseudopregnant female. Chimeric pups carrying Agouti coat color were crossed with C57BL/6J mice to identify offspring that transmitted the Idua-W392X allele through the germ line. The Idua-W392X allele was detected by PCR-amplifying the region from Idua exon 8 to the 3′ end of the neo cassette (oriented 3′ to 5′ relative to the Idua allele) using primers DB1109 5′-GCCTGGCACA TCCTGTATTG-3′ and DB1101 5′-GCGAATGGGC TGACCGCTTC-3′. The floxed neo cassette was excised by crossing Idua-W392X mice with C57BL/6-Tg(Zp3-cre)93Knw/J mice. The 34 bp loxP site that remained after the excision was utilized to confirm removal of the neo cassette and to distinguish the Idua-W392X allele from the wild-type Idua allele by PCR using primers DB1109 5′-GCCTGGCACA TCCTGTATTG-3′ and DB0943 5′-GTAGGGGCTG CTGAGTTGAT-3′. Heterozygous Idua-W392X mice were backcrossed six more generations (eight generations total) against the C57BL/6J background to obtain congenic Idua-W392X mice. All animal protocols used in this study were reviewed and approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Enzymatic Assays
α-L-iduronidase assays were performed using a protocol described by Hopwood et al. [25]. β-D-glucuronidase and β-D-hexosaminidase assays were performed as described by Birkenmeier et al. [26] and Wendeler & Sandhoff [27], respectively. Tissue was homogenized in T-Per Tissue Protein Extraction Reagent (Pierce) using a rotor-stator homogenizer (Biospec). α-L-iduronidase assays were performed using 4-methyl-umbelliferyl-α-L-iduronide (Glycosynth) and D-saccharic acid 1,4-lactone monohydrate (Sigma). β-D-glucuronidase and β-D-hexosaminidase reactions were performed using 4-methyl-umbelliferyl-β-D-glucuronide and 4-methyl-umbelliferyl-N-acetyl-β-D-glucosaminide, respectively (Sigma).
Urine GAG Assays
Animals were fed with Peptamen Liquid Elemental Diet (Nestle) for one week before being individually housed in a diuresis cage (Nalgene) to obtain urine for a 24-hour period. Urine was collected, filtered through a sterile 0.45μM filter (Whatman), and stored at 4°C until assay. Urinary GAG excretion was quantitated using a 1,9-dimethylmethylene blue chloride (DMB) (Sigma) assay as previously described [28, 29] and normalized to creatinine. 50-100 microliters of each urine sample was used to perform the assay.
Tissue GAG Assays
Tissue was homogenized and delipidated in chloroform: methanol (2:1) using a rotor-stator homogenizer (Biospec) and then dried in a speed vac. The GAGs were released from the defatted, dried tissue by incubation with papain at 60°C overnight. The level of sulfated GAGs was determined by the Alcian Blue precipitation method described by Kobayashi et al. [30]. 50 microliters of each cleared tissue lysate was used for the assay.
Quantitive Real Time PCR (qRT-PCR)
Total RNA was isolated from wild-type and mutant mouse tissues using TRI reagent (Sigma) followed by Qiagen RNeasy columns. The RNA was reverse transcribed into cDNA using AMV reverse transcriptase (Promega). Equal amounts of cDNA were used to perform qRT-PCR with the iQ5 Real Time PCR Detection System using the iQ SYBR Green Supermix (Bio-Rad). Primers used to amplify iduronidase were: DB3192 5′-TGACAATGCC TTCCTGAGCT ACCA-3′ and DB3193 5′-TGACTGTGAG TA CTGGCTTT CGCA-3′. Mouse actin mRNA was used as a reference gene to normalize gene expression and amplified using DB2783 5′-CTTCTGCATC CTGTCAGCAA T-3′ and DB2784 5′-GAGGCTCTTT TCCAGCCTTC C-3′. PCR efficiency was found to be between 95-105% for both iduronidase and actin reactions. The relative gene expression was determined by the Livak method [31].
Histology
Mice were sacrificed by exsanguination under deep anesthesia induced by intraperitoneal injection of 100 mg/kg pentobarbital. Liver, spleen, kidneys, heart, lungs and brain were collected and fixed in neutral buffered 10% formalin or buffered ethanolic formalin containing 75% ethanol and 10% formalin. Tissues were trimmed thus: Liver, center of the left lobe and right portion of the median lobe; spleen, hemisectioned longitudinally; kidneys, hemisectioned longitudinally through the pelvis; heart, hemisectioned through the long axis and perpendicular to the septum; lung, multiple cross sections across all lobes; and brain, entire organ cut into 2 mm blocks including cuts at the optic chiasm and the midpoint of the cerebellum. Tissues were processed routinely for paraffin sectioning, sectioned at 5 μm thickness, and stained with hematoxylin and eosin (HE). Duplicate sections were stained with periodic acid-Schiff and hematoxylin (PASH). Selected tissues were digitally photographed using a Nikon E600 microscope and SPOT Insight® digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI).
Transmission Electron Microscopy
Mice were perfused first with PBS and then with a modified Karnovsky's solution (2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer). Tissue specimens were fixed with the modified Karnovsky's solution, post fixed with 1% osmium tetroxide in phosphate buffer and embedded (Embed 812, EMS, Fort Washington, PA). The embedded specimens were thick sectioned and stained with Toluidine Blue, thin sectioned and stained with uranyl acetate-lead citrate and placed on copper grids. After drying the grids were viewed on a Tecnai Twin 120kv TEM (FEI, Hillsboro, OR) and digital images were taken with an AMT CCD camera.
Radiography
Mice were anesthetized with isoflurane and examined using a Specimen Radiography System MX-20 (Faxitron X-ray Corporation, Lincolnshire, IL) at 26KV for 10 second.
Dual-energy X-ray Absorptiometry (DXA)
Mice were anesthetized with isoflurane (3% in oxygen) and placed prostrate on the imaging plate. The legs were extended away from the body. Scanning was performed using the GE-Lunar PIXImus (Madison, WI) with software version 1.45. Due to the small imaging area, the heads of the mice were excluded from all analysis.
Statistical Analysis
All statistical analyses were performed using the two-tailed t-test (InStat software) with the exception of the survival curves. The survival curves were constructed using the Kaplan-Meier method and were compared using the two-tailed log-rank test (GraphPad Prism software).
RESULTS
Replacement targeting of the Idua locus in murine ES cells
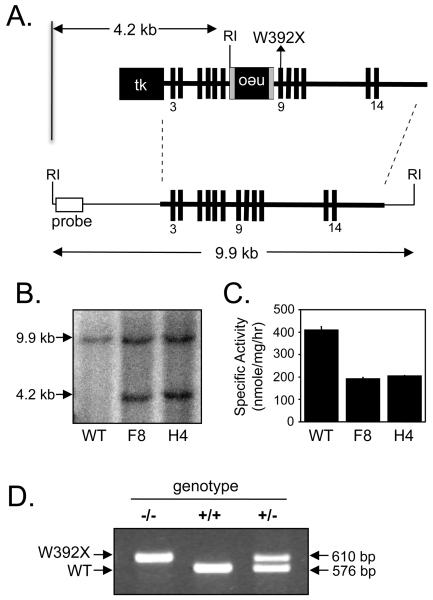
A replacement-targeting construct was generated using a 129/Sv mouse genomic DNA fragment containing Idua exons 3-14 (Figure 1A) [11]. The W392X mutation (TGG→TAG), which corresponds to the W402X mutation found in MPS I-H patients, was introduced into exon 9 of the Idua gene. Thymidine kinase and neomycin resistance genes were introduced into the targeting construct to provide a means of identifying ES cell clones that had undergone homologous recombination. After transfecting 129/Sv ES cells with the linearized targeting construct and culturing in the presence of gancyclovir and G418, several resistant clones were isolated. Southern blotting was used to verify the integration of the Idua-W392X targeting construct into the Idua locus. An EcoRI digest of mouse genomic DNA that resulted in a 9.9 kb fragment from the wild-type Idua allele and a 4.2 kb fragment from the Idua-W392X targeted allele (Figure 1A, 1B) was used to identify several ES cell clones that showed recombination of the Idua-W392X allele into the genomic Idua locus (the F8 and H4 clones are shown). In order to verify that a duplication of the Idua locus had not occurred during the recombination of the Idua-W392X allele, ES cell clones were further assayed biochemically to confirm the level of α-L-iduronidase activity. The F8 and H4 ES cells were observed to produce only 50% of the enzymatic activity present in wild-type ES cells (Figure 1C), confirming that a proper recombination event had occurred. The F8 and H4 targeted ES cell clones were identified as heterozygous for the Idua-W392X allele and used to generate Idua-W392X mice.
Figure 1.
Generation of the Idua-W392X knock-in mouse. (A) The Idua-W392X targeting construct (upper) was integrated into the mouse Idua locus (lower) by homologous recombination (dashed lines). The bold horizontal lines indicate the targeted region. The bold vertical lines indicate exons (not shown to scale). Exons 3, 9, and 14 are numbered. The W392X mutation was introduced into exon 9. The sizes of the genomic DNA fragments after EcoRI digestion in both wild-type and targeted Idua loci are indicated as well as the region of probe binding used in Southern blotting. tk: thymidine kinase gene; neo: neomycin resistance gene (flanked by loxP sites shown as gray bars). (B) Southern blot of wild-type and two targeted ES cell clones (denoted as F8 and H4) resulted in a 9.9 kb fragment from the wild-type Idua locus and a 4.2 kb fragment from the targeted Idua locus. (C) α-L-iduronidase specific activity determined in wild-type and two targeted ES cell clones. (D) PCR of the Idua allele using genomic DNA derived from tail snips (following Cre recombinase-mediated excision of the neo cassette) results in a 576 bp product from the wild-type Idua allele and a 610 bp product from the W392X allele.
Generation of Idua-W392X mice
Idua-W392X mice were generated from two ES cell lines, F8 and H4. Preliminary characterization showed that mice derived from the two independent ES cell lines had an identical phenotype. All further characterization was done using the Idua-W392X mouse derived from the H4 ES cell line. The floxed neo resistance cassette in the targeted allele was removed by Cre-mediated recombination to avoid the possibility that a cryptic splice site inside the cassette might cause the Idua-W392X allele to be alternatively spliced. The 34 bp loxP site that remained within the intron between exons 8 and 9 after excision of the neo cassette was used to distinguish the targeted Idua-W392X allele (610 bp) from the wild-type Idua allele (576 bp) by PCR (Figure 1D). The Idua-W392X mice derived from the targeted H4 ES cell clone were crossed against C57BL/6J mice for eight generations and were used in all subsequent experiments. Since heterozygotes were phenotypically normal, we used both homozygous (Idua+/+) and heterozygous (Idua+/−) mice as wild-type controls for all characterization assays with the exception of α-L-iduronidase enzymatic assays and mRNA quantitation.
Biochemical characterization of Idua-W392X mice
α-L-iduronidase activity
The W402X nonsense mutation identified in MPS I-H patients has been reported to eliminate the activity of α-L-iduronidase since the mutation leads to the premature termination of α-L-iduronidase protein synthesis [32]. We predicted that the mouse Idua-W392X mutation, located at the analogous codon as the human W402X mutation, would also severely reduce the function of α-L-iduronidase. To evaluate α-L-iduronidase function, we performed assays to determine the specific activity of α-L-iduronidase in brain and liver homogenates from 5-, 10- and 30-week-old homozygous wild-type (Idua+/+) and homozygous Idua-W392X (Idua−/−) mice (Table 1). While we could reproducibly detect a normal level of α-L-iduronidase activity in homogenates from wild-type mice, the level of enzyme activity in corresponding samples from homozygous mutant mice was below the level of detection of our fluorescent assay. Heterozygous (Idua+/−) mice were found to have approximately 50% of the α-L-iduronidase activity found in homozygous wild-type (Idua+/+) mice (data not shown).
Table 1.
Lysosomal Enzyme Activity in Mouse Tissue Lysates.
| 5 weeks |
10 weeks |
30 weeks |
|||||
|---|---|---|---|---|---|---|---|
| WT (n=5) | W392X (n=5) | WT (n=6) | W392X (n=5) | WT (n=4) | W392X (n=4) | ||
| α-Idua | Brain | 8.4±1.7 | bls | 9.6±1.3 | bls | 11.3±0.4 | bls |
| Liver | 4.6±1.8 | bls | 7.1±1.0 | bls | 5.6±0.9 | bls | |
| β-Gluc | Brain | 17.0±2.8 | #55.7±9.1 | 8.8±1.5 | *13.4±3.0 | 8.4±0.5 | #18.7±2.3 |
| Liver | 154.5±34.9 | #372.0±88.2 | 119.8±12.5 | #394.0±42.6 | 117.4±4.2 | #635.8±29.8 | |
| β-Hex | Brain | 2303.5±284.3 | #4397.1±736.0 | 431.3±52.0 | #1486.8±231.4 | 413.2±36.9 | #2087.5±151.1 |
| Liver | 435.7±159.5 | #3071.0±718.7 | 160.6±31.1 | #1547.3±309.2 | 140.5±10.3 | #2417.5±226.5 | |
Data expressed as the mean ± SD of the enzymatic specific activity (nanomoles FMU released per milligram total protein per hour);
n = number of mice; bls = below level of sensitivity for assay.
p < 0.001,
p < 0.02.
Urine GAG excretion
Since α-L-iduronidase is required for the degradation of heparan sulfate and dermatan sulfate, the primary biochemical defect in homozygous Idua-W392X mice was expected to be GAG accumulation. Accumulation of GAGs in MPS diseases often results in an increase in the amount of GAGs excreted in the urine [2]. To determine if urinary GAG levels were elevated in the Idua-W392X mouse, a dimethylmethylene blue assay was performed to measure the level of sulfated GAGs that were excreted in the mouse urine [28, 29]. The level of urine GAGs was normalized to urine creatinine levels to correct for kidney function. In all three age groups, urine GAG levels in homozygous Idua-W392X mice were significantly elevated relative to age-matched wild-type mice (Figure 2A). Urine GAG levels were elevated 2.5-fold, 3.6-fold, and 5.6-fold in 5-, 10-, and 30-week-old mutant mice, respectively. This progressive increase in GAG excretion in mutant mice suggests that the GAG storage and MPS I-H disease is progressive in the Idua-W392X mouse.
Figure 2.
GAG accumulation in Idua-W392X mice. (A) Increased urinary GAG excretion in Idua-W392X mice (W392X) compared to wild-type mice (WT). (B) Fold-increase in tissue GAG levels. Each bar represents the mean ± SD of the fold-increase in tissue GAGs in mutant mice relative to wild-type controls. The dashed line indicates the normalized wild-type levels. Each assay was performed using at least 5 mice. *p < 0.001 (W392X vs. WT in each age group); #p < 0.001 (W392X at 30-weeks-old vs. W392X at 5- or 10-weeks-old).
Tissue GAG accumulation
In order to directly measure the level of sulfated GAGs in mouse tissues, an Alcian Blue GAG binding assay was performed [30]. GAGs isolated from brain, heart, kidney, liver, lung, and spleen were quantitated in mice from three age groups. Evidence of abnormal GAG storage was present in all tissues assayed from all three age groups of Idua-W392X mice (Figure 2B and Table 2). The increase in GAG accumulation among the various tissues from mutant mice ranged from 1.2- to 7.5-fold for 5-week-old mice, 1.5- to 9.6-fold for 10-week-old mice, and 1.6- to 17.7-fold for 30-week-old mice. For all three age groups, the organ with the lowest increase in GAG accumulation was the brain and the organ with the highest increase in GAG accumulation was the liver. While the increase in GAGs observed in 5-week-old mutant mice remained relatively constant for brain, kidney, and lung tissues as the mice aged, a progressive increase in GAGs was observed in the heart, liver, and spleen as the mice became older (Figure 2B). These data suggest a continual accumulation of GAGs in these organs with the MPS I-H disease progression.
Table 2.
Tissue GAG Levels.
| 5 weeks |
10 weeks |
30 weeks |
||||
|---|---|---|---|---|---|---|
| WT (n=5) | W392X (n=5) | WT (n=7) | W392X (n=7) | WT (n=8) | W392X (n=8) | |
| Brain | 3.24±0.24 | #4.01±0.36 | 2.53±0.45 | *3.86±1.25 | 2.80±0.27 | #4.50±0.51 |
| Heart | 1.38±0.31 | #3.14±0.40 | 1.60±0.98 | #5.08±1.95 | 0.84±0.12 | #5.16±0.45 |
| Kidney | 3.02±0.53 | #11.23±2.14 | 3.05±0.71 | #10.95±1.26 | 2.59±0.51 | #10.03±1.53 |
| Liver | 1.38±0.20 | #10.38±0.86 | 1.23±0.32 | #11.79±2.04 | 1.11±0.13 | #19.62±2.80 |
| Lung | 4.39±1.08 | #13.75±1.17 | 3.92±1.04 | #13.31±2.84 | 3.49±0.32 | #8.50±0.49 |
| Spleen | 4.87±2.20 | *8.90±1.96 | 6.87±1.28 | #13.78±2.33 | 4.21±1.02 | #13.50±1.80 |
Tissue GAG values are represented as the mean ± SD of the micrograms of GAGs per milligram of defatted, dried tissue. n = number of mice.
p < 0.004,
p ≤ 0.02.
Idua mRNA quantitation
Premature stop mutations have been found to lead to a reduction in steady state mRNA levels [33]. The W402X nonsense mutation has previously been shown to cause a reduction in steady state IDUA mRNA levels in human cells [34]. In order to determine if the W392X mutation leads to a reduction in steady state mouse Idua mRNA levels, we isolated total mRNA from brain, liver, and spleen tissues in homozygous wild-type and mutant mice. The mRNA was reverse-transcribed to cDNA and then subjected to real-time quantitative PCR analysis using primers specific for the mouse Idua mRNA. We found that the steady state Idua mRNA level was reduced in mutant mice by 30-50%, indicating that the W392X mutation leads to a reduction in the steady state level of mouse Idua mRNA (Table 3).
Table 3.
Idua mRNA levels in Idua-W392X mouse tissues.
| Tissue: | Percent of wild-type Idua mRNA levels: |
|---|---|
| Brain | 54.3±6.3 |
| Liver | 65.9±8.5 |
| Spleen | 69.3±5.6 |
Quantitation of steady state Idua mRNA levels in tissues from homozygous wild-type and mutant mice was performed using quantitative real time PCR. Each reaction was performed in quadruplicate using tissues from at least two mice. The data is expressed as the average ± SD of the percent of Idua mRNA levels from Idua-W392X mice relative to Idua mRNA levels from wild-type mice.
Microscopic analysis of tissues
Histology
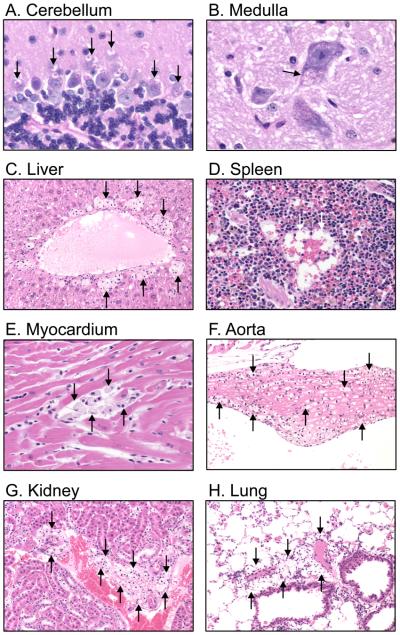
In order to determine the effects of GAG storage in tissues at the cellular level, histological staining was performed using tissues from age-matched Idua-W392X and wild-type mice. The tissues examined included the brain (cerebellum and medulla), liver, spleen, heart (myocardium and aorta), kidney, and lung. Representative histological samples from 30-week-old Idua-W392X mice are shown in Figure 3. Cytoplasmic inclusions representing lysosomal storage materials were found in the Purkinje cells of the cerebellum (A), and in neurons of the medulla (B). In Purkinje cells, we also consistently observed the retention of stained material inside the cytoplasmic inclusions (A). Foamy macrophages distended by GAG accumulation were observed in the liver (C), spleen (D), myocardium (E), aorta (F), kidney (G), and lung (H) tissues from homozygous mutant mice. None of these abnormalities were found in wild-type tissues. Consistent with the progressive nature of MPS I-H, vacuolation and foamy macrophage infiltration was markedly less severe in younger mice (data not shown).
Figure 3.
Representative histopathology of tissues from 30-week-old Idua-W392X mice. Arrows indicate inclusions representing GAG accumulation in (A) the cytoplasm of Purkinje cells in the cerebellum; and (B) the cytoplasm of a neuron in the medulla. Foamy macrophages distended by GAG accumulation are observed in (C) liver, (D) spleen, (E) myocardium, (F) aorta, (G) kidney, and (H) lung. Panels A and B were stained with PAS-hematoxylin, original magnification 80X; Panels C, F, G, and H were stained with HE, original magnification 20X; Panels D and E were strained with HE, original magnification 40X.
Electron microscopy
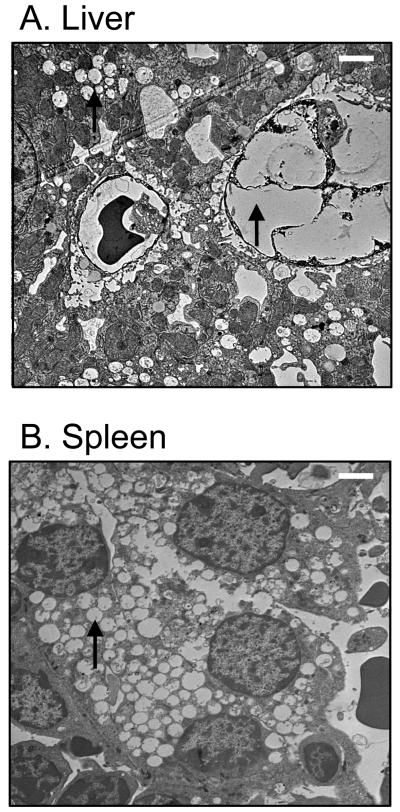
While histological analysis of spleen and liver tissues from older Idua-W392X mice revealed substantial foamy macrophage infiltration and cytoplasmic inclusions, these features were not as readily detected in tissues from younger MPS I-H mice. We therefore employed electron microscopy (EM) to compare spleen and liver tissues from 14-week-old wild-type and mutant mice at the cellular level to determine the extent of vacuolation caused by GAG storage. EM analysis of liver and spleen tissues from homozygous Idua-W392X mice revealed significant vacuolation (Figure 4). Foamy macrophage infiltration could also be detected in these samples as shown in the liver specimen in Panel A.
Figure 4.
Representative electron micrographs of tissues from 14-week-old Idua-W392X mice. Arrows indicate vacuolation and a foamy macrophage in (A) liver and vacuolation in (B) spleen. White bars (in upper right of each panel) represent 2 μm.
Additional Lysosomal markers
It was previously shown that the depletion of one lysosomal hydrolase often leads to increased activity of other lysosomal enzymes through poorly understood mechanisms [12, 13, 35]. For example, increased β-D-hexosaminidase activity was documented in an Idua knock-out mouse model [12, 35]. We investigated whether β-D-glucuronidase and β-D-hexosaminidase activities are elevated in brain and liver tissue lysates in the Idua-W392X mouse. β-D-glucuronidase degrades dermatan sulfate and heparan sulfate (the same substrates as α-L-iduronidase) as well as chondroitin-4-sulfate and chondroitin-6-sulfate. β-D-hexosaminidase degrades GM2 gangliosides. We found increases in the activities of both enzymes in Idua-W392X mice (Figure 5 and Table 1). The β-D-glucuronidase activity increased 1.5- to 3.3-fold in the brain and 2.4- to 5.4-fold in the liver tissues of mutant mice. The β-D-hexosaminidase activity increased 1.9- to 5.1-fold in the brain and 7.0- to 17.2-fold in the liver tissues among the mutant mice. In general, the fold-increase in enzyme activity correlated with increasing age, suggesting disease progression. The only exception to this observation occurred with the β-D-glucuronidase activity in brain, which showed a higher level in 5-week-old mice than in 10- or 30-week-old mice (Figure 5 and Table 1).
Figure 5.
Additional lysosomal enzyme activities in Idua-W392X mouse tissue lysates. β-D-glucuronidase (β-Gluc) and β-D-hexosaminidase (β-Hex) activities were measured. Each bar indicates the mean +/− standard deviation of the fold-increase in lysosomal enzyme specific activities in Idua-W392X tissue lysates relative to wild-type. Each assay was performed using at least four mice. The dashed, horizontal line indicates the normalized levels of wild-type activity.
Morphological characterization of Idua-W392X mice
Facial features
Bone abnormalities are often associated with lysosomal storage diseases, including MPS I-H [2]. We therefore monitored homozygous Idua-W392X mice for evidence of morphologic abnormalities and bone defects. At birth, the facial features of homozygous Idua-W392X mice were morphologically normal and indistinguishable from wild-type littermates (data not shown). At 10-15 weeks of age, mutant mice gradually showed a slightly broadened snout, which became more prominent at 35-weeks-old (Figure 6A, upper panels). Radiography revealed a mild thickening of the zygomatic arch in mice at 15-weeks-old (data not shown) and a more pronounced zygomatic arch thickening in 35-week-old Idua-W392X mice (Figure 6A, lower panels). This abnormal appearance was consistent with other MPS I-H animal models and the coarse facial features observed in MPS I-H patients [11].
Figure 6.
Morphological assessment of Idua-W392X mice. (A) Photographs showing a broadened snout (upper) and radiographs showing a thickened zygomatic arch (lower, white arrows) in a 35-week-old Idua-W392X mouse. (B) Radiographs showing a shortened femur in a 5-week-old Idua-W392X mouse, and a widened femur in a 35-week-old Idua-W392X mouse. Arrows indicate the regions of bone length (left) and width (right) measured, respectively. (C) Quantitation of femur measurements. Each bar indicates the mean +/− standard deviation of the length or width of the femur in Idua-W392X mice relative to wild-type. The dashed line indicates normalized wild-type values. Each measurement contains at least 5 mice. *p < 0.01.
Bone development
Additional abnormalities were observed in the long bones of homozygous Idua-W392X mice. We were able to quantitate age-dependent differences in the femur length and width in homozygous Idua-W392X mice compared to age-matched wild-type controls. In 5-week-old W392X mice, the femur was 15% shorter than in age-matched wild-type controls (Figure 6B,C); however, this decrease in femur length is less evident in 15-week-old mutant mice and was no longer detectable in 35-week-old mutant mice. No difference was found in the femur width between wild-type and mutant 5-week-old mice. However, the femur was found to be wider than normal in both 15- and 35-week-old mutant mice with a 1.13-fold and 1.37-fold increase in femur width, respectively (Figure 6B,C).
Dual-energy X-ray absorptiometry (DXA) analysis
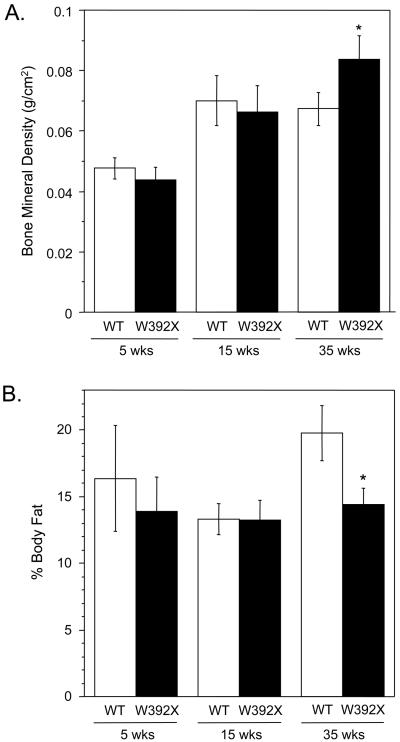
To identify other disease markers suitable for non-invasive evaluation of therapeutic effects, we performed DXA analysis to examine bone mineral density (BMD) and body composition. Previous studies showed that in an Idua knockout mouse model, BMD increased [19, 36] while percent body fat decreased [37]. While we found no significant differences in the BMD or percent body fat between mutant mice and wild-type controls at 5- or 15-weeks of age, we did observe that 35-week-old homozygous Idua-W392X mice showed a 24% increase in femur BMD (Figure 7A) and a 28% decrease in the percent body fat (Figure 7B). In general, these results agree with data obtained with previous MPS I-H animal models.
Figure 7.
DXA analysis of Idua-W392X mice. (A) Increased bone mineral density of femur in 35-week-old Idua-W392X mice. (B) Decreased percent body fat in 35-week-old Idua-W392X mice. Each bar represents mean +/− standard deviation from at least 5 mice. *p < 0.01.
Survival
Previous studies have shown that the lifespan of a MPS I-H knock-out mouse model is abbreviated compared to wild-type control mice [16]. In order to determine whether Idua-W392X mice have a reduced lifespan, we observed the survival of 14 wild-type and 15 Idua-W392X mice (Figure 8). We observed that Idua-W392X mice have a significantly reduced survival rate compared to wild-type controls (p = 0.0011), with a median lifespan estimated to be 69 weeks.
Figure 8.
Survival curves. 14 wild-type (WT) (indicated by the thick line) and 15 Idua-W392X (W392X) (indicated by the thin line) mice were monitored to calculate the percent survival of each group, respectively. The circles represent spontaneous death events. The squares represent live mice that are counted for percent survival calculations during the time of their current ages.
DISCUSSION
In this study, we report the generation and characterization of an Idua-W392X knock-in mouse model of MPS I-H. The Idua-W392X mutation is analogous to the IDUA-W402X mutation found in MPS I-H patients. We found that this mouse has a phenotype similar to other MPS I-H animal models and to human MPS I-H. The Idua-W392X mutation results in loss of α-L-iduronidase activity, leading to a significant increase in GAG levels in multiple tissues and an increase in urine GAG excretion. Abnormal tissue and cell morphology was observed in mutant samples using histological analysis and electron microscopy. In addition, abnormal metabolism in mutant mice was evidenced by an increase in other lysosomal enzymes, a decrease in body fat, and an increase in bone density in mutant mice. Bone abnormalities such as thickening of the zygomatic arch and aberrations in the length and width of the femur were also observed.
By analyzing the Idua-W392X mouse at three ages, we also observed a distinct progression in the severity of the MPS I-H phenotype in this animal model. This included a progression in the primary biochemical defect of GAG accumulation in the heart, liver, and spleen and an increase in urine GAG excretion. We also saw a distinct progression in the severity of the morphological abnormalities of the mice that included broadening of the face and thickening of the zygomatic arch. An increase in the level of lysosomal enzyme activity was also observed with age progression.
Small animal models with mutations that differ from those found in patients can pose a problem for testing novel therapeutic approaches in vivo. For example, the previous MPS I-H mouse models were generated by the insertion of a neomycin cassette into exon 6 of the Idua gene [10, 11]. This provided a mouse model that allowed investigators to learn about the pathophysiology of MPS I-H. While these animal models were helpful to examine the effectiveness of some therapies, they are not useful to investigate novel therapeutic approaches designed to target a specific mutation (or a specific type of mutation). By introducing the W392X point mutation into the mouse Idua locus, we have made a small animal model that contains a mutation analogous to the W402X nonsense mutation frequently found in MPS I-H patients. Since this mouse model contains a more physiologically relevant mutation, it can be used to test a variety of therapeutic approaches that target specific mutations such as oligonucleotide mutation repair strategies [38] or nonsense suppression strategies [39-41].
A large number of studies suggest that nonsense suppression therapy may be a useful therapeutic approach to treat genetic diseases caused by nonsense mutations [39-41]. Suppression therapy utilizes compounds that induce the ribosome to suppress translation termination specifically at nonsense mutations to restore at least a fraction of normal protein function. Several drugs known to induce suppression of premature stop codons have been found to be effective in restoring α-L-iduronidase function in human cells that carry nonsense mutations in the IDUA gene [42-44]. Current treatments for MPS I-H that include stem cell transplantation and enzyme replacement therapy have been able to alleviate the disease phenotype in many tissues. However, stem cell transplantation is currently the only therapy shown to prevent neurological deterioration in MPS I-H patients, since exogenously supplied α-L-iduronidase cannot cross the blood-brain barrier [45, 46]. Because some of the compounds previously shown to suppress premature stop mutations can cross the blood-brain barrier to some extent [47-52], this therapeutic approach might be a way to alleviate the neurological phenotype in MPS I-H patients that carry nonsense mutations. Since the Idua-W392X mouse has several quantifiable phenotypes that could be conveniently monitored, this mouse model may provide a useful tool to evaluate the effectiveness of suppression therapy for MPS I-H, either alone or in conjunction with other therapeutic approaches.
We also have evidence that the W392X mutation leads to a decrease in the level of mouse Idua mRNA. This decrease in Idua mRNA is most likely due to an mRNA degradation pathway called nonsense-mediated mRNA decay (NMD). NMD is a conserved eukaryotic mRNA surveillance mechanism that targets mRNAs containing premature stop codons for degradation [53]. NMD has been shown to reduce IDUA mRNA levels in fibroblasts from MPS I-H patients that carry the common Q70X and W402X nonsense mutations [34]. NMD may contribute to the severity of the MPS I-H phenotype since it reduces the level of Idua transcript available for translation. Thus, strategies to inhibit the degradation of Idua transcripts that contain nonsense mutations may also modulate the severity of the MPS I-H disease. In addition, strategies to inhibit NMD may also enhance the efficacy of suppression therapy, since it would make more nonsense-containing mRNA available for translation and subsequent suppression. Thus, the Idua-W392X mouse will provide a good model to investigate the relationship between NMD and the severity of the MPS I-H phenotype, as well as the relationship between NMD and nonsense suppression in vivo.
CONCLUSIONS
The Idua-W392X mouse was found to have biochemical, metabolic, and morphologic defects that are consistent with the MPS I-H disease phenotype. These quantitative phenotypes are highly reproducible and relatively convenient to measure, thus serving as excellent markers to monitor therapeutic effects. Most importantly, the Idua-W392X mouse is suitable for evaluating therapeutic interventions that target premature stop mutations and NMD.
ACKNOWLEDGEMENTS
This study was supported by NIH grant R01 NS057412. The authors also acknowledge the assistance of the UAB Transgenic Mouse Facility (NIH P30 CA13148, P30 AR048311, and P30 AR046031) and the UAB Small Animal Phenotyping Laboratory (NIH P30 DK56336, P30 AR046031, P30 NS057098, and P60 DK079626).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Walkley SU. Pathogenic cascades in lysosomal disease-Why so complex? J Inherit Metab Dis. 2009;32:181–189. doi: 10.1007/s10545-008-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neufeld EF, Muenzer J. The Mucopolysaccharidoses. The McGraw-Hill Companies; 2001. pp. 3421–3452. The Online Metabolic & Molecular Bases of Inherited Disease ( www.ommbid.com) [Google Scholar]
- 3.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 4.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 5.He X, Li CM, Simonaro CM, Wan Q, Haskins ME, Desnick RJ, Schuchman EH. Identification and characterization of the molecular lesion causing mucopolysaccharidosis type I in cats. Mol Genet Metab. 1999;67:106–112. doi: 10.1006/mgme.1999.2860. [DOI] [PubMed] [Google Scholar]
- 6.Haskins ME, Jezyk PF, Desnick RJ, McDonough SK, Patterson DF. Alpha-L-iduronidase deficiency in a cat: a model of mucopolysaccharidosis I. Pediatr Res. 1979;13:1294–1297. doi: 10.1203/00006450-197911000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Spellacy E, Shull RM, Constantopoulos G, Neufeld EF. A canine model of human alpha-L-iduronidase deficiency. Proc Natl Acad Sci U S A. 1983;80:6091–6095. doi: 10.1073/pnas.80.19.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shull RM, Munger RJ, Spellacy E, Hall CW, Constantopoulos G, Neufeld EF. Canine alpha-L-iduronidase deficiency. A model of mucopolysaccharidosis I. Am J Pathol. 1982;109:244–248. [PMC free article] [PubMed] [Google Scholar]
- 9.Menon KP, Tieu PT, Neufeld EF. Architecture of the canine IDUA gene and mutation underlying canine mucopolysaccharidosis I. Genomics. 1992;14:763–768. doi: 10.1016/s0888-7543(05)80182-x. [DOI] [PubMed] [Google Scholar]
- 10.Ohmi K, Greenberg DS, Rajavel KS, Ryazantsev S, Li HH, Neufeld EF. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc Natl Acad Sci U S A. 2003;100:1902–1907. doi: 10.1073/pnas.252784899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke LA, Russell CS, Pownall S, Warrington CL, Borowski A, Dimmick JE, Toone J, Jirik FR. Murine mucopolysaccharidosis type I: targeted disruption of the murine alpha-L-iduronidase gene. Hum Mol Genet. 1997;6:503–511. doi: 10.1093/hmg/6.4.503. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Rozengurt N, Ryazantsev S, Kohn DB, Satake N, Neufeld EF. Treatment of the mouse model of mucopolysaccharidosis I with retrovirally transduced bone marrow. Mol Genet Metab. 2003;79:233–244. doi: 10.1016/s1096-7192(03)00116-1. [DOI] [PubMed] [Google Scholar]
- 13.Shull RM, Helman RG, Spellacy E, Constantopoulos G, Munger RJ, Neufeld EF. Morphologic and biochemical studies of canine mucopolysaccharidosis I. Am J Pathol. 1984;114:487–495. [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Rivera MF, Colvin-Wanshura LE, Nelson MS, Nan Z, Khan SA, Rogers TB, Maitra I, Low WC, Gupta P. Characterization of an immunodeficient mouse model of mucopolysaccharidosis type I suitable for preclinical testing of human stem cell and gene therapy. Brain Res Bull. 2007;74:429–438. doi: 10.1016/j.brainresbull.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartung SD, Frandsen JL, Pan D, Koniar BL, Graupman P, Gunther R, Low WC, Whitley CB, McIvor RS. Correction of metabolic, craniofacial, and neurologic abnormalities in MPS I mice treated at birth with adeno-associated virus vector transducing the human alpha-L-iduronidase gene. Mol Ther. 2004;9:866–875. doi: 10.1016/j.ymthe.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Russell C, Hendson G, Jevon G, Matlock T, Yu J, Aklujkar M, Ng KY, Clarke LA. Murine MPS I: insights into the pathogenesis of Hurler syndrome. Clin Genet. 1998;53:349–361. doi: 10.1111/j.1399-0004.1998.tb02745.x. [DOI] [PubMed] [Google Scholar]
- 17.Desmaris N, Verot L, Puech JP, Caillaud C, Vanier MT, Heard JM. Prevention of neuropathology in the mouse model of Hurler syndrome. Ann Neurol. 2004;56:68–76. doi: 10.1002/ana.20150. [DOI] [PubMed] [Google Scholar]
- 18.Graupman P, Pan D, Konair B, Hartung S, McIvor S, Whitley C, Low W, Lam CH. Craniofacial abnormalities in a murine knock-out model of mucopolysaccharidosis I H: a computed tomography and anatomic study. J Craniofac Surg. 2004;15:392–398. doi: 10.1097/00001665-200405000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Xu L, Hennig AK, Kovacs A, Fu A, Chung S, Lee D, Wang B, Herati RS, Mosinger Ogilvie J, Cai SR, Parker Ponder K. Liver-directed neonatal gene therapy prevents cardiac, bone, ear, and eye disease in mucopolysaccharidosis I mice. Mol Ther. 2005;11:35–47. doi: 10.1016/j.ymthe.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Pan D, Sciascia A, 2nd, Vorhees CV, Williams MT. Progression of multiple behavioral deficits with various ages of onset in a murine model of Hurler syndrome. Brain Res. 2008;1188:241–253. doi: 10.1016/j.brainres.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reolon GK, Braga LM, Camassola M, Luft T, Henriques JA, Nardi NB, Roesler R. Long-term memory for aversive training is impaired in Idua(−/−) mice, a genetic model of mucopolysaccharidosis type I. Brain Res. 2006;1076:225–230. doi: 10.1016/j.brainres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Braunlin E, Mackey-Bojack S, Panoskaltsis-Mortari A, Berry JM, McElmurry RT, Riddle M, Sun LY, Clarke LA, Tolar J, Blazar BR. Cardiac functional and histopathologic findings in humans and mice with mucopolysaccharidosis type I: implications for assessment of therapeutic interventions in hurler syndrome. Pediatr Res. 2006;59:27–32. doi: 10.1203/01.pdr.0000190579.24054.39. [DOI] [PubMed] [Google Scholar]
- 23.Jordan MC, Zheng Y, Ryazantsev S, Rozengurt N, Roos KP, Neufeld EF. Cardiac manifestations in the mouse model of mucopolysaccharidosis I. Mol Genet Metab. 2005;86:233–243. doi: 10.1016/j.ymgme.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakkis ED, McEntee MF, Schmidtchen A, Neufeld EF, Ward DA, Gompf RE, Kania S, Bedolla C, Chien SL, Shull RM. Long-term and high-dose trials of enzyme replacement therapy in the canine model of mucopolysaccharidosis I. Biochem Mol Med. 1996;58:156–167. doi: 10.1006/bmme.1996.0044. [DOI] [PubMed] [Google Scholar]
- 25.Hopwood JJ, Muller V, Smithson A, Baggett N. A fluorometric assay using 4-methylumbelliferyl alpha-L-iduronide for the estimation of alpha-Liduronidase activity and the detection of Hurler and Scheie syndromes. Clin Chim Acta. 1979;92:257–265. doi: 10.1016/0009-8981(79)90121-9. [DOI] [PubMed] [Google Scholar]
- 26.Birkenmeier EH, Davisson MT, Beamer WG, Ganschow RE, Vogler CA, Gwynn B, Lyford KA, Maltais LM, Wawrzyniak CJ. Murine mucopolysaccharidosis type VII. Characterization of a mouse with beta-glucuronidase deficiency. J Clin Invest. 1989;83:1258–1266. doi: 10.1172/JCI114010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendeler M, Sandhoff K. Hexosaminidase assays. Glycoconj J. 2008 doi: 10.1007/s10719-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 28.Whitley CB, Ridnour MD, Draper KA, Dutton CM, Neglia JP. Diagnostic test for mucopolysaccharidosis. I. Direct method for quantifying excessive urinary glycosaminoglycan excretion. Clin Chem. 1989;35:374–379. [PubMed] [Google Scholar]
- 29.de Jong JG, Wevers RA, Liebrand-van Sambeek R. Measuring urinary glycosaminoglycans in the presence of protein: an improved screening procedure for mucopolysaccharidoses based on dimethylmethylene blue. Clin Chem. 1992;38:803–807. [PubMed] [Google Scholar]
- 30.Kobayashi H, Carbonaro D, Pepper K, Petersen D, Ge S, Jackson H, Shimada H, Moats R, Kohn DB. Neonatal gene therapy of MPS I mice by intravenous injection of a lentiviral vector. Mol Ther. 2005;11:776–789. doi: 10.1016/j.ymthe.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Scott HS, Bunge S, Gal A, Clarke LA, Morris CP, Hopwood JJ. Molecular genetics of mucopolysaccharidosis type I: diagnostic, clinical, and biological implications. Hum Mutat. 1995;6:288–302. doi: 10.1002/humu.1380060403. [DOI] [PubMed] [Google Scholar]
- 33.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 34.Menon KP, Neufeld EF. Evidence for degradation of mRNA encoding alpha-L-iduronidase in Hurler fibroblasts with premature termination alleles. Cell Mol Biol (Noisy-le-grand) 1994;40:999–1005. [PubMed] [Google Scholar]
- 35.Chung S, Ma X, Liu Y, Lee D, Tittiger M, Ponder KP. Effect of neonatal administration of a retroviral vector expressing alpha-L-iduronidase upon lysosomal storage in brain and other organs in mucopolysaccharidosis I mice. Mol Genet Metab. 2007;90:181–192. doi: 10.1016/j.ymgme.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Ma X, Liu Y, Tittiger M, Hennig A, Kovacs A, Popelka S, Wang B, Herati R, Bigg M, Ponder KP. Improvements in mucopolysaccharidosis I mice after adult retroviral vector-mediated gene therapy with immunomodulation. Mol Ther. 2007;15:889–902. doi: 10.1038/sj.mt.6300112. [DOI] [PubMed] [Google Scholar]
- 37.Woloszynek JC, Coleman T, Semenkovich CF, Sands MS. Lysosomal dysfunction results in altered energy balance. J Biol Chem. 2007;282:35765–35771. doi: 10.1074/jbc.M705124200. [DOI] [PubMed] [Google Scholar]
- 38.Kmiec EB. Targeted gene repair -- in the arena. J Clin Invest. 2003;112:632–636. doi: 10.1172/JCI19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24:552–563. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Keeling KM, Bedwell DM. Pharmacological suppression of premature stop mutations that cause genetic diseases. Current Pharmacogenomics. 2005;3:259–269. [Google Scholar]
- 41.Keeling KM, Du M, Bedwell DM. Therapies of Nonsense-Associated Diseases. In: Maquat LE, editor. Nonsense-Mediated mRNA Decay. Landes Bioscience; Georgetown, Texas: 2006. pp. 121–136. [Google Scholar]
- 42.Keeling KM, Brooks DA, Hopwood JJ, Li P, Thompson JN, Bedwell DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10:291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 43.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J Mol Med. 2002;80:367–376. doi: 10.1007/s00109-001-0317-z. [DOI] [PubMed] [Google Scholar]
- 44.Brooks DA, Muller VJ, Hopwood JJ. Stop-codon read-through for patients affected by a lysosomal storage disorder. Trends Mol Med. 2006;12:367–373. doi: 10.1016/j.molmed.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Aldenhoven M, Boelens JJ, de Koning TJ. The clinical outcome of Hurler syndrome after stem cell transplantation. Biol Blood Marrow Transplant. 2008;14:485–498. doi: 10.1016/j.bbmt.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Miebach E. Enzyme replacement therapy in mucopolysaccharidosis type I. Acta Paediatr Suppl. 2005;94:58–60. doi: 10.1111/j.1651-2227.2005.tb02114.x. discussion 57. [DOI] [PubMed] [Google Scholar]
- 47.Strausbaugh LJ, Brinker GS. Effect of osmotic blood-brain barrier disruption on gentamicin penetration into the cerebrospinal fluid and brains of normal rabbits. Antimicrob Agents Chemother. 1983;24:147–150. doi: 10.1128/aac.24.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith AL, Daum RS, Siber GR, Scheifele DW, Syriopoulou VP. Gentamicin penetration into cerebrospinal fluid in experimental Haemophilus influenzae meningitis. Antimicrob Agents Chemother. 1988;32:1034–1039. doi: 10.1128/aac.32.7.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman RL, Holt RJ. Intrathecal gentamicin in treatment of ventriculitis in children. Br Med J. 1967;2:539–542. [PMC free article] [PubMed] [Google Scholar]
- 50.McCracken GH, Jr., Chrane DF, Thomas ML. Pharmacologic evaluation of gentamicin in newborn infants. J Infect Dis. 1971;124(Suppl):124–214. doi: 10.1093/infdis/124.supplement_1.s214. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Samblas AM, Torres CL, Wang H, Feuer WJ, Goldberg RN. Effectiveness of a gentamicin dosing protocol based on postconceptional age: comparison to published neonatal guidelines. Ann Pharmacother. 1992;26:534–538. doi: 10.1177/106002809202600416. [DOI] [PubMed] [Google Scholar]
- 52.Ho KK, Bryson SM, Thiessen JJ, Greenberg ML, Einarson TR, Leson CL. The effects of age and chemotherapy on gentamicin pharmacokinetics and dosing in pediatric oncology patients. Pharmacotherapy. 1995;15:754–764. [PubMed] [Google Scholar]
- 53.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]