Abstract
Translocations resulting in ectopic expression of the TLX1 homeobox gene (previously known as HOX11) are recurrent events in human T-cell acute lymphoblastic leukemia (T-ALL). Transduction of primary murine hematopoietic stem/progenitor cells with retroviral vectors expressing TLX1 readily yields immortalized hematopoietic progenitor cell lines. Understanding the processes involved in TLX1-mediated cellular immortalization should yield insights into the growth and differentiation pathways altered by TLX1 during the development of T-ALL. In recent clinical gene therapy trials, hematopoietic clonal dominance or T-ALL-like diseases have occurred as a direct consequence of insertional activation of the EVI1, PRDM16 or LMO2 proto-oncogenes by the retroviral vectors used to deliver the therapeutic genes. Additionally, the generation of murine hematopoietic progenitor cell lines due to retroviral integrations into Evi1 or Prdm16 has also been recently reported. Here we determined by linker-mediated nested polymerase chain reaction the integration sites in 8 TLX1-immortalized hematopoietic cell lines. Notably, no common integration site was observed among the cell lines. Moreover, no insertions into the Evi1 or Prdm16 genes were identified although insertion near Lmo2 was observed in one instance. However, neither Lmo2 nor any of the other genes examined surrounding the integration sites showed differential vector-influenced expression compared to the cell lines lacking such insertions. While we cannot exclude the possibility that insertional side effects transiently provided a selective growth/survival advantage to the hematopoietic progenitor populations, our results unequivocally rule out insertions into Evi1 and Prdm16 as being integral to the TLX1-initiated immortalization process.
INTRODUCTION
TLX1 (T-cell leukemia homeobox 1, previously known as HOX11) is an evolutionarily conserved member of the non-clustered NKL subclass of homeobox genes that orchestrates spleen organogenesis and specification of neuronal cell fates (Roberts et al., 1994; Qian et al., 2002; Cheng et al., 2004; Holland et al., 2007). Although TLX1 is not expressed during normal hematopoietic development, it is frequently activated in T-cell acute lymphoblastic leukemia (T-ALL) by chromosomal translocations which place the TLX1 coding region downstream of T cell receptor (TCR) gene regulatory elements (Lu et al., 1991; Dube et al., 1991; Hatano et al., 1991; Kennedy et al., 1991; Owens and Hawley, 2002). The recurrent TLX1-associated translocations, which occur during thymocyte differentiation, are presumed to be initiating genetic lesions in this disease. In support of this notion, enforced expression of TLX1 in bone marrow-derived stem/progenitor cells induced T-cell tumors in transplanted mice, albeit at long latency (Hawley et al., 1994a; Hawley et al., 1997).
The potential of TLX1 to disrupt normal hematopoietic processes has also been demonstrated in a number of in vitro studies, where retroviral expression of TLX1 promoted the immortalization of murine progenitor cells derived from various hematopoietic sources (Hawley et al., 1994a; Hawley et al., 1997; Keller et al., 1998; Yu et al., 2002; Owens et al., 2003; Yu et al., 2003; reviewed in Hawley et al., 2008). Structure-function analysis of the TLX1 protein showed that an intact homeodomain is necessary for hematopoietic progenitor cell immortalization but regions near the NH2 and COOH termini of TLX1 are not essential for immortalizing activity (Hawley et al., 1997; Owens et al., 2003). Several lines of evidence indicate that TLX1 induces hematopoietic progenitor immortalization by increasing replicative capacity while concomitantly blocking differentiation (Su et al., 2006; Riz et al., 2007). A better understanding of the molecular mechanisms involved in TLX1-mediated immortalization should shed light on the growth and differentiation pathways subverted by this oncogene during leukemic transformation (Hahn, 2002; Riz and Hawley, 2005; Owens et al., 2006; Riz et al., 2009a).
Genome-wide analyses of integration sites have revealed that murine leukemia virus (MLV)-based retroviral vectors do not integrate at random throughout the genome. Rather, they preferentially insert into open chromatin regions, frequently in close proximity to or within genes (Wu et al., 2003; Lewinski et al., 2006). It is clear therefore that gene activation or disruption as a consequence of retroviral integration is greater than what would be predicted based on the assumption of a random distribution of sites throughout the genome. In this context, integration site analyses in recent preclinical and clinical hematopoietic stem cell gene transfer studies have confirmed that retroviral vector integrations can unfortunately result in the activation of proto-oncogenes (Baum et al., 2006; Nienhuis et al., 2006). These findings first came dramatically to light with the development of T-ALL in a number of patients after retrovirus-mediated gene therapy of X-linked severe combined immunodeficiency disease (SCID-X1), where in several instances the vector was found to have integrated near the LMO2 proto-oncogene (Hacein-Bey-Abina et al., 2003; Hacein-Bey-Abina et al., 2008). LMO2, like TLX1, was initially discovered as a result of its activation by chromosomal translocations involving the TCR loci in a subset of T-ALL patients (Nam and Rabbitts, 2006). Additionally, clonal expansion of myeloid lineage cells was observed in two patients in a gene therapy trial for chronic granulomatous disease due to activating insertions into the functionally-related EVI1 or PRDM16 proto-oncogenes (Ott et al., 2006).
The immortalization of murine hematopoietic progenitor cells due to integrations of retroviral vector backbones alone has been recently reported (Du et al., 2005; Modlich et al., 2006; Ott et al., 2006). Using MLV-derived retroviral vectors expressing fluorescent protein reporter genes, one study found that almost all of the transduced primary murine bone marrow cells that could be clonally expanded after growth in short-term cultures supplemented with a cocktail of stem cell factor (SCF), Flt-3 ligand, IL-11 and IL-3, contained insertions within Evi1 (Modlich et al., 2006). All of the expanding clones expressed varying amounts of the cell surface antigens CD11b and Ly-6G/Gr-1, indicating that they were of myeloid origin. In another study, ~50% of the IL-3/SCF-supplemented long-term cultures of murine bone marrow cells transduced with an MLV-based murine stem cell virus (MSCV) vector expressing the neomycin phosphotransferease (neo) gene (MSCVneo; Hawley et al., 1994b) produced immortalized cell lines, more than half of which contained integrations into either Evi1 or Prdm16 (Du et al., 2005). The immortalized cell lines were composed largely of immature myeloid cells with biphenotypic differentiation potential for the monocyte/macrophage and granulocyte lineages.
In view of these findings, we were interested in investigating the potential contribution of retroviral insertional mutagenesis to TLX1-induced murine bone marrow progenitor cell immortalization (Hawley et al., 1994a; Hawley et al., 1997). Similar to the retrovirally-immortalized murine bone marrow-derived cell lines (Du et al., 2005; Modlich et al., 2006), cell lines established following transduction of primary murine bone marrow cells with an MSCV retroviral vector expressing TLX1 in addition to neo (MSCVneo-TLX1) represent an early myeloid precursor that can be stimulated to partially differentiate along the monocyte/macrophage (CD11b+) and granulocyte (Ly-6G/Gr-1+) lineages (Owens et al., 2003). These TLX1-immortalized cell lines, historically referred to as HX cell lines to reflect the utilization of HOX11 (TLX1) as an immortalizing gene, are strictly dependent on IL-3 for their survival and proliferation. In this study, we characterized the MSCVneo-TLX1 integration sites in 8 HX cell lines —including 3 independent clones —derived in 5 separate experiments. Notably, no common vector integration sites were found among the HX cell lines, indicating that retroviral-mediated insertional mutagenesis of a specific locus is not required for cell line establishment by ectopic TLX1 expression.
MATERIALS AND METHODS
Cell Lines
The TLX1-immortalized HX cell lines used in this study were established previously (Hawley et al., 1994a; Hawley et al., 1997). In brief, bone marrow, enriched for hematopoietic stem/progenitor cells by 5-fluorouracil pretreatment (administered by intravenous injection 4 days previously at 150 mg/kg body weight), was flushed from the hind limbs of 6 to 8 week-old female BALB/c mice with ice-cold Iscove’s Modified Dulbecco’s Medium (IMDM) containing 10% heat-inactivated fetal bovine serum (FBS). After erythrocyte lysis in 0.17 M ammonium chloride, nucleated cells were added to 100 mm petri dishes at a density of 5 × 105 cells/ml in IMDM containing 10% heat-inactivated FBS plus 2% pokeweed mitogen-stimulated spleen-conditioned medium (PWM-SCM, a source of general hematopoietic growth factors including GM-CSF and IL-3) or 10% conditioned medium from X630-rlL3 cells (X630-rIL3 CM, a source of recombinant IL-3) (see below for details). After 48 hours, the bone marrow cells were collected and added to subconfluent monolayers of GP+E-86 packaging cells producing helper-free ecotropic MSCVneo-TLX1 retroviral vector particles (with a titer of 8 × 106 geneticin (G418)-resistant colony-forming units/ml when assayed on NIH3T3 fibroblasts) at a density of 5 × 105 cells/ml in fresh complete medium supplemented with 8 µg/ml polybrene. After a further 48 hours, nonadherent bone marrow cells were harvested and transiently selected by addition of 0.75 mg/ml G418 to the medium. The cells were collected 24 hours later and seeded at 1 × 103 and 1 × 104 cells/ml in 1 ml cultures containing 0.8% methylcellulose in the presence or absence of G418. The transduction efficiency was ~50% as determined by the respective number of colonies that formed by day 7 (range, 30–80%; n = 5 experiments). The G418-resistant colonies that arose in primary methylcellulose cultures in the first three experiments that were performed were separately pooled and placed in liquid cultures containing 0.75 mg/ml G418 and 2% PWM-SCM. The cells continued to propagate and could be serially passaged under G418 selection as bulk suspension cultures (for greater than 3 months), giving rise to the HX1, HX2 and HX3 cell lines, respectively. When tested for survival and proliferation in cultures supplemented with conditioned medium or purified recombinant factors, including IL-1, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-11, GM-CSF, G-CSF, M-CSF or SCF, only IL-3 supported their continued growth. Thereafter, the cells were maintained in culture medium supplemented with 10% X630-rIL3 CM instead of 2% PWM-SCM. A clone of the HX3 cell line (HX3 c32) was obtained following expansion of a single cell after limiting dilution in 96-well plates 9 days after the pooled primary colonies had been transferred to 1 ml liquid culture. Three additional cell lines were established in a fourth experiment from G418-resistant colonies obtained from three primary methylcellulose cultures carried out in parallel: HX4 was generated from pooled colonies (from plate 1) while HX4 c4 (from plate 2) and HX4 c13 (from plate 3) were derived from single colonies that were individually transferred to 0.5 ml liquid cultures using a fine glass pipette. Finally, the HX5 cell line was derived in a fifth experiment from pooled G418-resistant colonies from primary methylcellulose cultures. The conditions employed in the latter two experiments were exactly the same as those described for the first three experiments except that the cultures were supplemented throughout with 10% X630-rlL3 CM instead of 2% PWM-SCM. Upon thawing of frozen stocks, the IL-3-dependent HX cell lines were routinely maintained in IMDM (Mediatech, Inc., Herndon, VA) containing 4 mM L-glutamine, 50 IU/ml penicillin, 50 µg/ml streptomycin, 10% heat-inactivated FBS (Cambrex BioScience Walkersville, Inc., Walkersville, MD), 10% X630-rIL3 CM and 0.75 mg/ml G418 (Invitrogen Corp., Carlsbad, CA).
The retrovirus-induced, IL-3-dependent M-NSF-60 cell line (ATCC No. CRL-1838; American Type Culture Collection, Manassas, VA) was derived from a myeloid leukemia that contains a proviral insertion in the Evi1 gene (Morishita et al., 1988). M-NSF-60 cells were cultured in IMDM containing 4 mM L-glutamine, 50 IU/ml penicillin, 50 µg/ml streptomycin, 10% heat-inactivated FBS and 10% X630-rlL3 CM.
NIH3T3 fibroblasts (ATCC No. CRL-1658) were cultured in Dulbecco’s Modified Eagle Medium (Mediatech Inc.) supplemented with 4.5 g/l glucose, 4 mM L-glutamine, 50 IU/mL penicillin, 50 µg/ml streptomycin and 10% heat-inactivated FBS.
Karyotypic Analysis
The metaphases were arrested by incubation with colcemid (KaryoMax® Colcemid Solution, Invitrogen, Carlsbad, CA) (10 µg/ml) 2 hours prior to harvest. Cells were collected and treated with hypotonic solution (0.075 M KCl) for 15 min at 37°C and fixed with methanol:acetic acid (3:1). Chromosomes were stained with a trypsin-Giemsa staining technique (Seabright, 1971). Analyses were performed under an Axioplan 2 microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) coupled with a CCD camera (Photometrics, Tucson, AZ); images were captured with Band View 5.5 karyotyping software (Applied Spectral Imaging Inc., Vista, CA). The karyotype was determined by comparison to the standard ideogram of banding patterns for mouse chromosomes (Nesbitt and Francke, 1973). A minimum of 20 mitoses was examined per cell line.
Southern Blot Analysis
Southern blot analysis was performed as described previously (Ramezani et al., 2000; Moayeri et al., 2004). Briefly, genomic DNA was extracted using the GenElute Mammalian Genomic DNA Kit (Sigma, St Louis, MO). Genomic DNA (10 µg) was EcoRI-digested (New England Biolabs, Beverly, MA), separated on 0.8% agarose gels, and transferred to Hybond-N+ membranes (Amersham Biosciences, Piscataway, NJ). Membranes were fixed by exposure to UV light and hybridized with a 32P-labeled randomly primed neo probe. Digital images were acquired using a Storm 860 PhosphorImager and radioactivity was quantitated using ImageQuant software (Amersham Biosciences).
LM-PCR
Linker-mediated nested polymerase chain reaction (LM-PCR) was performed as previously described (Wu et al., 2003; Bauer, Jr. et al., 2008) with modifications that allowed the identification of genomic regions adjacent to MSCVneo-TLX1 vector sequences in a murine background. Briefly, genomic DNA was isolated from cell lines using the GenElute Mammalian Genomic DNA Kit and digested with Tsp509I or BstYI (New England Biolabs). The restriction enzyme was removed by phenol chloroform extraction and the DNA was ligated to the linker specific for the Tsp509I overhang (5’-GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG-3 ’ and 5 ’-AATTCCTAACTGCTGTGCCACTGAATTCAGATC-3’) or BstYI overhang (5’-GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG-3’ and 5’- GATCCCTAACTGCTGTGCCACTGAATTCAGATC-3’). Primary PCR was performed using primers complementary to the linker (5’-GACCCGGGAGATCTGAATTC-3’) and the MSCV vector sequence in the 5’ LTR (5’-AACCTTGATCTGAACTTCTC-3’) under the following conditions: 30 cycles at 95°C for 15 sec, 60°C for 30 sec, and 72°C for 1 min. Secondary PCR was performed using nested primers (5’-AGTGGCACAGCAGTTAGG-3’ and 5’-CCATGCCTTGCAAAATGGC-3’) after a 1:50 dilution of the primary PCR product under the same conditions as above. The MSCV PCR primers were modified from primers previously used in a retroviral vector insertional mutagenesis study in a mouse model (Kustikova et al., 2005) to specifically detect the MSCV 5’ LTR sequence. Nested PCR products were separated by gel electrophoresis on 1% agarose gels in TAE buffer and bands excised and purified using the Qiagen MinElute Gel Extraction Kit (Qiagen, Valencia, CA). All PCR amplifications were performed using Platinum Taq (Invitrogen). PCR products were cloned into the pCR4-TOPO vector and transformed into TOP10 cells (Invitrogen). DNA was isolated from ampicillin resistant colonies and sequenced using an M13 reverse primer. Proviral junctions from LM-PCR clones were examined for the presence of the linker cassette primer, LTR primer, and 5’ LTR sequences. The integrations were considered valid if the end of the 5’ LTR was less than two base pairs from the matching genomic sequence within the integration and if there was at least 90% homology to the mouse genome. Sequences were analyzed with CLC Free Workbench (CLCBio USA, Cambridge, MA). Integration sites were identified by matching sequences to the mouse genomic build of July 2007 (National Center for Biotechnology Information Build 37) using the UCSC Genome Browser BLAT alignment program (http://genome.ucsc.edu/) (Kent, 2002; Kuhn et al., 2009). Vector orientation and location of the vector with respect to the nearest gene were determined. Transcription start sites for genes were identified using the UCSC Table Browser data retrieval tool (Karolchik et al., 2004). Hits in the Mouse Retrovirus Tagged Cancer Gene Database (RTCGD; http://rtcgd.abcc.ncifcrf.gov/) (Akagi et al., 2004) and the Insertional Dominance Database (IDDb; http://www99.mh-hannover.de/kliniken/zellth/method.html) associated with hematopoietic clonal expansion (Kustikova et al., 2007) were identified for the nearest gene and genes within 100 kb of the integration sites.
Northern Blot Analysis
Northern blot analysis was performed as described previously (Ramezani et al., 2000; Moayeri et al., 2004). Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (20 µg) was separated on 1.2% agarose-formaldehyde gels, transferred to Hybond-N+ membranes in 20X SSC buffer (Ambion, Inc. Austin, TX), and hybridized with a 32P-labeled probe specific for each candidate gene according to standard protocols. Probes were prepared from coding regions of cDNA clones: Ap1g2 (ATCC No. 10955586), Evi1 (pBluescript KS(+)-Evi1; (Morishita et al., 1988), Foxl1 (ATCC No. 10866590), Gpr39 (Clone ID No. 30745457, Invitrogen), Itsn1 (ATCC No. 10699443), Lmo2 (ATCC No. 10470162), Lrrc33 (ATCC No. MGC-36838), Pitpnm2 (ATCC No. 10699600), Prdm16 (Clone ID No. 6409778, Invitrogen), and Vps37b (ATCC No. 10324522). As a control for equal loading of RNA, each blot was rehybridized with a glyceraldehyde-3-phosphate dehydrogenase (Gapdh) probe.
RT-PCR
Total RNA was extracted using Trizol reagent. RNA (1 µg) was reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). PCR was performed using Taq DNA polymerase (Roche Diagnostics, Indianapolis, IN) under the following conditions: 35 cycles at 94°C for 30 sec, 65°C for 1 min, and 72°C for 1 min. RT-PCR products were separated on a 1.5 % agarose gel. Primers were designed using Primer 3 software (http://primer3.sourceforge.net/) to span across introns such that PCR amplification of genomic DNA sequences results in a product that is larger in size than the product amplified from the corresponding cDNA templates. In addition, the primers were designed so that the size of the RT-PCR product was similar in size to the Gapdh RT-PCR product to ensure similar amplification efficiency: Abcb9 (NM_019875.2) forward 5'-AAGCATGGATCAGTTCACCA-3', reverse 5'-TGTGCGGTTCTCATCAAAGA-3'; Foxl1 (NM_008024.2) forward 5'-CGGCATCTACCAGTTCATCA-3', reverse 5'-CCGTTCTCAAACATGTCCAA-3'; Foxc2 (NM_013519.1) forward 5'-TGAGTGCTTCGTGAAAGTGC-3', reverse 5'-GACTTTCTTCTCGGCCTCCT-3'; Fbxo3 (NM_212433.1) forward 5'-GTGCTACTTTTACCGACTGGTTT-3', reverse 5'-CGGTATGTGAAGAAATAGTGTGG-3'; Cirh1a (NM_011574.2) forward 5’-GCACAGTGCCTGCTTACAAC-3’, reverse 5’-GTAGGCATCATGGAGGAGGA-3’ and Gapdh (NM_008084.2) forward 5'-ATCCCAGAGCTGAACG-3', reverse 5'-GAAGTCGCAGGAGACA-3'. All primers were validated using genomic DNA.
Quantitative RT-PCR
RNA was extracted from the HX cell lines using Trizol reagent. RNA (1 µg) was reverse transcribed into cDNA and qRT-PCR was performed on a 7300 Real Time PCR System by Taqman Gene Expression Assay using a primer and probe set (Mm00450301_m1) for the Tnk2 gene (Applied Biosystems, Foster City, CA).
Western Blot Analysis
Whole cell lysates were prepared as described previously (Owens et al., 2003; Riz and Hawley, 2005; Akimov et al., 2005). TLX1/HOX11 was detected using a polyclonal rabbit antibody (C-18: sc-880, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and α-tubulin was detected using a mouse monoclonal antibody (sc-5286, Santa Cruz Biotechnology, Inc.). An alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology, Inc.) was used. Chemifluorescence detection was performed using ECF substrate and digital images were acquired on a Storm 860 PhosphorImager using ImageQuant software (Amersham Biosciences).
RESULTS
No Common “Immortalizing” Genes Were Identified by Mapping of Retroviral Integration Sites in TLX1-Immortalized Cell Lines
IL-3-dependent HX cell lines, which had been established previously by immortalization of murine bone marrow cells following transduction with the MSCVneo-TLX1 retroviral vector (Hawley et al., 1994a; Hawley et al., 1997), were thawed from cryopreserved stocks and reinitiated as suspension cultures. The cell lines examined consisted of 5 independent bulk cultures (HX1 to HX5) derived in separate experiments from pooled colonies from primary methylcellulose cultures (within 8 days of transduction) as well as 3 clones (HX3 c32, HX4 c4, and HX4 c13): HX3 c32 is a single-cell clone of HX3 isolated from the bulk culture 9 days after the primary methylcellulose colonies were pooled, whereas HX4 c4 and HX4 c13 were established from individual colonies arising in primary methylcellulose cultures in the same experiment as the polyclonal HX4 cell line (all from different dishes) (Hawley et al., 1994a). It was previously demonstrated by Southern blotting that the HX4 cell line contained multiple integrants whereas the HX4 c4 and HX4 c13 cell lines contained single retroviral integrations (Hawley et al., 1994a; Hawley et al., 1997). Vector copy numbers had not been determined for the other cell lines.
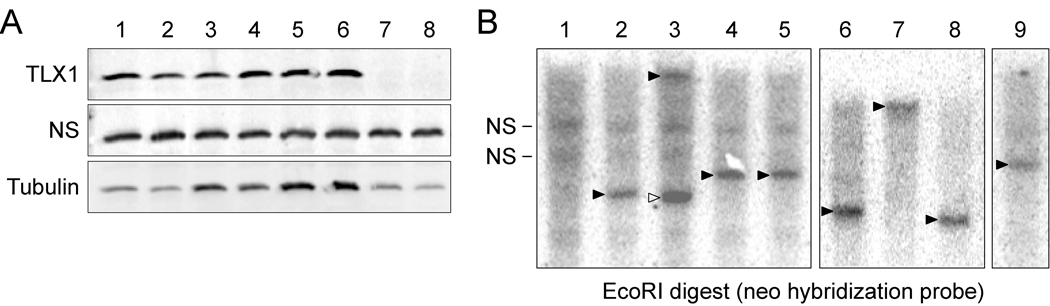
We first confirmed that all of the HX cell lines expressed TLX1 by Western and Northern blot analyses (Fig. 1A; and data not shown). We next carried out Southern blot analyses on EcoRI-digested genomic DNA extracted from the HX cell lines (EcoRI cuts the MSCVneo-TLX1 vector once) using a neo probe (Fig. 1B). As expected, unique bands representative of single MSCVneo-TLX1 vector insertions were observed for the HX4 c4 and HX4 c13 clonal cell lines as previously reported (Hawley et al., 1994a; Hawley et al., 1997). A single integrated vector was also detected in genomic DNA from the HX3 c32 clonal cell line and this corresponded to the prominent band present in HX3 genomic DNA. The HX1, HX2, HX4 and HX5 cell lines had also become predominantly clonal or oligoclonal during continued propagation in culture, exhibiting one (HX1, HX4 and HX5) or two (HX2) prominent vector junction fragments hybridizing to the neo probe.
Figure 1.
Expression and integration of MSCVneo-TLX1 retroviral vectors in immortalized HX cell lines. (A) TLX1 expression was detected in whole cell extracts (10 µg) from HX cell lines using an anti-TLX1 antibody (C-18, Santa Cruz). NS, nonspecific cross-reacting protein demonstrating equal loading. α-Tubulin (B-7, Santa Cruz) levels were also used as a loading control. Lanes are as follows: 1. HX1, 2. HX2, 3. HX3, 4. HX4, 5. HX4 c4, 6. HX4 c13, 7. HX5, 8. SPL136 (TLX1+ T-cell tumor). Note that the TLX1 gene in HX5 and SPL136 has a deletion which removed the last 40 amino acids, the region containing the epitope recognized by the anti-TLX1 antibody (Hawley et al., 1997; Riz et al., 2009a). In these cases, TLX1 expression was confirmed at the level of RNA by Northern blot analysis. (B) Southern blot analysis of EcoRI-digested genomic DNA with a neo probe demonstrated integration of the MSCVneo-TLX1 retroviral vector in each of the cell lines. Integration sites that were recovered by LM-PCR whose predicted sizes corresponded to the prominent vector-specific bands are indicated (►). In the case of HX2, the integration site corresponding to a prominent ~3.8 kb band (▻) was not obtained. NS, nonspecific cross-reacting endogenous sequences. Lanes: 1, NIH3T3 (negative control); 2, HX1; 3, HX2; 4, HX3; 5, HX3 c32; 6, HX4; 7, HX4 c4; 8, HX4 c13; 9, HX5.
To identify the retroviral vector integration sites in the HX cell lines, we performed LM-PCR to amplify the MSCVneo-TLX1/genomic junctions. The procedure was initially carried out with genomic DNA that had been digested with Tsp509I, which recognizes a 4-base restriction site (▼AATT). For some of the samples, the screen was repeated with genomic DNA that had been cleaved with BstYI, which has a degenerate 6-base recognition site with a different 4-base core sequence (R▼GATCY). Sequences for the integration sites in the HX cell lines were matched to the mouse genome using the UCSC Genome Browser BLAT alignment program (Table 1). In this manner, with one exception, we were able to recover all of the integration sites whose predicted sizes for EcoRI-digested genomic DNA corresponded to the prominent vector-specific bands revealed by Southern blotting. In the case of HX2, the integration site corresponding to only one of the two prominent bands was recovered (~18 kb band; Fig. 1B, lane 3). Strikingly, none of the integration sites identified in the HX cell lines were within the loci of the known hematopoietic progenitor cell immortalizing genes Evi1 or Prdm16. Moreover, no common integration site (CIS) was identified among the cell lines, indicating that neither immortalization nor clonal expansion of the cell lines was due to retroviral integration into a particular gene or locus.
TABLE 1.
MSCVneo-TLX1 Integration Sites in Immortalized Cell Lines
| Cell line | Gene at integration sitea | Gene ID | Chromosome | LTR junction | Location and distanceb |
Retrovirus orientation |
|
|---|---|---|---|---|---|---|---|
| HX1 | Abcb9 | ATP-binding cassette sub-family B member 9 |
56325 | mu5qF | 124501873 | 3’, 10 kb | R |
| HX2 | Foxl1 | forkhead box L1 | 14241 | mu8qE1 | 123707088 | 3’, 56 kb | R |
| HX3 | Lrrc33 | leucine rich repeat containing 33 | 224109 | mu16qB2 | 32164358 | intron 1, 21 kb | F |
| HX3 c32 | Lrrc33 | leucine rich repeat containing 33 | 224109 | mu16qB2 | 32164358 | intron 1, 21 kb | F |
| HX4 | Gpr39 | G protein-coupled receptor 39 | 71111 | mu1qE3 | 127573910 | 5’ UTR, <1 kb | R |
| Akap13 | A kinase (PRKA) anchor protein 13 | 75547 | mu7qD2 | 82728485 | intron 5, 26 kb | R | |
| Lyrm7 | LYR motif containing 7 | 75530 | mu11qB1.3 | 54621217 | 3’, 19 kb | R | |
| Itm2a | integral membrane protein 2A | 16431 | muXqD | 104681297 | 5’, 89 kb | R | |
| HX4 c4 | Thtpa | thiamine triphosphatase | 105663 | mu14qC3 | 55714396 | exon 1, <1 kb | F |
| HX4 c13 | Itsn1 | intersectin 1 (SH3 domain protein 1A) |
16443 | mu16qC4 | 91794474 | intron 5, 12 kb | R |
| HX5 | Lmo2 | LIM domain only 2 | 16909 | mu2qE2 | 103800793 | 5’, 10 kb | R |
R, reverse; F, forward.
MSCVneo-TLX1 insertion within gene or nearest gene; four MSCVneo-TLX1 integration sites were recovered for the HX4 cell line.
Location and distance of MSCVneo-TLX1 integration site with respect to transcription start site of gene (identified using UCSC table browser).
To characterize the vector integration sites, we next searched the Mouse Retrovirus Tagged Cancer Gene Database (RTCGD) for CISs previously identified in multiple independent murine tumor models (Akagi et al., 2004). Three of the genes closest to the integration sites identified in the HX cell lines were listed as CISs in the RTCGD. Most notably, the murine ortholog of the LMO2 proto-oncogene inadvertently activated in the SCID-X1 clinical trials (Hacein-Bey-Abina et al., 2003; Hacein-Bey-Abina et al., 2008) was identified 10 kb downstream of the prominent integration site in the HX5 cell line (6 hits in the RTCGD). The second CIS, Foxl1 (3 hits in the RTCGD), encoding a forkhead box transcription factor that serves as a point of crosstalk between the hedgehog and Wnt/β-catenin signaling networks (Katoh, 2007), was identified 56 kb upstream of the one prominent integration site recovered for the HX2 cell line. The third CIS, Akap13 (4 hits in the RTCGD), a guanine nucleotide exchange factor involved in the osmoprotective response (Aramburu and Lopez-Rodriguez, 2009) was identified in HX4, where the vector integrated into intron 5 of the gene (although it was not the prominent band revealed by Southern blot analysis). Two other genes were identified that were listed as rare hits in the RTCGD: Abcb9 (2 entries), a lysosomal peptide transporter which is member of the superfamily of ATP-binding cassette transporters was located 10 kb upstream of the prominent HX1 integration site; and the MSCVneo-TLX1 vector was found to have integrated into intron 1 of Lrrc33 (2 entries), a gene encoding a leucine rich repeat-containing protein of unknown function, corresponding to the only vector integration in the HX3 c32 clone (and the prominent band in parental HX3 cells).
No Differential Expression of Genes Surrounding MSCVneo-TLX1 Integration Sites
Although no CIS was shared among the cell lines, there is a possibility that an overlap in gene function exists among the targeted loci. Therefore, we compiled lists of genes whose transcription start sites were within 100 kb upstream or downstream from each integration site (Table 2). An additional RTCGD CIS, Foxc2 (3 entries), another forkhead box transcription factor gene, which is involved in specifying mesenchymal cell fate during embryogenesis (Hannenhalli and Kaestner, 2009), was identified 11 kb upstream of the Foxl1 CIS in the HX2 cell line. Extensive searching of the literature, including the Insertional Dominance Database (IDDb) associated with hematopoietic clonal expansion (Kustikova et al., 2007), revealed additional genes that had been reported on at least one occasion to have been targeted by retroviral vectors: Bex6, Fbxo3, Gpr39, Itsn1, Pitpnm2, and Vps37b. Gene ontology analysis did not reveal overrepresentation of a particular functional group (Ashburner et al., 2000). There were, however, some common functions shared among the encoded proteins of these and additional genes located within the 100-kb integration site windows; these include defined and putative roles in intracellular trafficking involving the golgi and clathrin-coated vesicles (Ap1g2, Itsn1, Vps37b), endoplasmic reticulum remodeling and stress (Gpr39, Pitpnm2), and as anti-apoptotic proteins (Gpr39, Itsn1).
TABLE 2.
Genes Within 100-Kb Genomic Windows Surrounding MSCVneo-TLX1 Integration Sites
| Cell line | Genea | Product | Distance to TSSb |
|---|---|---|---|
| HX1 | Abcb9* | ATP-binding cassette, sub-family B (MDR/TAP), member 9 | 10 kb |
| Vps37b* | vacuolar protein sorting 37B | −47 kb | |
| Ogfod2 | 2-oxoglutarate and iron-dependent oxygenase domain containing 2 | 60 kb | |
| Arl6ip4 | ADP-ribosylation factor-like 6 interacting protein 4 | 64 kb | |
| Pitpnm2* | phosphatidylinositol transfer protein, membrane-associated 2 | 67 kb, 80 kb | |
| HIP1R | huntingtin interacting protein 1 related | −78 kb | |
| HX2 | Foxl1* | forkhead box L1 | −56 kb |
| Foxc2* | forkhead box C2 | −67 kb | |
| Mthfsd | methenyltetrahydrofolate synthetase domain containing | −85 kb, −86kb | |
| Foxf1a | forkhead box F1a | −99 kb | |
| HX3 / HX3 c32 | Bex6* | brain expressed gene 6 | 16 kb |
| Lrrc33* | leucine rich repeat containing 33 | −21 kb | |
| 1500031L02Rik | hypothetical protein LOC66994 | −64 kb | |
| Fbxo45 | F-box protein 45 | 66 kb | |
| Pigx | phosphatidylinositol glycan anchor biosynthesis | −80 kb | |
| Wdr53 | WD repeat domain 53 | 83 kb | |
| HX4 | Gpr39† | G protein-coupled receptor 39 | <−1 kb |
| Akap13* | A kinase (PRKA) anchor protein 13 | 26 kb, 28kb, 118 kb −127 kb, −128 kb |
|
| Lyrm7 | LYR motif containing 7 | 19 kb | |
| Hint1* | histidine triad nucleotide binding protein 1 | 59 kb | |
| Cdc42se2 | CDC42 small effector 2 | −90 kb | |
| Gpx3 | glutathione peroxidase 3 | 95 kb | |
| Itm2a | integral membrane protein 2A | −89 kb | |
| HX4 c4 | Thtpa | thiamine triphosphatase | <−1 kb, −40 kb |
| Ap1g2 | adaptor protein complex AP-1, gamma 2 subunit | 3 kb | |
| Jph4 | junctophilin 4 | 11 kb, 15 kb | |
| Zfhx2 | zinc finger homeobox 2 | −32 kb, −34 kb | |
| Ngdn | neuroguidin, EIF4E binding protein | −80 kb | |
| HX4 c13 | Itsn1† | intersectin 1 (SH3 domain protein 1A) | −12 kb, −65 kb, 93 kb |
| Cryzl1 | crystallin zeta (quinone reductase)-like 1 | −99 kb, −105 kb | |
| HX5 | Lmo2* | LIM domain only 2 | 9 kb, 10 kb |
| Fbxo3* | F-box only protein 3 | 67 kb | |
TSS, transcription start site identified using UCSC table browser.
Four MSCVneo-TLX1 integration sites were recovered for the HX4 cell line; genes previously reported as hits in the Mouse Retrovirus Tagged Cancer Gene Database (RTCGD) are indicated by asterisks; genes previously reported as hits in other studies reported in the literature are indicated by daggers.
Pitpnm2, Mthfsd, Akap13, Thtpa, Jph4, Zfhx2, Itsn1, Cryzl1 and Lmo2 have multiple transcription start sites.
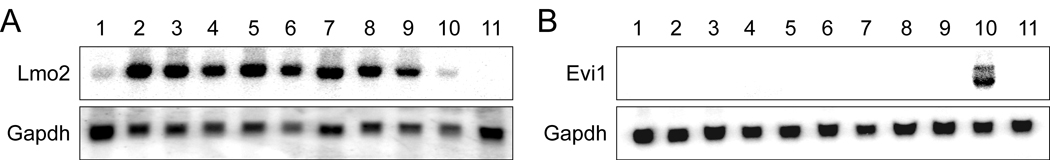
Based on the compiled data, the expression levels of a panel of 12 candidate target genes surrounding the vector integration sites — corresponding to the single vector integrants in the HX3 c32, HX4 c4, and HX4 c13 clonal cell lines and the dominant clones in the other HX cell lines —were examined by Northern blotting and/or RT-PCR. This analysis failed to uncover any integration-specific differences in the expression of any gene tested among the various HX cell lines. For example, Northern blot analysis revealed that Lmo2 is similarly expressed in all of the HX cell lines (Fig. 2A). Moreover, in many cases, the expression levels of the genes surrounding the integration sites were below detection limits (Supplementary Fig. 1; summarized in Table 3). Finally, no upregulation in Evi1 or Prdm16 expression was detected in any of the HX cell lines (Fig. 2B; and data not shown).
Figure 2.
Northern blot analysis of Lmo2 or Evi1 expression in TLX1-immortalized HX cell lines. (A) RNA (20 µg) was hybridized with a probe for Lmo2 or Gapdh. Lanes: 1, MSCV-immortalized bone marrow cell line (empty vector); 2, HX1; 3, HX2; 4, HX3; 5, HX3 c32; 6, HX4; 7, HX4 c4; 8, HX4 c13; 9, HX5; 10, SPL136 (TLX1+ T-cell tumor); 11, THY137 (TLX1+ T-cell tumor) (Hawley et al., 1997). (B) RNA (20 µg) was hybridized with a probe for Evi1 or Gapdh. Lanes: 1, primary murine bone marrow progenitor cells cultured in IL-3 for 3 weeks; 2, HX1; 3, HX2; 4, HX3; 5, HX4; 6, HX4 c4; 7, HX4 c13; 8, HX5; 9, SPL136 (TLX1+ T-cell tumor); 10, M-NFS-60 (Evi1+ myeloid leukemia); 11, MSCV-immortalized murine bone marrow cell line (empty vector).
TABLE 3.
Expression of Genes Surrounding MSCVneo-TLX1 Integration Sites
| Gene | Cell line-specific integration |
Expressiona | Detection method |
|---|---|---|---|
| Abcb9 | HX1 | + | RT-PCR |
| Vps37b | − | Northern | |
| Pitpnm2 | − | Northern | |
| Foxl1 | HX2 | − | RT-PCR |
| Foxc2 | − | RT-PCR | |
| Lrrc33 | HX3 / HX3 c32 | − | Northern |
| Tnk2b | + | qRT-PCR | |
| Gpr39 | HX4 | − | Northern |
| Ap1g2 | HX4 c4 | − | Northern |
| Itsn1 | HX4 c13 | − | Northern |
| Lmo2 | HX5 | + | Northern |
| Fbxo3 | + | RT-PCR | |
No differential expression was observed for any gene near or containing an MSCVneo-TLX1 insertion compared to the other cell lines lacking the insertion; + or −, indicates whether mRNA was detectable or below detection, respectively, by the method used.
Tnk2 has 3 transcription start sites located ~481 kb, 504 kb, and 509 kb away from the MSCVneo-TLX1 integration site in HX3 and HX3 c32 cells; its expression in HX3 cells was initially detected by genome-wide expression analysis (I. Riz and R.G. Hawley, unpublished observations).
No Recurring Chromosomal Abnormalities Were Identified in TLX1-Immortalized Cell Lines
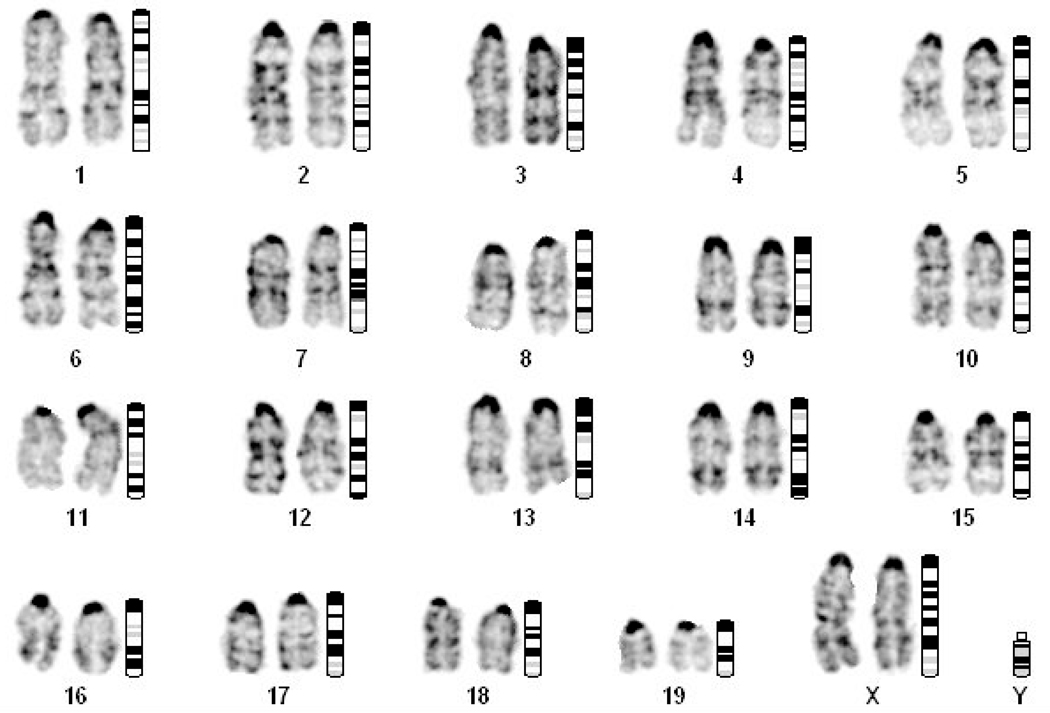
It had been reported that the IL-3-dependent myeloid progenitor cell line 32D clone 3 (32D Cl3), which was established from long-term murine bone marrow cultures, exhibits a highly variable and unstable karyotype characterized by several complex chromosomal rearrangements (Agliano et al., 2000). To determine whether secondary chromosomal abnormalities might contribute to TLX1-mediated hematopoietic progenitor cell immortalization, we performed cytogenetic analyses of the HX1, HX2, HX3, HX4, HX4 c4, HX4 c13 and HX5 cell lines. With the exception of HX2, all cell lines had a diploid 40,XX karyotype by G-banding of chromosome spreads, consistent with their derivation from normal bone marrow cells of female mice (Fig. 3 and Supplementary Fig. 2). In the case of HX2, although we did not detect evidence of any chromosomal rearrangements, some numerical alterations were observed. The karyotype of this cell line was 40,X0,+16[15] (2 cells were 40,XX and 3 cells had a hypodiploid karyotype), with loss of one copy of the X chromosome and trisomy 16 confirmed by fluorescence in situ hybridization (Supplementary Fig. 3).
Figure 3.
Cytogenetic analysis of the TLX1-immortalized HX3 cell line. Shown is a G-banded chromosome preparation demonstrating a normal diploid karyotype, with chromosome pairs lined up next to their respective ideogram representations.
DISCUSSION
We and others have reported that enforced expression of TLX1 promotes the immortalization of murine hematopoietic progenitors derived from bone marrow as well as from fetal liver, yolk sac and in vitro differentiated embryonic stem cells from various mouse strains at relatively high frequency (reviewed in Hawley et al., 2008). One caveat of these studies is that gene delivery via MLV-based MSCV retroviral vectors was used to introduce the TLX1 transgene into the target cell populations. Large-scale mapping of MLV retroviral integration events has shown that approximately 25% are near transcription start sites (Wu et al., 2003; Lewinski et al., 2006), potentially resulting in insertional activation of proto-oncogenes or other cell fate signaling genes (summarized in Baum et al., 2006). Indeed, Copeland and colleagues took advantage of this observation and used MLV retroviral insertional mutagenesis (with the MSCVneo backbone) as a tool to identify hematopoietic progenitor cell immortalization genes (Du et al., 2005). To this end, they cloned over 80% of the MSCV vector integrations present in 37 immortalized myeloid progenitor cell lines derived from murine bone marrow: 7 cell lines contained integrations in the first or second intron of Evi1, while another 13 cell lines contained integrations in the first intron of Prdm16. Northern blot analysis of 29 of the immortalized cell lines showed that 8 expressed high levels of Evi1 and 17 expressed high levels of Prdm16. Interestingly, overexpression of Evi1 was observed in 1 cell line without a retroviral integration in Evi1 and overexpression of Prdm16 was detected in 4 cell lines without a retroviral integration in that gene. The authors speculated that the cell lines in question may contain activating integrations at other genes that function upstream of Evi1 or Prdm16, respectively. In another study, Baum and colleagues characterized the integration sites of MLV retroviral vectors (based on the SF91 backbone) in clones of hematopoietic progenitors that showed a dominant growth advantage during short-term culture (4–5 weeks) under myeloid-supportive conditions, continued expansion of which led to immortalization in one instance (Modlich et al., 2006). In 6 of 8 expanded clones examined, an integration in Evi1 was detected, and strong upregulation of Evi1 expression was found in all cases including the 2 clones without detectable Evi1 integrations. In contrast to these findings, none of the 8 HX cell lines analyzed in this study (from 5 separate experiments), which were established following retroviral transduction of murine bone marrow with an MSCVneo retroviral vector constitutively expressing TLX1, contained integrations in Evi1 or Prdm16; nor was overexpression of Evi1 or Prdm16 observed in any case. Thus, our results exclude retroviral insertional activation of Evi1 or Prdm16 as playing an obligatory role in the series of events leading to cell line establishment of hematopoietic progenitors ectopically expressing TLX1. These observations raise the possibility that there are shared features of the pathways subverted by TLX1 and Evi1 (Prdm16) leading to immortalization, as suggested from previous studies investigating the ability of these oncogenes to interfere with hematopoietic differentiation programs (Morishita et al., 1992; Kreider et al., 1993; Hawley et al., 1994a; Greene et al., 2002; Riz et al., 2007).
Another significant difference between the TLX1-expressing HX cell lines and the hematopoietic progenitor cell lines established by retroviral vector insertional mutagenesis in the previous experiments is that the clonal HX lines (HX3 c32, HX4 c4, and HX4 c13) contain single MSCV integrations whereas the latter lines contained multiple retroviral vector integrations (typically 3 to 10 insertions per cell line) (Du et al., 2005; Modlich et al., 2006). Notably, no common loci were found to have been targeted by the MSCVneo-TLX1 vector among the HX cell lines. Together, these observations are in accord with the supposition that enforced TLX1 expression is sufficient to promote cellular immortalization and reinforces the notion that retroviral insertional mutagenesis is not playing the principal role (Hawley, 2008; Varas et al., 2008). Indeed, examination of the hematopoietic “fingerprint” expression database of hematopoietic stem cells and their differentiated progeny (Chambers et al., 2007) reveals that the loci targeted were most likely expressed in the immature hematopoietic cell populations giving rise to the HX cell lines and thus favored for retroviral integration (Lewinski et al., 2006). For example, the human ortholog of Gpr39, an orphan G-protein-coupled receptor, corresponding to the prominent integration site in the HX4 cell line, was previously identified as a rare insertion in the peripheral blood sample of one of the patients in the SCID-X1 clinical trials who did not develop T-ALL or preneoplastic myeloid clonal expansions (Deichmann et al., 2007). However, we cannot rule out the possibility that the targeted integration sites do contribute to the early stages of cell line establishment but become dispensable following immortalization, as reported previously in the case of retrovirally-activated c-myb during in vitro passage of a T-lymphoma cell line (Hanlon et al., 2003). It is interesting in this regard that the single vector insertion in the HX4 c13 clonal cell line was found to have occurred within intron 5 of Itsn1, a multidomain adaptor protein implicated in endothelial cell survival (Predescu et al., 2007); retroviral vector integration into intron 3 of Itsn1 was previously reported as one of 7 characterized insertion sites out of 9 in a dominant hematopoietic clone characterized by Baum and colleagues, which also contained insertions in Evi1 and HoxA7 (Modlich et al., 2006). We recently developed a self-inactivating retroviral vector, RMSinOFB, in which a novel enhancer-blocking element derived from chromatin insulators flanks the retroviral vector backbone upon integration, virtually eliminating hematopoietic immortalizing potential due to insertional mutagenesis (Ramezani et al., 2008). It will therefore be of much interest to evaluate the ability of this vector backbone expressing TLX1 to immortalize hematopoietic progenitor cells.
An especially intriguing observation to come from this study was the finding that the MSCVneo-TLX1 vector had integrated near the Lmo2 proto-oncogene in the HX5 cell line. We cannot exclude a role for Lmo2 in TLX1-mediated hematopoietic progenitor cell immortalization at this time. However, Lmo2 expression was found in all of the HX cell lines, ruling out selective transcriptional upregulation by insertional mutagenesis in HX5 cells. Lmo2 was only weakly expressed (SPL136) or below detection (THY137) in two cell lines derived from TLX1-induced T-cell tumors, arguing against Lmo2 being a universal downstream target gene of TLX1. Finally, Lmo2 expression was previously documented to be mainly expressed in myeloid cells — including the IL-3-dependent 32D, FDCP-mix and FDC-P1 myeloid cell lines — implying that the pattern of Lmo2 expression in the HX cell lines is most likely reflective of their myeloid phenotype (Hansson et al., 2007).
The molecular mechanism by which TLX1 induces immortalization remains an open question. The accumulated evidence from our laboratory and others indicates that TLX1 functions as a transcriptional regulator that can either activate or repress gene expression (Dear et al., 1993; Greene et al., 1998; Zhang et al., 1999; Allen et al., 2000; Owens et al., 2003; Hoffmann et al., 2004; Riz and Hawley, 2005; Riz et al., 2007; Dixon et al., 2007; Rice et al., 2008a; Rice et al., 2008b; Riz et al., 2009a; Riz et al., 2009b). Drawing parallels between TLX1 and the pluripotent stem cell factor Nanog (Pan and Thomson, 2007), another member of the non-clustered NKL subclass of homeobox genes with which it shares 66% amino acid sequence similarity (46% homology) (Holland et al., 2007), may provide clues to the “reprogramming” process of hematopoietic progenitor immortalization. It is noteworthy in this regard that TLX1-mediated immortalization resulted in a very low incidence of chromosomal abnormalities. Therefore, with the exception of the HX2 cell line, these results rule out aneuploidy as being an important contributor to any secondary genetic changes that might be required for the immortalization process. Nonetheless, when several of the TLX1-immortalized HX cell lines studied here were tested previously, including HX2, they were not leukemogenic in vivo (Hawley et al., 1994a). These observations are consistent with findings in many other experimental systems showing that acquisition of an immortal state is required but not sufficient for the malignant transformation of normal mammalian cells in culture (Newbold and Overell, 1983; Rangarajan et al., 2004).
In conclusion, we believe that additional studies into the process of TLX1-mediated immortalization in this murine model, which occurs in the absence of widespread genomic instability, will yield important insights into the pathogenetic contribution of TLX1 to human TALL (Hahn, 2002; Aifantis et al., 2008).
Supplementary Material
ACKNOWLEDGMENTS
We thank Sara Karandish for technical assistance and Mehreen Hai for advice on LM-PCR. This work was supported in part by National Institutes of Health Grants R01HL66305 and R01HL65519, by an Elaine H. Snyder Cancer Research Award and the King Fahd Endowment Fund from The George Washington University Medical Center, and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research. Portions of this study were performed by L.A.Z. in partial fulfillment of the requirements for the Ph.D. degree in Biochemistry and Molecular Genetics from the Institute for Biomedical Sciences, The George Washington University.
REFERENCES
- Agliano AM, Santangelo C, Silvestri I, Gazzaniga P, Giuliani L, Naso G, Frati L, Castiglia R. On chromosomal instability: what is the karyotype of your 32D CI3 cell line. Blood. 2000;95:3636–3637. [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimov SS, Ramezani A, Hawley TS, Hawley RG. Bypass of senescence, immortalization and transformation of human hematopoietic progenitor cells. Stem Cells. 2005;23:1423–1433. doi: 10.1634/stemcells.2005-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Zhu Y-X, Hawley TS, Hawley RG. TALE homeoproteins as HOX11-interacting partners in T-cell leukemia. Leuk Lymphoma. 2000;39:241–256. doi: 10.3109/10428190009065824. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Lopez-Rodriguez C. Brx shines a light on the route from hyperosmolarity to NFAT5. Sci.Signal. 2009;2:e20. doi: 10.1126/scisignal.265pe20. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. NatGenet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer TR, Jr, Allen JM, Hai M, Tuschong LM, Khan IF, Olson EM, Adler RL, Burkholder TH, Gu YC, Russell DW, Hickstein DD. Successful treatment of canine leukocyte adhesion deficiency by foamy virus vectors. Nat Med. 2008;14:93–97. doi: 10.1038/nm1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. HumGene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, Merchant MG, Fisk CJ, Shaw CA, Goodell MA. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Dear TN, Sanchez-Garcia I, Rabbitts TH. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc Natl Acad Sci USA. 1993;90:4431–4435. doi: 10.1073/pnas.90.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, Glimm H, Gyapay G, Prum B, Fraser CC, Fischer N, Schwarzwaelder K, Siegler ML, de RD, Pike-Overzet K, Howe SJ, Thrasher AJ, Wagemaker G, Abel U, Staal FJ, Delabesse E, Villeval JL, Aronow B, Hue C, Prinz C, Wissler M, Klanke C, Weissenbach J, Alexander I, Fischer A, Von KC, Cavazzana-Calvo M. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DN, Izon DJ, Dagger S, Callow MJ, Taplin RH, Kees UR, Greene WK. TLX1/HOX11 transcription factor inhibits differentiation and promotes a non-haemopoietic phenotype in murine bone marrow cells. Br J Haematol. 2007;138:54–67. doi: 10.1111/j.1365-2141.2007.06626.x. [DOI] [PubMed] [Google Scholar]
- Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube ID, Kamel-Reid S, Yuan CC, Lu M, Wu X, Corpus G, Raimondi SC, Crist WM, Carroll AJ, Minowada J, Baker JB. A novel human homeobox gene lies at the chromosome 10 breakpoint in lymphoid neoplasias with chromosomal translocation t(10;14) Blood. 1991;78:2996–3003. [PubMed] [Google Scholar]
- Greene WK, Bahn S, Masson N, Rabbitts TH. The T-cell oncogenic protein HOX11 activates Aldh1 expression in NIH 3T3 cells but represses its expression in mouse spleen development. Mol Cell Biol. 1998;18:7030–7037. doi: 10.1128/mcb.18.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene WK, Ford J, Dixon D, Tilbrook PA, Watt PM, Klinken SP, Kees UR. Enforced expression of HOX11 is associated with an immature phenotype in J2E erythroid cells. Br J Haematol. 2002;118:909–917. doi: 10.1046/j.1365-2141.2002.03704.x. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, Macintyre E, Dal CL, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint BG, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le DF, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hahn WC. Immortalization and transformation of human cells. Mol Cells. 2002;13:351–361. [PubMed] [Google Scholar]
- Hanlon L, Barr NI, Blyth K, Stewart M, Haviernik P, Wolff L, Weston K, Cameron ER, Neil JC. Long-range effects of retroviral insertion on c-myb: overexpression may be obscured by silencing during tumor growth in vitro. J Virol. 2003;77:1059–1068. doi: 10.1128/JVI.77.2.1059-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Zetterblad J, van Duren C, Axelson H, Jonsson JI. The Lim-only protein LMO2 acts as a positive regulator of erythroid differentiation. Biochem Biophys Res Commun. 2007;364:675–681. doi: 10.1016/j.bbrc.2007.10.071. [DOI] [PubMed] [Google Scholar]
- Hatano M, Roberts CW, Minden M, Crist WM, Korsmeyer SJ. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- Hawley RG. Does retroviral insertional mutagenesis play a role in the generation of induced pluripotent stem cells? Mol Ther. 2008;16:1354–1355. doi: 10.1038/mt.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Fong AZC, Lu M, Hawley TS. The HOX11 homeobox-containing gene of human leukemia immortalizes murine hematopoietic precursors. Oncogene. 1994a;9:1–12. [PubMed] [Google Scholar]
- Hawley RG, Fong AZC, Reis MD, Zhang N, Lu M, Hawley TS. Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res. 1997;57:337–345. [PubMed] [Google Scholar]
- Hawley RG, Hawley TS, Cantor AB. TLX1 (HOX11) immortalization of embryonic stem cell-derived and primary murine hematopoietic progenitors. Curr Protoc Stem Cell Biol. 2008;7:1F.7.1–1F.7.19. doi: 10.1002/9780470151808.sc01f07s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Lieu FHL, Fong AZC, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994b;1:136–138. [PubMed] [Google Scholar]
- Hoffmann K, Dixon DN, Greene WK, Ford J, Taplin R, Kees UR. A microarray model system identifies potential new target genes of the proto-oncogene HOX11. Genes Chromosomes Cancer. 2004;41:309–320. doi: 10.1002/gcc.20104. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 2007;3:30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- Keller G, Wall C, Fong AZC, Hawley TS, Hawley RG. Overexpression of HOX11 leads to the immortalization of embryonic precursors with both primitive and definitive hematopoietic potential. Blood. 1998;92:877–887. [PubMed] [Google Scholar]
- Kennedy MA, Gonzalez-Sarmiento R, Kees UR, Lampert F, Dear N, Boehm T, Rabbitts TH. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc.Natl.Acad.Sci.USA. 1991;88:8900–8904. doi: 10.1073/pnas.88.20.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreider BL, Orkin SH, Ihle JN. Loss of erythropoietin responsiveness in erythroid progenitors due to expression of the Evi-1 myeloid-transforming gene. Proc Natl Acad Sci USA. 1993;90:6454–6458. doi: 10.1073/pnas.90.14.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, von NN, Schlegelberger B, Li Z, Baum C. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, Pike-Overzet K, de Ridder D, Staal FJ, von Keudell G, Cornils K, Nattamai KJ, Modlich U, Wagemaker G, Goodell MA, Fehse B, Baum C. Retroviral vector insertion sites associated with dominanthematopoietic clones mark "stemness" pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Gong Z, Shen W, Ho AD. The tcl-3 proto-oncogene altered by chromosomal translocation in T-cell leukemia codes for a homeobox protein. EMBO J. 1991;10:2905–2910. doi: 10.1002/j.1460-2075.1991.tb07840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Ramezani A, Morgan RA, Hawley TS, Hawley RG. Sustained phenotypic correction of hemophilia A mice following oncoretroviral-mediated expression of a bioengineered human factor VIII gene in long-term hematopoietic repopulating cells. Mol Ther. 2004;10:892–902. doi: 10.1016/j.ymthe.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, Von KC, Knoss S, Schambach A, Baum C. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita K, Parganas E, Matsugi T, Ihle JN. Expression of the Evi-1 zinc finger gene in 32Dc13 myeloid cells blocks granulocytic differentiation in response to granulocyte colony-stimulating factor. Mol Cell Biol. 1992;12:183–189. doi: 10.1128/mcb.12.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- Nam CH, Rabbitts TH. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Mol Ther. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Nesbitt MN, Francke U. A system of nomenclature for band patterns of mouse chromosomes. Chromosoma. 1973;41:145–158. doi: 10.1007/BF00319691. [DOI] [PubMed] [Google Scholar]
- Newbold RF, Overell RW. Fibroblast immortality is a prerequisite for transformation by EJ c-Ha-ras oncogene. Nature. 1983;304:648–651. doi: 10.1038/304648a0. [DOI] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, Naundorf S, Brinkmann A, Deichmann A, Fischer M, Ball C, Pilz I, Dunbar C, Du Y, Jenkins NA, Copeland NG, Luthi U, Hassan M, Thrasher AJ, Hoelzer D, Von KC, Seger R, Grez M. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Owens BM, Hawley RG. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- Owens BM, Hawley TS, Spain LM, Kerkel KA, Hawley RG. TLX1/HOX11-mediated disruption of primary thymocyte differentiation prior to the CD4+CD8+ double-positive stage. Br J Haematol. 2006;132:216–229. doi: 10.1111/j.1365-2141.2005.05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens BM, Zhu YX, Suen TC, Wang PX, Greenblatt JF, Goss PE, Hawley RG. Specific homeodomain-DNA interactions are required for HOX11-mediated transformation. Blood. 2003;101:4966–4974. doi: 10.1182/blood-2002-09-2857. [DOI] [PubMed] [Google Scholar]
- Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Predescu SA, Predescu DN, Knezevic I, Klein IK, Malik AB. Intersectin-1s regulates the mitochondrial apoptotic pathway in endothelial cells. J Biol Chem. 2007;282:17166–17178. doi: 10.1074/jbc.M608996200. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS, Hawley RG. Combinatorial incorporation of enhancer blocking components of the chicken β-globin 5'HS4 and human T-cell receptor α/δ BEAD-1 insulators in self-inactivating retroviral vectors reduces their genotoxic potential. Stem Cells. 2008;26:3257–3266. doi: 10.1634/stemcells.2008-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Rice KL, Izon DJ, Ford J, Boodhoo A, Kees UR, Greene WK. Overexpression of stem cell associated ALDH1A1, a target of the leukemogenic transcription factor TLX1/HOX11, inhibits lymphopoiesis and promotes myelopoiesis in murine hematopoietic progenitors. Leuk Res. 2008a;32:873–883. doi: 10.1016/j.leukres.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Rice KL, Kees UR, Greene WK. Transcriptional regulation of FHL1 by TLX1/HOX11 is dosage, cell-type and promoter context-dependent. Biochem Biophys Res Commun. 2008b;367:707–713. doi: 10.1016/j.bbrc.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Riz I, Akimov SS, Eaker SS, Baxter KK, Lee HJ, Marino-Ramirez L, Landsman D, Hawley TS, Hawley RG. TLX1/HOX11-induced hematopoietic differentiation blockade. Oncogene. 2007;26:4115–4123. doi: 10.1038/sj.onc.1210185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riz I, Hawley RG. G1/S transcriptional networks modulated by the HOX11/TLX1 oncogene of T-cell acute lymphoblastic leukemia. Oncogene. 2005;24:5561–5575. doi: 10.1038/sj.onc.1208727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riz I, Hawley TS, Johnston H, Hawley RG. Role of TLX1 in T-cell acute lymphoblastic leukaemia pathogenesis. Br J Haematol. 2009a;145:140–143. doi: 10.1111/j.1365-2141.2008.07556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riz I, Lee HJ, Baxter KK, Behnam R, Hawley TS, Hawley RG. Transcriptional activation by TLX1/HOX11 involves Gro/TLE corepressors. Biochem Biophys Res Commun. 2009b;380:361–365. doi: 10.1016/j.bbrc.2009.01.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;2:971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Su X, Drabkin H, Clappier E, Morgado E, Busson M, Romana S, Soulier J, Berger R, Bernard OA, Lavau C. Transforming potential of the T-cell acute lymphoblastic leukemia-associated homeobox genes HOXA13, TLX1, and TLX3. Genes Chromosomes Cancer. 2006;45:846–855. doi: 10.1002/gcc.20348. [DOI] [PubMed] [Google Scholar]
- Varas F, Stadtfeld M, De Andres-Aguayo L, Maherali N, di TA, Pantano L, Notredame C, Hochedlinger K, Graf T. Fibroblast derived induced pluripotent stem cells show no common retroviral vector insertions. Stem Cells. 2008;27:300–306. doi: 10.1634/stemcells.2008-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yu WM, Hawley TS, Hawley RG, Qu CK. Catalytic-dependent and -independent roles of SHP-2 tyrosine phosphatase in interleukin-3 signaling. Oncogene. 2003;22:5995–6004. doi: 10.1038/sj.onc.1206846. [DOI] [PubMed] [Google Scholar]
- Yu WM, Hawley TS, Hawley RG, Qu CK. Immortalization of yolk sac-derived precursor cells. Blood. 2002;100:3828–3831. doi: 10.1182/blood-2002-03-0937. [DOI] [PubMed] [Google Scholar]
- Zhang N, Shen W, Hawley RG, Lu M. HOX11 interacts with CTF1 and mediates hematopoietic precursor cell immortalization. Oncogene. 1999;18:2273–2279. doi: 10.1038/sj.onc.1202545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.