Summary
We investigated miRNA expression changes associated with stress-induced premature senescence (SIPS) in primary cultures of human diploid fibroblasts (HDF) and human trabecular meshwork (HTM) cells. Twenty-five miRNAs were identified by miRNA microarray analysis and their changes in expression were validated by TaqMan realtime RT-PCR in three independent cell lines of HTM and HDF. SIPS in both HTM and HDF cell types was associated with significant down-regulation of four members of the miR-15 family and five miRNAs of the miR-106b family located in the oncogenic clusters miR-17–92, miR-106a-363, and miR-106b-25. SIPS was also associated with up-regulation of two miRNAs (182 and 183) from the miR-183-96-182 cluster. Transfection with miR-106a agomir inhibited the up-regulation of p21CDKN1A associated with SIPS while transfection with miR-106a antagomir led to increased p21CDKN1A expression in senescent cells. In addition, we identified retinoic acid receptor gamma (RARG) as a target of miR-182 and showed that this protein was down-regulated during SIPS in HDF and HTM cells. These results suggest that changes in miRNA expression might contribute to phenotypic alterations of senescent cells by modulating the expression of key regulatory proteins such as p21CDKN1A as well as by targeting genes that are down-regulated in senescent cells such as RARG.
Keywords: cellular senescence, miRNAs, gene-targeting, trabecular meshwork
Introduction
Cellular senescence is a state of permanent proliferative arrest that can result from telomere shortening after multiple rounds of cell division (replicative senescence) or from various stresses such as oncogenes or oxidative stress (stress-induced premature senescence, SIPS).
Senescent cells display phenotypic characteristics distinct from those of young cells and are associated with alterations in gene expressions (Borlon et al., 2007), protein processing (Noyan-Ashraf et al., 2008), and altered metabolic processes (A et al., 2008). While cellular senescence is considered a protective mechanism against cancer, it has also been hypothesized that the progressive accumulation of senescent cells in some tissues may contribute to several age-related diseases and organismal aging (Aikata et al., 2000; Castro et al., 2003; Castro et al., 2004; Flanary et al., 2007; Kitada et al., 1995; Minamino et al., 2002; Muller, 2006; Vasile et al., 2001). Specifically, senescence of human diploid fibroblasts (HDF) has been implicated in several pathologic conditions (Hasenmaile et al., 2003; Kojima et al., 2006; Lee et al., 2009; Lim et al., 2001; Lim et al., 2000; Song et al., 2008; Stein and Dulic, 1998). In addition, senescence of human trabecular meshwork (HTM) cells has been proposed to contribute to the decreased ability of the HTM to maintain normal levels of aqueous humor outflow resistance, leading thus to abnormal increase in intraocular pressure that is frequently associated with primary open angle glaucoma (Liton et al., 2006; Sacca and Izzotti, 2008).
A great deal of progress has been made in elucidating the major regulatory pathways that control the senescence response (Kim et al., 2009; Ren et al., 2009; Sakuma et al., 2008). However, the specific mechanisms regulating the senescent phenotype have not been completely elucidated.
MicroRNAs (miRNAs) have emerged recently as a new class of small evolutionarily conserved non-coding RNAs that negatively regulate gene expression (Bartel, 2009; Cannell et al., 2008; Sun et al., 2008). miRNAs inhibit the expression of target genes by affecting the translation and/or stability of mRNA by binding to their target sites in the 3’UTR of the mRNA. More than 700 human miRNAs have already been identified, and over 1000 miRNAs are predicted to exist in humans. Although the specific biological functions of most miRNAs remain largely unknown, they are believed to constitute a large gene regulatory network that can modulate the expression of up to 30% of total cellular proteins. There is increasing experimental evidence supporting the role of miRNAs in the regulation of a range of physiological responses, including apoptosis (Lynam-Lennon et al., 2009), cellular differentiation (Zhang et al., 2009), and cancer (Kaddar et al., 2009; Uziel et al., 2009). Several miRNAs have been shown to be involved in the regulation of pathways involved in cellular senescence and exert important effects on cell cycle progression (Brosh et al., 2008; He et al., 2007; Kim et al., 2007a; Kim et al., 2007b; Kumamoto et al., 2008; Lafferty-Whyte et al., 2009; Poliseno et al., 2008; Tazawa et al., 2007; Wagner et al., 2008; Wang, 2007). In addition, elimination of miRNA expression by knockdown of Dicer has been recently shown to result in the induction of cellular senescence through activation of the p53 pathway (Mudhasani et al., 2008). However, there is still little information regarding the potential involvement of miRNAs in modulating the alterations of gene expression observed in senescent cells.
To gain insight on the potential role of miRNAs in cellular senescence we investigated whether SIPS mediated by oxidative stress is associated with changes in expression of miRNAs in two different cell types, HDF and HTM cells. We also investigated whether these changes in miRNA expression contribute to the alteration in gene expression associated with SIPS.
Materials and methods
Cell Culture of Primary Human TM (HTM) cells
Post-mortem human eyes or cornea rings were obtained from the N.Y. eye bank within 7 days post-mortem according to the tenants of the Declaration of Helsinki. HTM from a single individual was dissected out from surrounding tissue, digested in 10 mg collagenase/20 mg bovine serum albumin (BSA)/5 ml phosphate buffer saline (PBS) solution. The cells were seeded on collagen I coated 3 cm Petri dishes and maintained at 37 °C in a humidified atmosphere of 5% CO2 in TM culture medium containing 20% fetal bovine serum (FBS). The TM culture medium was low glucose Dulbecco's Modified Eagle Medium (DMEM) with L-glutamine and 110 mg/l sodium pyruvate, supplemented), 100 µM nonessential amino acids, 100 units/ml penicillin, and 100 µg/ml streptomycin sulfate. All reagents were obtained from Invitrogen Corporation (Carlsbad, CA). These primary cells subsist for 9–14 passages, and experiments in this study were carried out with passages 4–7. The three primary cell lines used in this study originated from a 22-year-old Caucasian female, 17-year-old African-American male and a 12-year-old donor eye (HTM636-07-22; HTM786-07-17 and HTM1723-07-12, respectively).
Human dermal fibroblast cells were commercially obtained from Cell Applications, INC. and grown in DMEM with high glucose medium containing 10% FBS.
Experimental Model for Chronic Oxidative Stress in HTM and HDF cells
Human TM and HDF cells (passage 4–7) were treated with H2O2 200 µM in DMEM containing 10% FBS, twice a day, for four days. To differentiate from acute stress responses to oxidative challenge, TM cells were allowed a recovery time of three days after the H2O2 treatment. The medium was changed with fresh DMEM containing 10% FBS on the first day of recovery. RNAs or proteins were extracted three days after the H2O2 treatment. For the time course study, HTM and HDF cells were subjected to the same H2O2 treatment for four days, and then RNAs were isolated at day 1, 3, 7 and 14, respectively after H2O2 treatment.
miRNA Array and Data Analysis
The miRNA array was conducted from total RNA (extracted using TRIzol reagent (Sigma) and hybridized to Ambion miRChip V1 by Asuragen Inc. The chip contains miRNAs from Sanger miRBase B9.2 (human: 475 probes; rat: 234 probes; mouse: 377 probes), DiscovArray content (human: 467 probes; rat 234 probes; mouse: 293 probes) and Exploratory content: 12,894 probes. The total number of probes on the chip was 13,349. The data were analyzed using miRInform™ software. Detection calls were based on a Wilcoxon rank-sum test of the miRNA probe signal compared to the distribution of signals from GC-content matching anti-genomic probes. Background value was estimated for each probe from the median signal of a set of GC-matched anti-genomic controls and subtracted. Arrays were normalized together according to the variance stabilization method (Huber et al., 2003). For statistical hypothesis testing, a two-sample t-test, with assumption of equal variance, was applied.
Affymetrix geneChip microarray and data analysis
HTM cell cultures were transfected with miR-182 or control mimic. Three days post-transfection, RNAs were isolated using RNeasy mini kit (QIAGEN) and hybridized to Human Genome U133 2.0 Array at the Duke University Microarray facility (Durham, NC). This array covers the Human Genome U133 Set plus 6,500 additional genes for analysis of over 47,000 transcripts. Raw data was normalized and analyzed using GeneSpring 10 (Silicon Genetics, Wilmington, DE). Genes were filtered to their intensities in the control channel. A paired student t-test was performed (p values ≤ 0.05 were considered significant) and fold changes were excluded below 1.5 (miR-182 vs. control mimic).
Amaxa nucleofector transfection
Transfection of miRNAs and siRNAs was performed using the Nucleofector System (Amaxa Inc. Gaithersburg, MD) according to the manufacturer’s instructions. MiR-106a, miR-182 mimics or inhibitors and negative miRNA control mimics or inhibitors (Dharmacon, Inc. Chicago, IL) were transfected 120 pmoles per 5 × 105 cells using programs U23 for HDF and T23 for HTM cells. Same amount of validated p53 siRNA or control scramble siRNA (Applied BioSystems, Austin, Texas) were co-transfected with miR-106a mimic or inhibitor and their individual controls to HDF and HTM cells using the same programs.
Quantitative real-time PCR analysis of miRNAs
For the quantitative analysis of miRNAs, small RNAs were isolated using a mirVanaTM miRNA Isolation Kit (Applied BioSystems, Austin, Texas). Two-step TaqMan realtime RT-PCR analysis was performed using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Austin, Texas). The reverse transcript (RT) reaction was performed by sequential incubation at 16°C for 30 min, 42°C for 30 min, and 85 ° C for 5 min. Each PCR reaction mixture (20 µl) contained 1.33 µl of RT product, 10 µl of TaqMan 2× Universal PCR Master Mix and 1 µl of the appropriate TaqMan MicroRNA Assay (20×) containing primers and probe for the miRNA of interest. The mixture was initially incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. PCR reactions were performed in triplicate. All RNA samples were normalized relative to human U6B miRNA.
Protein Extraction and Immunoblot
Cells were washed twice in cold PBS. Total protein was extracted using RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 5 mM EDTA, pH8.0) containing 1x protease inhibitor cocktail (Coche, Inc). Protein concentration was determined using Micro BCA Protein Assay Kit (Pierce, Rockford, IL). Total protein extracts (40 µg) were separated by 10% SDS-PAGE and transferred to PVDF membrane (Bio-Rad, Hercules, CA). Membranes were blocked with 5% nonfat dry milk and incubated overnight with anti-RARG (sc-550, Santa Cruz Biotechnology, INC.), a primary antibody, and then with a secondary anti-rabbit antibody conjugated to horseradish peroxidase (Sigma). Immunoreactive proteins were visualized using chemiluminescence substrate (ECL Plus, GE Healthcare, Pittsburgh, PA). For detection of endogenous control, the membrane was stripped with stripping buffer (25 mM glycine pH 3.0 plus 1% sodium dodecyl sulfate (SDS)) and then incubated with anti β-tubulin (SC-9935, Santa Cruz Biotechnology (Sta. Cruz, CA)).
Luciferase reporter assay
The 3’-UTR fragment of RARG gene, including or excluding the predicted target site for miR-182 was amplified by PCR from a full length image clone (pCMV.SPORT6, ATCC, Manassas, VA) using primers: F (5’-TATCTCGAGCTGAAGTCCCCAGCCTGACCAG), F (5’-TATCTCGAGGTTTCTAGGGGTGCCTCTGTGTTCA) and R (5’-TCAGCGGCCGCGAGTTTCCATCACTTTATTTTGC) that create XhoI and NotI sites, respectively. The 3’UTR fragment of p21CDKN1A was amplified from human cDNA using primers: F (5’-GTACTCGAGTGATCTTCTCCAAGAGGAAGCCCT) and R (5’-TATGCGGCCGCATTCAGCATTGTGGGAGGAGCTGT). The XhoI-NotI-digested product was cloned into the psiCheck-2 vector, which included both renilla and firefly luciferase reporter genes (Promega Corporation, Madison, WI). The H293A cells were co-transfected in 12-well plates using Effectene reagent (QIAGEN Inc, Valencia, CA) with 300 ng of the 3’UTR-luciferase report vector and 7 ng miRNA mimics or negative control mimic (Dharmacon, Lafayette, CO). Twenty-four hrs after transfection, firefly and renilla luciferase activities were measured consecutively using dual-luciferase assays (Promega Corporation, Madison, WI) according to the manufacturer’s protocol. A RARG 3’UTR control vector was generated by a deletion of 157nt from the RARG 3’UTR containing the predicted target sequence for miR-182. The control vector for p21CDKN1A 3’UTR was generated by inserting reverse 3’UTR fragment of p21CDKN1A to the psiCHECK2 reporter vector.
Assay of intracellular reactive oxygen species (iROS)
The production of iROS was measured by DCFH oxidation. Briefly, 10 mM H2DCFDA (Invitrogen Corporation, Carlsbad, CA) was dissolved in methanol and was diluted 500 fold in HBSS to give a 20 µM concentration of H2DCFDA. H2O2 treated and control cells in 12-well plates were exposed to H2DCFDA for 15min, and then cells were collected. The cells were pelleted and washed once with PBS, then resuspended in 200 µl of PBS. The cells were analyzed by flow cytometry (Becton Dickinson, San Jose, CA) with 488 nm excitation using emission filters appropriate for Alexa Fluor 488 dye.
Measurement of mitochondrial membrane potential (Δψm)
Mitochondrial membrane potential was monitored in cells loaded with JC-1 dye (Invitrogen Corporation, Carlsbad, CA) as described previously, with minor modifications (Li et al., 2007). Briefly, cells in six-well plates treated with or without H2O2 were trypsinized and pelleted by centrifuge. The cells were then washed with PBS once, loaded with JC-1 in 1 ml of PBS to final concentration of 2 µM, and incubated at 37 °C, 5% CO2 for 15 min. The cells were pelleted and washed again with PBS, then resuspended in 200 µl of PBS. The cells were analyzed on a flow cytometer with 488 nm excitation using emission filters appropriate for Alexa Fluor 488 dye and R-phycoerythrin.
Measurement of Endogenous Cellular Autofluorescence and SA-β-gal
Endogenous cellular autofluorescence was detected under the FITC filter by fluorescence microscopy and quantified by flow cytometry using a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA). For this, the fluorescence emitted by 10,000 cells in the FL-2 channel (563–607 nm) was recorded and analyzed with CELLQuest software (Becton Dickinson, San Jose, CA). Activity of SA-β-gal was also measured by flow cytometry using the fluorogenic substrate C12FDG (Invitrogen Corporation, Carlsbad, CA) as previously described (Fiering et al., 1991). Briefly, cells were were incubated with chloroquine 300 µM for 1 h at 37°C to modulate the intracellular pH. Cells were then trypsinized and washed once with PBS, loaded 2 mM FDG, incubated 1 min at 37 °C. Cells were then diluted 10 times with cold PBS, incubated on ice for 30 min and analyzed by flow cytometer in FL-1 channel (Alexa Fluor 488 nm). The average number of cells analyzed for each experiment was 10,000.
Cell Proliferation
Cell proliferation was quantified using the BrdU Cell Proliferation Assay (Calbiochem, San Diego, CA) according to the manufacturer’s instructions. Briefly, 100 µl of cells (con, H2O2 treated) at 2 ×105 cells/ml were seeded into a 96 well culture dish and incubated for 24 hrs. Culture medium was then replaced with 100 µl fresh DMEM containing 10% FBS and BrdU 1:10000 dilution. After overnight incubation, the cells were fixed with Fixative/Denaturing solution. The cells were consititutely incubated with Anti-BrdU Antibody, reconstituted Peroxidase Goat Anti-Mouse IgG HRP Conjugate. The color was then developed by adding Substrate Solution to each well. After 15 minutes of incubation in the dark at room temperature, Stop Solution was added to each well, and absorbance was measured using a spectrophotometric plate reader at dual wavelengths of 450–540 nm.
Statistical Analysis
The data were presented as the mean ±SD. For multiple comparisons of groups, ANOVA was used. Statistical significance between groups was assessed by Mann-Whitney U Test. A value of p<0.05 was considered statistically significant.
Results
Confirmation of oxidative stress induced cellular senescence
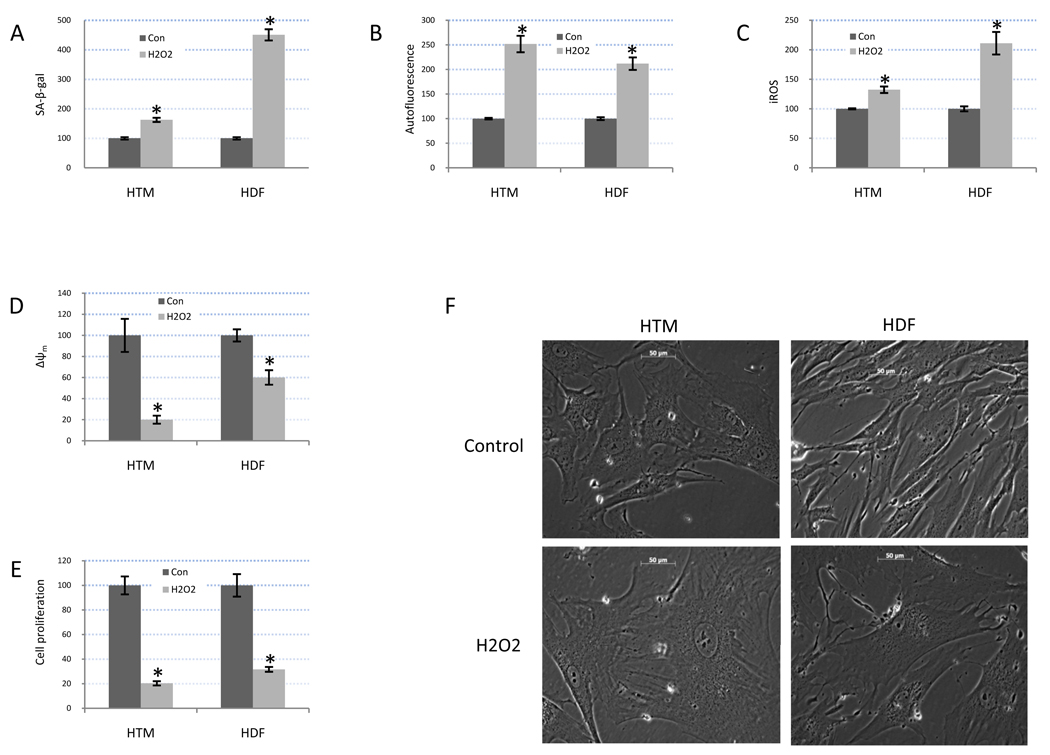
Several parameters were evaluated to confirm that treatment of primary cultures of both HTM and HDF cells with H2O2 (200 µM) twice a day for four days induced a senescent phenotype. This treatment resulted in significant increases in SA-β-gal activity, autofluorescence, and endogenous ROS production. The increased production of ROS was associated with a decrease of mitochondrial membrane potential measured by JC-1. There was also a significant decrease in cell proliferation rate quantified by BrdU incorporation. Finally, treated cells displayed the characteristic morphological changes associated with cellular senescence, including enlarged size and autofluorescence granules in the peri-nuclear region (Figure 1).
Figure 1. Cellular senescence markers in HTM and HDF cells under chronic oxidative stress conditions.
Three individual of HTM (636-07-22; 786-07-17; 1723-08-12) and HDF (1365; 1454; 1248) cell lines were treated with H2O2 200 µM twice a day for four days and then they were allowed three days to recover. Induction of beta-galactosidase (SA-β-gal) (A), autofluorescence (B), iROS (C), Δψm (D) were quantified by flow cytometry. Cell proliferation rate (E) was quantified by BrdU incorporation. Graphs represent the mean ± SD of the percentile change compared to controls from triplicate experiments conducted in the individual cell lines HTM-636-07-22 and HDF-1248 (n=3, * p<0.05). Morphology of HTM (636-07-22) and HDF (1248) cells following H2O2 treatment is shown in figure 1F. The images are representative of the overall morphological changes observed in the three individual cell lines of HTM (636-07-22; 786-07-17; 1723-08-12) and HDF (1365; 1454; 1248).
Changes in miRNA expression associated with SIPS in HTM and HDF cells
To search for miRNAS potentially up- or down-regulated during stress-induced senescence, total RNA, including small RNA samples from three biological replicas of senescent HDF, HTM and their respective non-senescent controls were hybridized to Ambion miRChip V1 (total 13,349 probes) by Asuragen Inc. Twenty-four (miR-15a, miR-15b, miR-16, miR-17-5p, miR-18a, miR-20a, miR-20b, miR-92, miR-106a, miR-106b, miR-139, miR-146b, miR-155, miR-182, miR-183, miR-192, miR-195, miR-199b, miR-200c, miR-204, miR-218, miR-342, miR-409-5p, and miR-493) miRNAs with average differences in intensity signals higher than 1.7 fold at P values lower than 0.03, or with intensity differences lower than −1.35 fold at P values lower than 0.04 in either HDF or HTM cells (Table 1), were selected to evaluate their potential up- or down-regulation by TaqMan realtime PCR in three primary cell lines of HDF and three lines of HTM cells from different donors. Fourteen miRNAS (miR-15a, miR-15b, miR-16, miR-17-5p, miR-18a, miR-20a, miR-92, miR-106a, miR-106b, miR-146b, miR-195, miR-199b, miR-204, and miR-342) were consistently down-regulated in both HDF and HTM cells, and three miRNAs (miR-200c, miR-182, miR-139) were consistently up-regulated in both cell types. In addition miRNAs miR-20b, miR-155, and miR-218 were down-regulated only in HDFs, and miRNAs miR-493 and miR-409-5p were up-regulated also only in HDFs. Finally, two miRNAS (miR-183 and miR-192) were found to be up-regulated in HTM cells but not in HDF (Figure 2).
Table 1.
miRNA array demonstrating changes of miRNA expressions in both senescent HTM and HDF cells
| Probe_ID | MicroRNA_Accession | HDF | HTM | ||
|---|---|---|---|---|---|
| Con vs H: ttest | H vs Con: Fold | con vs h: ttest | H vs Con: fold | ||
| hsa-miR-15a_st1 | MIMAT0000068 | 0.021 | −2.650 | 0.004 | −2.016 |
| hsa-miR-15b_st1 | MIMAT0000417 | 0.010 | −2.325 | ||
| gga-miR-15b_st1 | MIMAT0001154 | 0.036 | −1.379 | ||
| tni-miR-15b_st1 | MIMAT0003086 | 0.013 | −2.303 | ||
| ssc-miR-15b_st1 | NA | 0.010 | −2.330 | ||
| gga-miR-15b_st1 | MIMAT0001154 | 0.004 | −2.458 | ||
| dre-miR-15b_st1 | MIMAT0001773 | 0.002 | −2.492 | 0.024 | −1.334 |
| lca-miR-16_st1 | NA | 0.013 | −1.815 | ||
| tni-miR-16_st1 | MIMAT0003108 | 0.008 | −2.063 | 0.011 | −1.356 |
| hsa-miR-17-5p_st1 | MIMAT0000070 | 0.009 | −2.074 | 0.019 | −1.628 |
| hsa-miR-18a_st1 | MIMAT0000072 | 0.001 | −2.271 | ||
| hsa-miR-18a-_st1 | MIMAT0002891 | 0.037 | −1.457 | ||
| hsa-miR-20a_st2 | MIMAT0000075 | 0.018 | −2.154 | 0.002 | −1.884 |
| mmu-miR-20b_st2 | MIMAT0003187 | 0.009 | −1.997 | 0.046 | −1.618 |
| hsa-miR-20b_st2 | MIMAT0001413 | 0.002 | −2.259 | 0.025 | −1.862 |
| rno-miR-20b_st2 | MIMAT0003211 | 0.004 | −2.028 | 0.018 | −1.828 |
| dre-miR-20b_st2 | MIMAT0001778 | 0.005 | −2.053 | 0.025 | −1.737 |
| hsa-miR-92_st1 | MIMAT0000092 | 0.001 | −2.537 | 0.005 | −1.539 |
| tni-miR-92_st2 | MIMAT0002925 | 0.000 | −2.565 | 0.001 | −1.566 |
| mmu-miR-106a_st2 | MIMAT0000385 | 0.009 | −1.944 | 0.031 | −1.736 |
| hsa-miR-106a_st1 | MIMAT0000103 | 0.016 | −2.083 | 0.038 | −1.618 |
| hsa-miR-106b_st2 | MIMAT0000680 | 0.028 | −2.174 | 0.008 | −1.840 |
| hsa-miR-139_st2 | MIMAT0000250 | 0.000 | 7.625 | 0.002 | 5.779 |
| dre-miR-139_st2 | NA | 0.017 | 2.586 | 0.040 | 2.875 |
| hsa-miR-146b_st1 | MIMAT0002809 | 0.000 | −3.584 | 0.011 | −2.308 |
| gga-miR-146b_st1 | MIMAT0003351 | 0.001 | −3.617 | 0.013 | −2.718 |
| hsa-miR-155_st1 | MIMAT0000646 | 0.012 | −3.138 | 0.043 | −1.385 |
| mmu-miR-155_st2 | MIMAT0000165 | 0.000 | −3.054 | ||
| hsa-miR-182_st1 | MIMAT0000259 | 0.001 | 7.462 | 1.39E−05 | 9.631 |
| hsa-miR-183_st1 | MIMAT0000261 | 0.024 | 4.494 | 0.008 | 3.912 |
| tni-miR-192_st1 | MIMAT0002942 | 0.016 | 1.783 | 0.005 | 2.683 |
| hsa-miR-192_st2 | MIMAT0000222 | 0.015 | 1.762 | 0.011 | 2.507 |
| hsa-asg-192_st1 | NA | 0.035 | 1.475 | ||
| mmu-miR-192_st2 | MIMAT0000517 | 0.006 | 1.629 | 0.008 | 2.247 |
| hsa-miR-195_st2 | MIMAT0000461 | 0.032 | −1.763 | 0.018 | −1.689 |
| hsa-miR-199b_st2 | MIMAT0000263 | 0.001 | −3.707 | ||
| mmu-miR-199b_st1 | MIMAT0000672 | 0.013 | −1.854 | 0.041 | −1.843 |
| ppy-miR-200c_st1 | NA | 0.014 | 6.480 | 0.001 | 5.695 |
| hsa-miR-200c_st1 | MIMAT0000617 | 0.000 | 4.035 | 0.002 | 6.087 |
| dre-miR-200c_st1 | MIMAT0001863 | 0.009 | 3.022 | ||
| hsa-miR-204_st2 | MIMAT0000265 | 0.003 | −2.548 | 0.025 | −2.527 |
| tni-miR-218b_st1 | MIMAT0002986 | 0.020 | −2.325 | 0.015 | −2.905 |
| hsa-miR-218_st2 | MIMAT0000275 | 0.001 | −2.595 | 0.002 | −2.894 |
| tni-miR-218a_st1 | MIMAT0002994 | 0.003 | −3.256 | ||
| hsa-cand342_st1 | MIMAT0000092 | 0.001 | −2.616 | 0.001 | −1.497 |
| hsa-miR-342_st1 | MIMAT0000753 | 0.005 | −2.796 | ||
| rno-miR-409-5p_st1 | MIMAT0003204 | 0.001 | 2.254 | ||
| hsa-miR-409-5p_st1 | MIMAT0001638 | 0.000 | 2.140 | ||
| mmu-miR-409_st2 | MIMAT0001090 | 0.024 | 1.383 | 0.038 | 2.291 |
| hsa-miR-493-3p_st1 | MIMAT0003161 | 0.005 | 2.208 | ||
| hsa-miR-493_st1 | MIMAT0002813 | 0.000 | 1.757 | 0.011 | 1.927 |
Figure 2. Validation of miRNA expression in senescent HTM and HDF cells.
Small RNAs (20 ng) isolated from control and H2O2-treated cells from three individual of HTM (636-07-22; 786-07-17; 1723-08-12) and HDF (1365; 1454; 1248) cell lines were reverse transcribed and amplified with miRNA-specific primers and TaqMan probes. Relative expression was calculated using the comparative cycle threshold method and miRNA abundance was normalized relative to human RNU6B miRNA. Results are expressed as the fold of miRNA levels relative to that of control cultures. (A) up- and down-regulated miRNAs in HTM cell lines; (B) up- and down-regulated miRNAs in HDF cell lines. Data represent the mean ± SD, n=3, p < 0.05 to 0.001 compared to the control. Note: * labeled miRNAs showed not consistent changes in all three cell lines.
To determine whether these changes in expression were part of a transient or sustained modification of miRNA expression in senescent cells, we analyzed the levels of expression two weeks after the induction of cellular senescence (Figure 3). With the exception of miR-200c and miR-192, all other miRNAs showed a sustained change in expression for at least two weeks.
Figure 3. miRNA expression time course in senescent HTM and HDF cells.
One individual HTM (636-07-22) and HDF (1454) cell lines were treated with H2O2 200 µM twice a day for four days, RNAs were then isolated at days 1, 3, 7 and 14 after the last day of H2O2 treatment. Results are expressed as the fold of miRNA levels relative to that of the untreated control cultures. (A) up- or down-regulated miRNAs in HTM cells; (B) up- or down-regulated miRNAs in HDF cells. Data represent the mean ± SD, n=3.
Many of the miRNAs that showed a decrease in expression associated with oxidative stress-induced senescence have been reported to be up-regulated in a variety of cancer cells and during tissue regeneration (Table 2).
Table 2.
Dysregulated miRNAs in senescent HDF and HTM cells that are also found in cancers
| MiRNAs Down-regulated in SIPS | ||||||
|---|---|---|---|---|---|---|
| MiRNA | Family | Cluster | Downregulation in cancer | Reference | Upregulation in cancer | Reference |
| HDF and HTM | ||||||
| miR-15a | miR-15 | 15a/16-1 | Pituitary Adenoma | (Bottoni et al., 2005) | Pancreatic Adenocarcinoma |
(Lee et al., 2007) |
| miR-15b | miR-15 | 15b/16-2 | Acute Myeloid Leukemia | (Dixon-McIver et al., 2008) | Pancreatic Adenocarcinoma |
(Lee et al., 2007) |
| miR-16* | miR-15 | 15a/16-1* | Pituitary Adenoma | (Bottoni et al., 2005) | Pancreatic Adenocarcinoma |
(Lee et al., 2007) |
| 15b/16-2* | ||||||
| miR-17-5p | miR-106b | 17-92a-1 | Prostate Carcinoma | (Porkka et al., 2007; Volinia et al., 2006) |
Squamous Cell Carcinoma | (Wong et al., 2008) |
| miR-18a | miR-106b | 17-92a-1 | ||||
| miR-20a | miR-106b | 17-92a-1 | Ovarian Carcinoma | (Nam et al., 2008) | ||
| miR-92** | miR-25 | 17-92a-1** | Prostate Carcinoma | (Porkka et al., 2007; Volinia et al., 2006) |
Squamous Cell Carcinoma | (Wong et al., 2008) |
| 106a-363** | ||||||
| miR-106a | miR-106b | 106a-363 | Gastric Adenocarcinoma | (Petrocca et al., 2008b) | ||
| miR-106b | miR-106b | 106b/93/25 | Gastric Adenocarcinoma | (Petrocca et al., 2008b) | ||
| miR-146b | miR-146 | NA | Hepatocellular Carcinoma | (Meng et al., 2007; Murakami et al., 2006) |
Papillary Carcinoma | (Pallante et al., 2006) |
| miR-195 | miR-15 | 497/195 | Hepatocellular Carcinoma | (Murakami et al., 2006) | Acute Myeloid Leukemia | (Dixon-McIver et al., 2008) |
| miR-199b | miR-199 | NA | Hepatocellular Carcinoma | (Murakami et al., 2006) | ||
| miR-204 | miR-204 | NA | Hodgkin Lymphoma | (Navarro et al., 2008) | ||
| miR-342 | miR-342 | NA | Esophageal Adenocarcinoma | (Feber et al., 2008) | Esophageal Squmous Cell Carcinoma |
(Feber et al., 2008) |
| HDF | ||||||
| miR-20b | miR-106b | 106a-363 | ||||
| miR-155 | miR-155 | NA | Acute Myeloid Leukaemia | (Dixon-McIver et al., 2008) | ||
| miR-218 | miR-218 | NA | Gastric Adenocarcinoma | (Petrocca et al., 2008b) | ||
| miRNAs up-regulated in SIPS | ||||||
| MiRNA | Family | Cluster | Downregulation in cancer | Reference | Upregulation in cancer | Reference |
| HDF and HTM | ||||||
| miR-139 | miR-139 | NA | Squamous Cell Carcinoma | (Wong et al., 2008) | ||
| miR-182 | miR-182 | 183/96/182 | Acute Myeloid Leukemia | (Dixon-McIver et al., 2008) | Hepatocellular Carcinoma | (Murakami et al., 2006) |
| miR-200c *** | miR-8 | 200c/141 | Esophageal Squmous Cell Carcinoma |
(Feber et al., 2008) | Esophageal Adenocarcinoma |
(Feber et al., 2008) |
| HDF | ||||||
| miR-409-5p | miR-154 | 485-656 | ||||
| miR-493 | miR-154 | 493/337/665 | Ovarian Carcinoma | (Nam et al., 2008) | ||
| HTM | ||||||
| miR-183 | miR-183 | 183/96/182 | Hodgkin Lymphoma | (Navarro et al., 2008) | Poorly Differentiated Carcinoma |
(Nikiforova et al., 2008) |
| miR-192*** | miR-192 | 194-2/192 | EsophagealSqumous Cell Carcinoma |
(Feber et al., 2008) | Esophageal Adenocarcinoma |
(Feber et al., 2008) |
identical sequences for 16-1 and 16-2
identical sequences for 92a-1 and 92a-2
miRNAs temporarily up-regulated during the induction of SIPS
Down-regulation of miR-106a contributes to the up-regulation of p21CDKN1A in senescent HDF and HTM cells
Since the specific mechanisms leading to the induction of p21CDKN1A in oxidative stress-induced SIPS are not completely clear and may be independent from its up-stream activator p53 (Zdanov et al., 2007), we hypothesized that the down-regulation of members of the miR-106b family may contribute to the increase of p21CDKN1A in expression in SIPS. Consistent with this hypothesis, we confirmed by 3’UTR dual-luciferase activity assay and by western blot that the member of this family more clearly down-regulated during SIPS in HDF and HTM cells, miR-106a, also targets the p21CDKN1A mRNA (Fig 4A, B). Furthermore, transfection with miR-106a mimic resulted in lower induction of p21CDKN1A after H2O2 treatment, and transfection with a miR-106a antagomir led to increased expression of the p21CDKN1A protein (Fig 4C). We investigated whether the changes in p21CDKN1A expression were dependent on p53 by inhibiting p53 expression using siRNA. As shown in figure 4D, while the down-regulation of p21CDKN1A mediated by miR-106a mimic was observed regardless of whether p53 was inhibited or not, the induction of p21CDKN1A expression mediated by mir-106a antagomir was prevented by inhibition of p53 by siRNA. Transfection of HTM and HDF cells with miR-106a mimic also resulted in increased cell proliferation measured by BrdU incorporation. However, no significant effects on cell proliferation were observed in cells transfected with miR-106a antagomir (Fig 4E). Finally, neither miR-106a mimic nor antagomir transfection led to a significant change in SA-beta-galactosidase activity (Fig 4E).
Figure 4. Determination of p21CDKN1A as miR-106a target.
(A) The complete 3’UTR of p21CDKN1A was cloned into the psiCHECK2 dual-luciferase reporter vector and cotransfected with either miR-106a mimic (106a) or miRNA control mimic (con) into HEK 193 cells. Negative controls were generated by cloning the same 3’UTRs in reverse orientation (3’-p21-rev). The luciferase activities of hRluc and hluc were analyzed 24 hrs post-transfection. (n=3; *, p< 0.05 Mann-Whitney U Test comparison between miR-106a mimic and negative miRNA mimic control). (B) HDF and HTM cells were transfected with either miR-106a mimic (Mir-106aM) or miRNA control mimic (Mir-ConM) and the expression of p21CDKN1A was evaluated three days post-transfection by western blot analysis in three individual experiments. (C) HDF and HTM cells were transfected with control mimic (ConM), miR-106a mimic (106aM), control inhibitor (ConI), or miR-106a inhibitor (106aI) for one day and then treated with H2O2 200 µM twice a day for four days and recovered for additional three days. Total proteins were separated with 10% SDS-page gel and immunostained with specific anti- p21CDKN1A antibody. (D) HDF and HTM cells were transfected with ConM, 106aM, ConI, or 106aI with p53 siRNA or control scramble siRNA, and the expression of p21CDKN1A was evaluated three days post-transfection by western blot. Total proteins were separated with 10% SDS-page gel and immunostained with specific anti-p21CDKN1A antibody. The same blot was stripped and re-stained with beta-tubuin. (E) HDF and HTM cells were transfected with ConM, 106aM, ConI, or 106aI, and cell proliferation measured by BrdU incorporation and SA-beta-gal activity were evaluated three days post-transfection (n=3; Mann-Whitney U Test comparison between miR-106a mimic and negative miRNA mimic control indicated as *, p< 0.05 in HTM cells and #, p < 0.05 in HDF cells.
Up-regulation of miR-182 in SIPS contributes to the down-regulation of RARG in HDF and HTM cells by direct post-transcriptional regulation
Affymetrix gene array showed that 37 genes were significantly up- or down-regulated by miR-182 in HTM cells (fold change > 1.5, p < 0.05) and 14 genes indicated potential miR-182 targets in one of the four miRNA databases (miRbase, MIRANDA, TARGETSCAN and PICTAR-VERT) (Table 3). Retinoic acid receptor gamma (RARG) was found as a potential target in all four databases and was selected for further gene target study. In order to test whether RARG was a target of miR-182, the entire 3’UTR of the gene and the 3’UTR with a deletion of 157nt in the region containing the putative binding site of miR-182 were inserted into the dual luciferase reporter vector psiCHECK2. Co-transfection of H293 cells with miR-182 and psiCHECK2 containing the complete RARG 3’UTR was significantly decreased compared to cells co-transfected either with miR-182 and psiCHECK2 containing RARG 3’UTR in which the region containing the predicted miR-182 target sequence was deleted (58% ± 0.54 (SD), p = 0.0039) (Figure 5A). In addition, western blot analysis of HDF and HTM cells transfected with miR-182 showed a marked decrease in RARG protein expression compared to cells transfected with a scrambled control (Figure 5B). Western blot also showed that RARG was highly down-regulated during SIPS in both senescent HTM and HDF cells (Figure 5C). Transfection of HTM and HDF cells with miR-182 mimic led to a significant increase in SA-beta-galactosidase activity that was not associated with decreased proliferation. No effects on either SA-beta-galactosidase activity or proliferation were observed in cells transfected with miR-182 antagomir (Fig 5D).
Table 3.
miR-182 regulated gene expression in HTM cells
| Unigene | Gene Symbol |
Fold Change ([182] vs [con]) |
Regulation ([182] vs [con]) |
miRbase | MIRANDA | TARGETSCAN | PICTAR- VERT |
Gene Title |
|---|---|---|---|---|---|---|---|---|
| Hs.476402 | ARHGEF3 | 1.50 | down | T | T | T | Rho guanine nucleotide exchange factor (GEF) 3 | |
| Hs.709513 | ARL4C | 1.61 | down | T | ADP-ribosylation factor-like 4C | |||
| Hs.435655 | ASPN | 1.60 | down | asporin | ||||
| Hs.700589 | C10orf116 | 1.56 | down | chromosome 10 open reading frame 116 | ||||
| Hs.376071 | CCND2 | 1.54 | down | T | T | cyclin D2 | ||
| Hs.654387 | CPM | 1.69 | up | carboxypeptidase M | ||||
| Hs.632466 | CTSK | 1.64 | down | cathepsin K | ||||
| Hs.468410 | EPAS1 | 1.59 | down | T | T | T | endothelial PAS domain protein 1 | |
| Hs.62192 | F3 | 1.50 | up | coagulation factor III (thromboplastin, tissue factor) | ||||
| Hs.616962 | GDF15 | 1.59 | down | growth differentiation factor 15 | ||||
| Hs.234896 | GMNN | 1.62 | down | geminin, DNA replication inhibitor | ||||
| Hs.98206 | GREM2 | 1.51 | down | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | ||||
| Hs.159226 | HAS2 | 1.51 | down | T | hyaluronan synthase 2 | |||
| Hs.517581 | HMOX1 | 1.60 | down | heme oxygenase (decycling) 1 | ||||
| Hs.296942 | KRT34 | 1.66 | up | keratin 34 | ||||
| Hs.631263 | PLGLA, PLGLB1, PLGLB2 |
1.59 | down | plasminogen-like B2 /// plasminogen-like B1 /// plasminogen-like A /// hypothetical protein |
||||
| Hs.13245 | LPPR4 | 2.13 | up | T | T | plasticity related gene 1 | ||
| Hs.567412 | LRRC17 | 1.93 | up | leucine rich repeat containing 17 | ||||
| Hs.189445 | MATN2 | 1.53 | up | matrilin 2 | ||||
| Hs.432818 | MFAP3 | 1.91 | down | T | T | T | Microfibrillar-associated protein 3 | |
| Hs.432818 | MFAP3 | 1.61 | down | T | T | T | Microfibrillar-associated protein 3 | |
| Hs.647370 | MT1M | 1.89 | up | metallothionein 1M | ||||
| Hs.21334 | NAGPA | 1.53 | down | T | T | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | ||
| Hs.503500 | OLFML1 | 1.61 | down | olfactomedin-like 1 | ||||
| Hs.458573 | 1.50 | down | T | T | platelet-derived growth factor receptor-like | |||
| Hs.409352 | PID1 | 1.57 | down | T | phosphotyrosine interaction domain containing 1 | |||
| Hs.654978 | RAB27A | 1.56 | down | T | RAB27A, member RAS oncogene family | |||
| Hs.1497 | RARG | 1.71 | down | T | T | T | T | retinoic acid receptor, gamma |
| Hs.388918 | RECK | 1.94 | down | T | T | T | T | reversion-inducing-cysteine-rich protein with kazal motifs |
| Hs.709405 | SEPP1 | 1.77 | down | selenoprotein P, plasma, 1 | ||||
| Hs.567236 | SHROOM2 | 1.54 | up | shroom family member 2 | ||||
| Hs.101307 | SLC14A1 | 1.54 | down | solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
||||
| Hs.419240 | SLC2A3 | 1.62 | down | solute carrier family 2 (facilitated glucose transporter), member 3 | ||||
| Hs.313 | SPP1 | 1.68 | down | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T- lymphocyte activation 1) |
||||
| Hs.437277 | SQSTM1 | 1.51 | down | T | sequestosome 1 | |||
| Hs.489922 | TUFT1 | 1.50 | up | tuftelin 1 | ||||
| Hs.533977 | TXNIP | 1.52 | down | thioredoxin interacting protein | ||||
| Hs.567356 | WNT2 | 1.50 | up | wingless-type MMTV integration site family member 2 |
Note: miR-182 and control mimic transfected into HTM cells for three days, RNAs were used for Affimatrix gene array. The data showed that fold changes is > 1.5; paired student t test p value < 0.05. T indicated potential target.
Figure 5. RARG in miR-182 transfected and stress-induced senescent cells.
(A) PsiCHECK2 vector containing complete 3’UTR of RARA with (3’-RARG) or without (3’-RARG control) miR-182 predicted target site co-transfected with either miR-182 mimic (182M) or miRNA control mimic (ConM). The luciferase activities of hRluc and hluc were analyzed 24 hrs post-transfection. (n=3; *, p< 0.05 Mann-Whitney U Test comparison between miR-182 mimic and negative miRNA mimic control). (B) HDF and HTM cells were transfected with either miR-182 mimic (182M) or miRNA control mimic (ConM) and the expression of RARG was evaluated three days post-transfection by western blot analysis in three individual experiments. (C) HDF and HTM cells were treated with H2O2 200 µM twice a day for four days and recovered for additional three days. Total proteins were separated with 10% SDS-page gel and immunostained with specific anti-RARG antibody. (D) HDF and HTM cells were transfected with ConM, 182M, ConI, or 182I, and SA-β-gal activity and BrdU incorporation (cell proliferation) were evaluated three days post-transfection. Graphs represent the percentage of increase or decrease compared to controls (n=3; Mann-Whitney U Test comparison between miR-182M and ConM indicated as *, p< 0.05 in HTM cells and #, p < 0.05 in HDF cells).
Discussion
Our results revealed extensive changes in miRNA expression associated with the induction of SIPS in two different cell types. Most of these changes were consistent in the two different cell types analyzed (HDF and HTM cells). With the exception of miRNASs miR-200c and miR-192, all other changes were stable for at least two weeks and may represent permanent changes associated with the senescent response. The initial induction of miR-200c observed in HDF and HTM cells and that of miR-192 in HTM cells could be associated with a response to the oxidative stress used to induce cellular senescence.
Most of the miRNAs down-regulated in SIPS have also been shown to experience alterations in expression in different types of cancers (Wang and Lee, 2009). Interestingly, with the exception of miR-92, miR-199b and miR-342, all miRNAs consistently down-regulated in both senescent HDF and HTM cells were members of either the miR-15 or the miR-106b families of miRNAs.
The four known members of the miR-15 family, miR-15a, miR-15b, miR-16, and miR-195, were down-regulated in both senescent HDF and HTM cells. The miR-15/16 cluster is known to target the anti-apoptotic factor BCL2, leading to increased susceptibility for apoptosis (Cimmino et al., 2005). This cluster is frequently deleted in B-cell chronic lymphocytic leukemia (CLL) (Calin and Croce, 2006) and is down-regulated in several cancers such as pituitary adenoma (Bottoni et al., 2007; Zatelli and degli Uberti, 2008) and prostate carcinoma (Porkka et al., 2007). Down-regulation of the miR-15/16 cluster is believed to contribute to cancer growth by inhibiting apoptosis. Increased resistance to apoptosis is a well known feature of senescent cells (Ahn et al., 2003; Naderi et al., 2006; Rochette and Brash, 2008; Ryu et al., 2006; Ryu and Park, 2009). Therefore, the down-regulation of miRNAs from this cluster observed in SIPS could be interpreted as a mechanism that would contribute to the prevention of apoptotic death in senescent cells.
Five members of the miR-106b family (miR-17-5p, miR-18a, miR-20a, miR-106a, and miR-106b) showed decreased expression during stress-induced senescence in both HDF and HTM cells, and one (miR-20b) was significantly down-regulated in HDFs. With the exception of miR-18a, which has a seed region with a divergent nucleotide, the other five miRNAs of the miR-106b family down-regulated in senescent cells share identical seed regions that differ from the other members of this family that have seed regions upset by one nucleotide. An additional common feature of these miRNAs is that they are located in the three paralog miRNA polycistronic clusters, 17-5p-92a1, 106b-25, and 106a-363. These three miRNA clusters, located in different chromosomes (Chr 13, Chr 7, and Chr X, respectively), share a relatively high degree of similarity and appear to be involved in similar cellular functions such as control of the cell cycle and TGF-beta signaling (Petrocca et al., 2008). These clusters also include miR-192, one of the miRNAs down-regulated in SIPS that does not belong to either the miR-15 or miR-106b families.
The 17-5p-92a1, 106b-25, and 106a-363 clusters are considered oncogenic and have been found to be up-regulated in various types of cancers (Connolly et al., 2008; Fontana et al., 2008; Lu et al., 2007; Ventura et al., 2008). The oncogenic effects of these clusters are mediated at least in part by members of the miR-106b family and are known to promote cell cycle progression, and have been implicated in the development of a variety of cancers (Ivanovska and Cleary, 2008). At least three members of the miR-106b family (miR17-5p, miR-20a, and miR-106b) have been shown to promote cell cycle progression by targeting the mRNA of the cyclin-dependent kinase inhibitor p21CDKN1A (Fontana et al., 2008; Ivanovska and Cleary, 2008; Kim and Lim, 2009).
Since the specific mechanisms leading to the induction of p21CDKN1A in oxidative stress-induced SIPS are not completely clear and may be independent from its up-stream activator p53 (Zdanov et al., 2007), we hypothesized that the down-regulation of members of the miR-106b family may contribute to p21CDKN1A increase in expression in SIPS. Our results suggest that the down-regulation of miR-106a and potentially other members of the miR-106b family of miRNAs might indeed contribute to the induction of p21CDKN1A observed in stress-induced senescence. However, while the down-regulation of p21CDKN1A induced by miR-106a mimic was independent of p53, the up-regulation of this protein mediated by miR-106a antagomir was dependent on the presence of p53. These results suggest that, although the down-regulation of miR-106a observed in senescent cells may facilitate the increase in p21CDKN1A during the induction of SIPS, down-regulation of this miRNA alone is not sufficient to drive the increase in p21CDKN1A associated with SIPS. Similarly, miR-106a mimic increased cell proliferation but miR-106a antagomir did not have any significant effects. These observations may suggest the presence of redundant mechanism that compensate for the loss of miR-106a function. Such mechanisms could involve other members of the miR-106b family that are known to share similar targets, some of which (miR-17-5p, miR-18a, miR-20a, and miR-106b) also showed a decrease in expression associated with SIPS. Therefore, it is possible that the collective down-regulation of these miRNAs may exert effects that cannot be reproduced by the individual down-regulation of each one of them.
Only a small number of miRNAs showed consistent up-regulation during SIPS, and the information available about their biological functions is limited. The best characterized miRNA consistently up-regulated in HDF and HTM cells was miR-182. This miRNA is part of a paralogous cluster located in chromosome 7q32.2 that includes miR-96, miR-182, and miR-183. It has been suggested that the high expression of this cluster observed in retina and olfactory epithelia has been associated with a functional role in sensory tissues. Mir-182 is frequently amplified in melanoma and its over-expression has been shown to promote melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor (Segura et al., 2009). Our search for additional targets of miR-182 that contribute to some specific change in gene expression associated with SIPS and aging, led to the identification of RARG as a novel target of this miRNA. The specific down-regulation of RARG during cellular senescence could have some physiological implications in some tissues such as skin. Pharmacologic activation of retinoic acid receptors is currently the only effective method to treat photo-aging of the skin, and it has been shown that the beneficial action of retinoid is mediated through the RARG (Sakuta and Kanayama, 2006). Therefore, the observed decreased expression of RARG in senescent cells could potentially contribute to skin aging and to an age-related decline in the effectiveness of treatment with retinoid. Although increased expression of miR-182 may not be the only regulatory mechanism responsible for driving down the expression of RARG, our results are consistent with the concept that miRNAs could contribute to changes in gene expression in SIPS. The observation that miR-182 mimic resulted in an increase in SA-beta-galactosidase activity without a significant effect in cell proliferation suggests that the up-regulation of this miRNA may also be implicated in the acquisition of some of the characteristics of the senescent response such as the alterations of lysosomal function. More extensive analysis of the biological function(s) of miR-182 and a more complete identification of its target genes will be necessary to elucidate the relevance of the up-regulation of this miRNA in the acquisition of the senescent phenotype.
In conclusion, cellular senescence induced by oxidative stress was associated with sustained changes in expression of several miRNAs in both HDF and HTM cells. While some of these changes may be cell type specific, most were observed in both cell types. Although there is still relatively little information about the biological roles of most of these miRNAs, the down-regulation of members of the miR-15 and miR-106b families likely contributes to some of the features of senescent cells, such as the increased resistance to apoptosis and activation of p21CDKN1A. Similarly, the up-regulation of miRNAs, such as miR-182, may contribute to specific changes in gene expression associated with senescence, such as the down-regulation of RARG. Given the extent of the observed changes in miRNA expression, our results suggest that miRNAs might play an important role in modulating gene expression modifications involved in the acquisition and maintenance of the senescent phenotype.
Acknowledgements
We thank the Flow Cytometry Core Facility at the Duke Cancer Center. We acknowledge Dr. Anna Hong Bordelon for her carefully review the manuscript.
This work was supported by NEI EY01894, NEI EY016228, NEI EY019137, NEI EY05722, and Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A F, C J, H-J A. Properties of the SR Ca-ATPase in an Open Microsomal Membrane Preparation. Open Biochem J. 2008;2:91–99. doi: 10.2174/1874091X00802010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JS, Jang IS, Rhim JH, Kim K, Yeo EJ, Park SC. Gelsolin for senescence-associated resistance to apoptosis. Ann N Y Acad Sci. 2003;1010:493–495. doi: 10.1196/annals.1299.090. [DOI] [PubMed] [Google Scholar]
- Aikata H, Takaishi H, Kawakami Y, Takahashi S, Kitamoto M, Nakanishi T, Nakamura Y, Shimamoto F, Kajiyama G, Ide T. Telomere reduction in human liver tissues with age and chronic inflammation. Exp Cell Res. 2000;256:578–582. doi: 10.1006/excr.2000.4862. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlon C, Debacq-Chainiaux F, Hinrichs C, Scharffetter-Kochanek K, Toussaint O, Wlaschek M. The gene expression profile of psoralen plus UVA-induced premature senescence in skin fibroblasts resembles a combined DNA-damage and stress-induced cellular senescence response phenotype. Exp Gerontol. 2007;42:911–923. doi: 10.1016/j.exger.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, Vignali C, Calin GA, Negrini M, Croce CM, Degli Uberti EC. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol. 2007;210:370–377. doi: 10.1002/jcp.20832. [DOI] [PubMed] [Google Scholar]
- Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, Goldstein I, Madar S, Goldfinger N, Borresen-Dale AL, Ginsberg D, Harris CC, Pilpel Y, Oren M, Rotter V. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- Castro P, Giri D, Lamb D, Ittmann M. Cellular senescence in the pathogenesis of benign prostatic hyperplasia. Prostate. 2003;55:30–38. doi: 10.1002/pros.10204. [DOI] [PubMed] [Google Scholar]
- Castro P, Xia C, Gomez L, Lamb DJ, Ittmann M. Interleukin-8 expression is increased in senescent prostatic epithelial cells and promotes the development of benign prostatic hyperplasia. Prostate. 2004;60:153–159. doi: 10.1002/pros.20051. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiering SN, Roederer M, Nolan GP, Micklem DR, Parks DR, Herzenberg LA. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- Flanary BE, Sammons NW, Nguyen C, Walker D, Streit WJ. Evidence that aging and amyloid promote microglial cell senescence. Rejuvenation Res. 2007;10:61–74. doi: 10.1089/rej.2006.9096. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenmaile S, Pawelec G, Wagner W. A lack of telomeric non-reciprocal recombination (TENOR) may account for the premature proliferation blockade of Werner's syndrome fibroblasts. Biogerontology. 2003;4:253–273. doi: 10.1023/a:1026297926975. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Stat Appl Genet Mol Biol. 2003;2 doi: 10.2202/1544-6115.1008. Article3. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Cleary MA. Combinatorial microRNAs: working together to make a difference. Cell Cycle. 2008;7:3137–3142. doi: 10.4161/cc.7.20.6923. [DOI] [PubMed] [Google Scholar]
- Kaddar T, Rouault JP, Chien WW, Chebel A, Gadoux M, Salles G, Ffrench M, Magaud JP. Two new miR-16 targets: Caprin-1 and HMGA1, proteins implicated in cell proliferation. Biol Cell. 2009 doi: 10.1042/BC20080213. [DOI] [PubMed] [Google Scholar]
- Kim HS, Lim IK. Phosphorylated extracellular signal-regulated protein kinases 1 and 2 phosphorylate Sp1 on serine59 and regulates cellular senescence via transcription of p21Sdi1/Cip1/Waf1. J Biol Chem. 2009 doi: 10.1074/jbc.M808734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007a;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kang KW, Seu YB, Baek SH, Kim JR. Interferon-gamma induces cellular senescence through p53-dependent DNA damage signaling in human endothelial cells. Mech Ageing Dev. 2009;130:179–188. doi: 10.1016/j.mad.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Kim KS, Seu YB, Baek SH, Kim MJ, Kim KJ, Kim JH, Kim JR. Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol Biol Cell. 2007b;18:4543–4552. doi: 10.1091/mbc.E07-03-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Seki S, Kawakita N, Kuroki T, Monna T. Telomere shortening in chronic liver diseases. Biochem Biophys Res Commun. 1995;211:33–39. doi: 10.1006/bbrc.1995.1774. [DOI] [PubMed] [Google Scholar]
- Kojima T, Nakahama K, Yamamoto K, Uematsu H, Morita I. Age- and cell cycle-dependent changes in EPC-1/PEDF promoter activity in human diploid fibroblast-like (HDF) cells. Mol Cell Biochem. 2006;293:63–69. doi: 10.1007/s11010-006-2680-0. [DOI] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792:341–352. doi: 10.1016/j.bbadis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Lee YH, Govinda B, Kim JC, Kim TI, Lee NH, Lee JC, Yi HK, Jhee EC. Oxidative stress resistance through blocking Hsp60 translocation followed by SAPK/JNK inhibition in aged human diploid fibroblasts. Cell Biochem Funct. 2009;27:35–39. doi: 10.1002/cbf.1531. [DOI] [PubMed] [Google Scholar]
- Li G, Luna C, Liton PB, Navarro I, Epstein DL, Gonzalez P. Sustained stress response after oxidative stress in trabecular meshwork cells. Mol Vis. 2007;13:2282–2288. [PMC free article] [PubMed] [Google Scholar]
- Lim IK, Hong KW, Kwak IH, Yoon G, Park SC. Translocational inefficiency of intracellular proteins in senescence of human diploid fibroblasts. Ann N Y Acad Sci. 2001;928:176–181. doi: 10.1111/j.1749-6632.2001.tb05647.x. [DOI] [PubMed] [Google Scholar]
- Lim IK, Won Hong K, Kwak IH, Yoon G, Park SC. Cytoplasmic retention of p-Erk1/2 and nuclear accumulation of actin proteins during cellular senescence in human diploid fibroblasts. Mech Ageing Dev. 2000;119:113–130. doi: 10.1016/s0047-6374(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Liton PB, Luna C, Challa P, Epstein DL, Gonzalez P. Genome-wide expression profile of human trabecular meshwork cultured cells, nonglaucomatous and primary open angle glaucoma tissue. Mol Vis. 2006;12:774–790. [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I. Endothelial cell senescence in human atherosclerosis: role of telomere in endothelial dysfunction. Circulation. 2002;105:1541–1544. doi: 10.1161/01.cir.0000013836.85741.17. [DOI] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. Premature cellular senescence induced by pyocyanin, a redox-active Pseudomonas aeruginosa toxin. Free Radic Biol Med. 2006;41:1670–1677. doi: 10.1016/j.freeradbiomed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Naderi J, Lopez C, Pandey S. Chronically increased oxidative stress in fibroblasts from Alzheimer's disease patients causes early senescence and renders resistance to apoptosis by oxidative stress. Mech Ageing Dev. 2006;127:25–35. doi: 10.1016/j.mad.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Noyan-Ashraf MH, Sadeghinejad Z, Davies GF, Ross AR, Saucier D, Harkness TA, Juurlink BH. Phase 2 protein inducers in the diet promote healthier aging. J Gerontol A Biol Sci Med Sci. 2008;63:1168–1176. doi: 10.1093/gerona/63.11.1168. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Pitto L, Simili M, Mariani L, Riccardi L, Ciucci A, Rizzo M, Evangelista M, Mercatanti A, Pandolfi PP, Rainaldi G. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS ONE. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Ren JL, Pan JS, Lu YP, Sun P, Han J. Inflammatory signaling and cellular senescence. Cell Signal. 2009;21:378–383. doi: 10.1016/j.cellsig.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette PJ, Brash DE. Progressive apoptosis resistance prior to senescence and control by the anti-apoptotic protein BCL-xL. Mech Ageing Dev. 2008;129:207–214. doi: 10.1016/j.mad.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SJ, Cho KA, Oh YS, Park SC. Role of Src-specific phosphorylation site on focal adhesion kinase for senescence-associated apoptosis resistance. Apoptosis. 2006;11:303–313. doi: 10.1007/s10495-006-3978-9. [DOI] [PubMed] [Google Scholar]
- Ryu SJ, Park SC. Targeting major vault protein in senescence-associated apoptosis resistance. Expert Opin Ther Targets. 2009;13:479–484. doi: 10.1517/14728220902832705. [DOI] [PubMed] [Google Scholar]
- Sacca SC, Izzotti A. Oxidative stress and glaucoma: injury in the anterior segment of the eye. Prog Brain Res. 2008;173:385–407. doi: 10.1016/S0079-6123(08)01127-8. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Akiho M, Nakashima H, Akima H, Yasuhara M. Age-related reductions in expression of serum response factor and myocardin-related transcription factor A in mouse skeletal muscles. Biochim Biophys Acta. 2008;1782:453–461. doi: 10.1016/j.bbadis.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Sakuta T, Kanayama T. Marked improvement induced in photoaged skin of hairless mouse by ER36009, a novel RARgamma-specific retinoid, but not by ER35794,an RXR-selective agonist. Int J Dermatol. 2006;45:1288–1295. doi: 10.1111/j.1365-4632.2006.02913.x. [DOI] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LL, Alimirah F, Panchanathan R, Xin H, Choubey D. Expression of an IFN-inducible cellular senescence gene, IFI16, is up-regulated by p53. Mol Cancer Res. 2008;6:1732–1741. doi: 10.1158/1541-7786.MCR-08-0208. [DOI] [PubMed] [Google Scholar]
- Stein GH, Dulic V. Molecular mechanisms for the senescent cell cycle arrest. J Investig Dermatol Symp Proc. 1998;3:14–18. [PubMed] [Google Scholar]
- Sun BS, Dong QZ, Ye QH, Sun HJ, Jia HL, Zhu XQ, Liu DY, Chen J, Xue Q, Zhou HJ, Ren N, Qin LX. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 2008;48:1834–1842. doi: 10.1002/hep.22531. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E, Tomita Y, Brown LF, Kocher O, Dvorak HF. Differential expression of thymosin beta-10 by early passage and senescent vascular endothelium is modulated by VPF/VEGF: evidence for senescent endothelial cells in vivo at sites of atherosclerosis. FASEB J. 2001;15:458–466. doi: 10.1096/fj.00-0051com. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, Ho AD. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS ONE. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. MicroRNA, the putative molecular control for mid-life decline. Ageing Res Rev. 2007;6:1–11. doi: 10.1016/j.arr.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatelli MC, degli Uberti EC. MicroRNAs and possible role in pituitary adenoma. Semin Reprod Med. 2008;26:453–460. doi: 10.1055/s-0028-1096125. [DOI] [PubMed] [Google Scholar]
- Zdanov S, Debacq-Chainiaux F, Toussaint O. Knocking down p53 with siRNA does not affect the overexpression of p21WAF-1 after exposure of IMR-90 hTERT fibroblasts to a sublethal concentration of H2O2 leading to premature senescence. Ann N Y Acad Sci. 2007;1100:316–322. doi: 10.1196/annals.1395.034. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B cell differentiation. Blood. 2009 doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]