Abstract
Silica nanoparticle bioreactors featuring thin films of enzymes and polyions were utilized in a novel high-throughput 96-well plate format for drug metabolism profiling. The utility of the approach was illustrated by investigating the metabolism of the drugs diclofenac (DCF), troglitazone (TGZ) and raloxifene, for which we observed known metabolic oxidation and bioconjugation pathways and turnover rates. A broad range of enzymes was included by utilizing human liver (HLM), rat liver (RLM) and bicistronic human-cyt P450 3A4 (bicis.-3A4) microsomes as enzyme sources. This parallel approach significantly shortens sample preparation steps compared to an earlier manual processing with nanoparticle bioreactors, allowing a range of significant enzyme reactions to be processed simultaneously. Enzyme turnover rates using the microsomal bioreactors were 2-3 fold larger compared to using conventional microsomal dispersions, most likely because of better accessibility of the enzymes. Ketoconazole (KET) and quinidine (QIN), substrates specific to cyt P450 3A enzymes, were used to demonstrate applicability to establish potentially toxic drug-drug interactions involving enzyme inhibition and acceleration.
INTRODUCTION
Rapidly and accurately predicting in vivo metabolism, pharmacokinetics and toxicity of drug candidates early in the development process is a major challenge in drug discovery. This fact has driven the development of high-throughput in vitro bioanalytical methodologies for drug metabolism and pharmacokinetic studies (DMPK).1 These methods must address all important metabolic enzymes including cytochromes (cyt) P450, uridine diphosphoglucuronosyl transferase (UGT) and gluathione S-transferase (GST), which together are responsible for nearly 80% of the metabolism of currently marketed drugs.2,3 Liver microsomes are an enriched source of these metabolic enzymes, and are widely used for in vitro metabolism and toxicity studies.2,4
Sample preparation and workup is a bottleneck in drug metabolism studies. Although many approaches have been developed in the past decade to shorten and/or multiplex these steps, they remain the rate limiting steps in drug discovery.5 Significant progress has been made in integrating DMPK studies into robotic, automated 96 or 384-well plate assays.6-8 These automated systems are relatively expensive, and require sophisticated instrumentation and high maintenance. The 384-well plate format is a promising technology providing good efficiency,6 but progress is limited by concerns of cross-contamination, lack of appropriate tools for 384-sample multiplexing, sample volume and sensitivity.6 Although of lower throughput, the 96 well-plate assay systems minimize many of the pitfalls of 384 well plate systems and are well established and routinely used in metabolism studies. These 96-well plate drug metabolism assays are generally performed with microsomal enzymes dispersed in 100-500 μL of solution, with significant cyt P450 enzyme consumption. Since a large number of assays are needed for comprehensive drug metabolism profiling, current high-throughput in vitro approaches to evaluate metabolic properties with microsomal dispersions are relatively expensive, especially when single enzyme (bicistronic) cyt P450 microsomes are employed.9 Additionally, understanding structure-metabolism relationships and determining enzymes responsible for metabolism is a particularly difficult task because of relatively wide and overlapping substrate specificities of cyt P450s.10 In existing high-throughput systems utilizing reactions in solution with microsomal dispersions coupled with LC-MS, the elucidation of metabolic pathways involves advanced separation and structural elucidation techniques and is highly labor-intensive.11 This complexity can extend analysis time and limit analytical performance. Thus, there is considerable need for new, faster, lower cost methodologies for in vitro metabolism studies that simplify sample workup while still providing high quality kinetic and structural data.
We recently developed enzyme films on nanoparticles as bioreactors12 to generate samples for LC-MS to investigate metabolism, genotoxicity and enzyme inhibition. 13-15 This approach provides rates of metabolite formation and structures of the metabolic products. Microsomes can be used as sources of metabolic enzymes on the nanoparticles and offer distinct advantages including insignificant number of metabolic enzymes, good stability, longer storage time, and rapid separation of reaction products from microsomal enzymes. Microsomes eliminate the need for pure enzymes while providing the ability to recover and reuse the microsomal enzymes.13b,c,d While our earlier use of microsome-nanoparticle bioreactors featured manual processing of single sample, the features described above facilitate integration into a high-throughput, low cost system.
Herein we describe the utilization of microsome-coated silica nanoparticles in a well plate format (Scheme 1) for low cost, semi-automated, high throughput drug metabolite profiling in combination with LC-MS/MS. The nanoparticle bioreactors are dispensed into the 96-well filtration plate whose wells also serve as reaction chambers, allowing up to 96 reactions (or more if a larger plate is used) to be processed simultaneously in relatively short times. Consequently this approach facilitates rapid sample preparation in a single step, followed by simultaneous filtration and transfer to an autosampler for LC-MS/MS analysis. With microsomes on the nanoparticles, separation of the enzymes in the filtration step leads to a clean sample for simpler, faster and more sensitive LC-MS analysis. To address substrate and enzyme specificity, we included an individual cyt P450 enzyme system in a bicistronic microsome to study single enzyme metabolite routes in addition to inhibition and induction studies.
Scheme 1.
Features of the bioanalytical system for in vitro metabolic profiling: (A) Bioreactor assembly: a layer of the cationic polymer polydiallyldimethylammonium chloride (PDDA) is initially deposited on silica nanoparticles, followed by a layer of oppositely charge microsomes; (B) Reaction/filtration 96 well plate equipped with 10,000 Da cutoff mass filters showing the LC-MS ready sample collection plate underneath; (C) Schematic illustration of simultaneous enzyme reaction design using 96 well plate; and (D) LC-MS/MS analysis with an autosampler.
For proof of concept, diclofenac (DCF), troglitazone (TGZ), and raloxifene were used as model drugs to demonstrate the capabilities of this approach to simultaneously elucidate different in vitro oxidation and conjugation reactions together with enzyme turnover rates. To demonstrate the versatility of the bioreactors we also studied the major glucuronidation pathway of raloxifene. In addition, we demonstrated the applicability to drug-drug interactions (DDI) in which two drugs act upon a single metabolic enzyme, by utilizing an inhibitor and an activator of cyt P450 3A4.
EXPERIMENTAL
Reagents and Materials
Bicistronic human cytochrome P450 3A4 (bicis.-3A4) were expressed in transformed DH5R Escherichia coli following established protocols.16 Rat liver microsomes (RLM), human liver microsomes (HLM) and human UGT1A10 supersomes (containing UGT1A10 isozymes, 5.0 mg mL−1 in 0.1 M Tris buffer, pH 7.5) were from BD Gentest. Diclofenac, caffeine, troglitazone, raloxifene, glutathione (GSH), nicotinamide adenine dinucleotide phosphate reduced (NADPH), uridine 5’-diphosphoglucuronic acid triammonium salt (UDPGA), polydiallyldimethylammonium chloride (PDDA, MW <200 000) and all other chemicals were from Sigma. Silica nanoparticles were from Polyscience, Inc (500±50 nm dia., ~10% solids, d = 1.96 g cm−3) and used after redispersing by agitation. The AcroPrep™ 96 filter plate (Omega 10K) was from Pall Corporation.
Bioreactor Film Fabrication
A previously established protocol was followed to coat the nanoparticles with microsomes.12 Briefly, silica nanospheres (4.0 mL in water) were dispersed in 2 mg mL−1 PDDA for 20 min, then centrifuged and washed. The silica-PDDA nanospheres were then dispersed in microsome dispersion (500 μL, 0.5 mg mL−1 in potassium phosphate buffer), allowed 30-min for adsorption followed by centrifugation and washing (3x). The final architecture was Silica-PDDA/microsome and these will be referred as microsome-bioreactors (HLM-, RLM-, and bicis.3A4-bioreactors).
Reaction Conditions
(1) Diclofenac and Troglitazone Metabolism. HLM-bioreactor particles were re-dispersed in 50 mM potassium phosphate buffer pH 7.4, then 117 μL were pipetted into each well of the AcroPrep™ 96-well filter plates. Troglitazone (6.5 μL, 2 mM) and diclofenac (6.5 μL, 2.0 mM) were added to selected wells to a final concentration of 50 μM. For drug-drug interaction studies, ketoconazole (1.5 μL, 20 mM) and quinidine (3.0 μL, 10 mM) were added to selected wells to a final concentration of 0.1 mM. NADPH (125 μL, 2.0 mM) was added to the wells to initiate oxidation reactions. Reactions were stopped by adding 15 μL cold acetonitrile with 6%(v/v) formic acid. Controls were done in the absence of HLM or NADPH. Caffeine (10 μM final concentration) was added as an internal standard before LC-MS analysis. Similar protocols were followed for experiments using RLM- and bicis.-3A4-bioreactors. Microsome dispersion experiments were done using the same protocols, using the same total amount of microsomes as on the particles.
(2) Raloxifene Metabolism. Enzyme reactions were started by addition of co-factors, i.e. GSH or UDPGA, into appropriate wells containing 200 μL PDDA/microsome nanoparticles dispersed in 10 mM potassium phosphate buffer (pH 7.0) and 50 μM raloxifene. GST or UGT mediated reactions were conducted in the presence of 0.5 mM GSH and 0.5 mg mL−1 NADPH and 1.5 mM UDPGA and 20 mM MgCl2, respectively. Silica nanospheres coated with UGT1A10 supersomes were prepared and reactions were conducted following a similar protocol, and stopped as described above. Control incubations contained the same amount of raloxifene but lacked co-factors.
LC-MS
Capillary (cap)LC-MS analysis was done after the filtration of the reaction mixture from the 96-well plate into a 96-well collection plate. Samples were manually loaded onto the capLC autosampler. Capillary LC (Waters) with analytical (150 mm, 300 μm I.D., 5 μm particle size) and trapping columns (23.5mm) was used following a similar procedure as reported earlier (See supporting information).17 The CapLC in tandem with MS were programmed to automatically analyze all the loaded samples. A Q-TRAP 4000 Applied Biosystems (Foster City, CA) mass spectrometer was used in-line with capLC (see supporting information).
RESULTS
Diclofenac (DCF) is a nonsteroidal anti-inflammatory drug that is widely used for the treatment of rheumatoid arthritis, osteoarthritis and acute muscle pain.18 In rare cases, diclofenac may cause potentially severe hepatotoxicity, associated with the formation of reactive metabolites.19 Cyt P450 3A and 2C9 are responsible for catalyzing the biotransformation of diclofenac into mono-hydroxylated metabolites 5’-hydroxydiclofenac and 4’-hydroxydiclofenac (Scheme 2a).20
Scheme 2.
Major metabolic pathways of (a) Diclofenac, its major metabolites 5’-hydroxydiclofenac and 4’-hydroxydiclofenac, and minor metabolite diclofenac-2,5-quinone imine. (b) Troglitazone and its major metabolite troglitazone quinone. c) Raloxifene and its glucuronide metabolites and the glutathione adducts following reactive intermediate formation under cyt P450 metabolism.
Troglitazone (TGZ) is an antidiabetic drug that decreases tissue resistance to insulin by acting as antagonist of peroxisome proliferator-activated receptor γ (PPARγ).21,22 In human and rat hepatocytes, oxidation of TGZ to TGZ-quinone is metabolized by cyt P450 3A and 2C8 isozymes (Scheme 2b) in a dose-dependent fashion.23
Raloxifene is a second-generation selective estrogen receptor modulator used to treat osteoporosis and to prevent breast cancer.24 The major metabolic pathway of raloxifene is glucuronidation, resulting in 4’-R-glucuronide (4-R-G) and 6-R-glucuronide (6-R-G).25 Alternatively, it can undergo cyt P450 3A4 mediated metabolic activation, leading to several reactive intermediates, observed using trapping reagents such as glutathione (GSH) or N-acetylcysteine (NAC).26 GSH adducts of raloxifene were identified in incubations with human liver microsomes. Substitution with GSH occurred at the 5- or 7-position of the benzothiophene moiety or at the 3’-position of the phenol ring (Scheme 2c).26
The 96-well plate system was used to monitor metabolic oxidation of diclofenac, troglitazone and bioconjugation of raloxifene. A typical experimental plan is illustrated in Scheme 1, designed to monitor the metabolism and metabolite formation rates of one drug at a time using all 3 microsomal bioreactors (HLM, RLM, bicis.-3A4). This experiment is designed to simultaneously compare rat and human metabolic enzymes, as well as to monitor activity of cyt P450 3A4, a known major metabolic enzyme for these drugs.20,23,26 In the same plate, we monitored enzyme inhibition by ketoconazole and activation by quinidine in the metabolism of the drug. This experimental arrangement enabled rapid completion of 96 reactions for a single drug. Once the reactions were completed, samples were filtered from the filtration plate and collected for automated LC-MS/MS, leaving the bioreactor nanoparticles behind. Parallel experiments with the same amount of microsomes in dispersions were done for comparison.
Metabolite Profiling
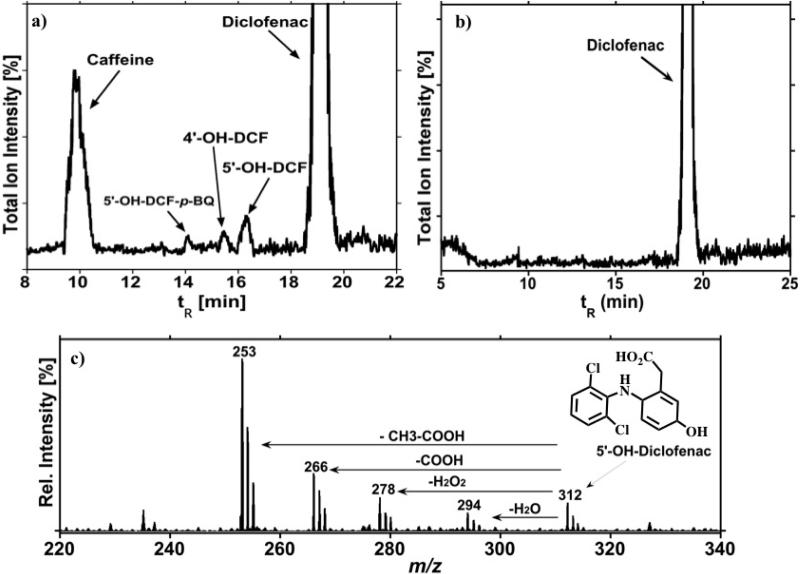
The total ion chromatogram in Figure 1 illustrates results for diclofenac (DFC) reacted with microsomal bioreactors activated by NADPH. When exposed to HLM or RLM, DCF is primarily hydroxylated to 5’-OH-DCF by cyt P450 3A and 4’-OH-DCF by cyt P450 2C9 (Scheme 2a).27 As illustrated in Figure 1, we detected both those metabolites. Diclofenac-2,5-quinone imine was also detected (DCF-2,5-QI, tR=14min) as a degradation product of 5’-OH-DCF. Results are in agreement with previous results on metabolic reactions of DFC catalyzed by human liver microsomes.28 When DCF was incubated with bicistronic cyt P450 3A4-bioreactors, 5’-OH-DCF and DCF-2,5-QI were detected (Scheme 2a.) This bioreactor permits direct monitoring of DCF metabolism by cyt P450 3A4 without the interference of other microsomal enzymes. The collision induced dissociation (CID) spectra were obtained to further confirm the structure of the metabolites (Figure 1c.). In the positive electrospray ionization (ESI+) mode, the product ion profile of the protonated molecule after CID, m/z 312, derived from the sequential loss of H2O (m/z 294), or cleavage of H2O2 (m/z 278). The predominant ion m/z 253 is generated from loss of CH3-COOH. The fragment m/z 266 is generated from loss of COOH. The 4’-OH-DCF metabolite generated from metabolism by CYP2C enzymes gave similar fragmentation (Supporting information, Figure S1).
Figure 1.
Mass Spectrometry Chromatogram in ESI+ ion mode illustrating: (a) metabolites of DCF generated after 5 min. incubation with microsome-bioreactors and NADPH; (b) Control incubation in absence of bioreactors; and (c) CID Spectra for 5’-OH-DCF.
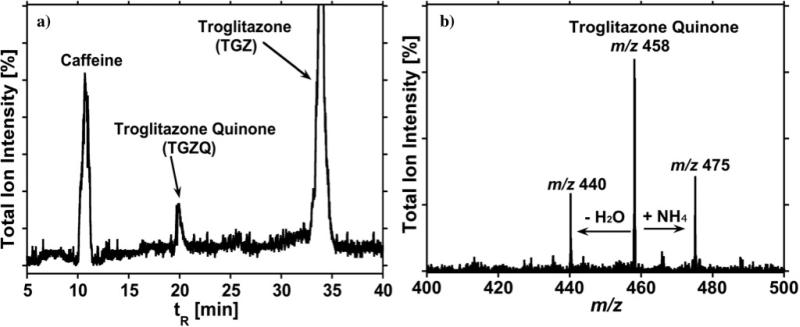
The metabolism of troglitazone (TGZ) was investigated using HLM-, RLM-, and bicistronic cyt P450 3A4-bioreactors. It was reported that troglitazone quinone (TGZQ) is the major metabolite formed by cyt P450 3A4 and 2C8 enzymes.21,29 Results of the TGZ reaction with HLM–bioreactors are shown in Figure 2. The formation of TGZQ was confirmed and the CID spectrum in Figure 2b is consistent with the structure of TGZQ, showing ion-current peaks at m/z 475 ([M+NH4]+) and m/z 458 [M]+ (where M = TGZQ) correspond to the metabolite at tR = 20 min from the bioactivation of TGZ by liver microsomes.30
Figure 2.
Mass Spectrometry Chromatogram in TIC mode illustrating: (a) Troglitazone metabolite generated by cyt. P450 3A4 and 2C8 present in the microsomes and NADPH, 5 min. incubation; and (b) Spectra for troglitazone quinone with tR = 20 min.
The fragment ion with m/z 440 in Figure 2b represents loss of water from the metabolite and the ion current peak at m/z 475 represents addition of NH4 from the ammonium acetate buffer used in the mobile phase. CID of TGZQ, m/z 458, in addition to m/z 440, produced a predominant fragment ion at m/z 222 and also ions at m/z 235, m/z 218 (corresponding to loss of -OH from m/z 235), m/z 149 (corresponding to loss of -C5H10O from m/z 235). The MS chromatogram in Figure 3 was obtained by single reaction monitoring (SRM) of the characteristic transition of the ion with m/z 458 to m/z 222, and confirms the presence of TGZQ.
Figure 3.
Mass Chromatogram in SRM mode confirming the presence of TGZQ by monitoring at the transition from m/z 458 to m/z 222.
Reaction rate studies illustrated in Figure 4 show that the generation of metabolites increased as a function of time for all systems. Metabolite formation rates (Supporting information Table S1) were estimated for all three drugs from these data. For comparison, a solution study was done using HLM microsomal dispersions, but no nanoparticles for diclofenac and raloxifene to obtain the metabolite formation rates. Results showed that metabolite formation rates were 2- to 3-fold smaller in solution reaction compared to the microsome-bioreactors containing the same amount of microsomes (Figure 4, Table S1).
Figure 4.
Influence of reaction time expressed as the ratio of the area of the metabolite peak vs. the internal standard caffeine in the LC chromatogram. for: (a) 5’-OH-DCF, by RLM-( ), HLM-(
), HLM-( ), bicis.-3A4-(
), bicis.-3A4-( )-bioreactors and HLM dispersed in solution (
)-bioreactors and HLM dispersed in solution ( ); (b) TGZQ by RLM-, HLM-, bicis.-3A4-bioreactors; (c) Bioactivation of raloxifene-GSH adduct via HLM-(
); (b) TGZQ by RLM-, HLM-, bicis.-3A4-bioreactors; (c) Bioactivation of raloxifene-GSH adduct via HLM-( )-bioreactors d) Monitor the formation of 6-R-G by HLM-(
)-bioreactors d) Monitor the formation of 6-R-G by HLM-( )-bioreactors, 6-R-G by HLM dispersed in solution (
)-bioreactors, 6-R-G by HLM dispersed in solution ( ); and 4-R-G by HLM-(
); and 4-R-G by HLM-( )-bioreactors & 4-R-G by HLM dispersed in solution (
)-bioreactors & 4-R-G by HLM dispersed in solution ( ).
).
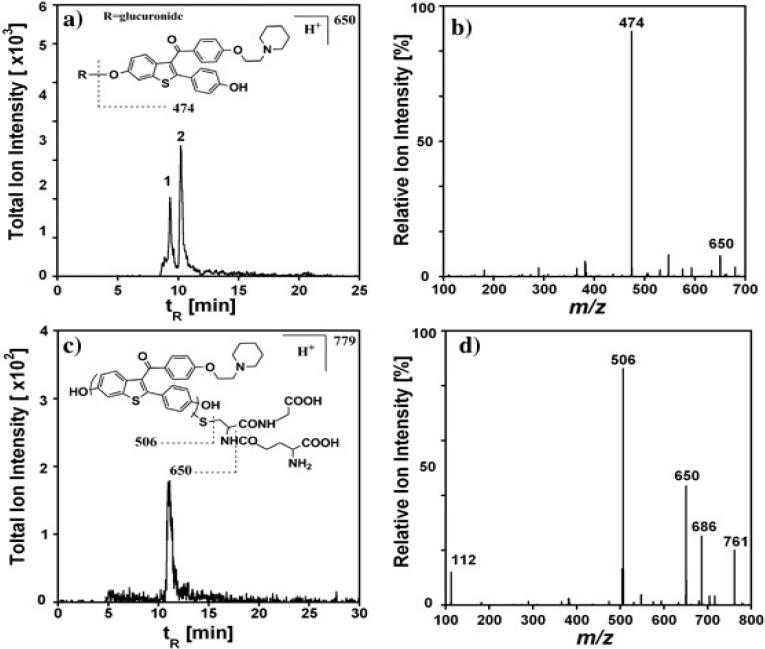
The glucuronidation pathway of raloxifene was activated by incubating HLM-bioreactors together with the cofactor UDP-glucuronic acid. Two glucuronic acid conjugates, i.e. 4-R-G and 6-R-G (see Scheme 2c), were identified using SRM by monitoring mass transition from singly charged raloxifene glucuronide (m/z 650) to raloxifene (m/z 474) (Figure 5a). The characteristic fragmentation pattern is illustrated in the inset of Figure 5a and the mass spectrum in Figure 5b. The assignment of 4-R-G was made by matching the retention time with the product peak when raloxifene was incubated with UGT1A10 supersome bioreactors (Supporting information, Figure S2). Based on previous reports that isozyme UGT1A10 catalyzes only the formation of 4-R-G.25 The relative formation rates for raloxifene glucuronides are shown in Figure 4d.
Figure 5.
Analysis of raloxifene reaction with HLM-bioreactors in the presence of GSH, 5 min. incubation: (a) SRM chromatogram of raloxifene-glucuronides with mass transition m/z 650 to 474, in which peak 1 and 2 represent 6-R-G and 4-R-G, respectively. (b) CID chromatogram of parent ion m/z 650. (c) SRM chromatogram of raloxifene-glutathione conjugate with mass transition m/z 779 to 650. (d) Product ion chromatogram of parent ion m/z 779.
Another conjugation pathway of raloxifene via cyt P450s activation followed by glutathione adduct formation was also investigated. The formation of raloxifene-glutathione adducts were identified by LC-MS with [M+H]+ m/z 779, when raloxifene reacted with HLM-bioreactors in the presence of NADPH and GSH. Subsequent CID of the parent ion m/z 779 produced fragments at m/z 506 and 650, corresponding to cleavage adjacent to the thioether moiety and the loss of pyroglutamate, respectively (Figure 5d), further confirming the formation of raloxifene-glutathione conjugates. Detailed fragmentation is illustrated in the inset of Figure 5c. The major GSH adduct observed here is likely to be 7-glutathionyl derivative based on previous studies,26 although we cannot exclude the possibility of other minor adducts. Similar relative formation rates of raloxifene-glutathione adducts were obtained, as shown in Figure 4c.
Drug-Drug Interactions: Enzyme inhibition and activation
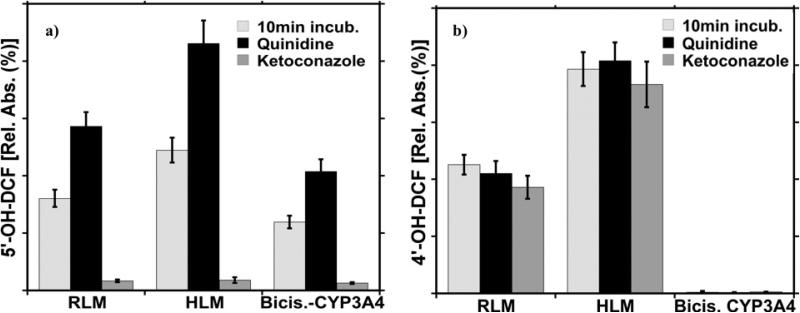
Microsome-bioreactors were incubated with either DCF or TGZ in the presence of KET or QIN. In the presence of KET, the amount of metabolite 5’-OH-DCF was greatly decreased compared to results without the inhibitor, indicating decreased cyt P450 3A activity (Figure 6a). Alternatively, in the presence of QIN the amount of the generated metabolites was elevated, suggesting an increase in cyt P450 3A catalytic activity. However, as illustrated in Figure 6b, neither KET nor QIN exerted any effect on the formation of 4’-OH-DCF, a DCF metabolite from cyt P450 2C9 metabolism.
Figure 6.
The effect of the KET and QIN on the metabolism of DCF by: a) Cyt P450 3A and b) Cyt P450 2C9 present in microsome bioreactors. Data collected after 10 min. incubation of bioreactors with DCF alone and DCF with KET or QIN, in the presence of NADPH.
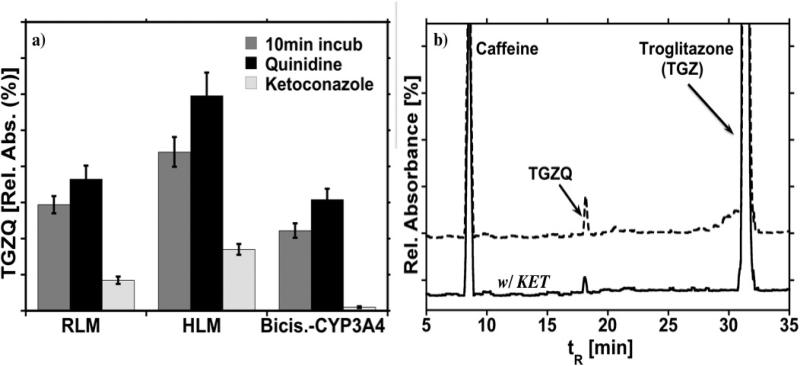
The effect of KET and QIN on the TGZ metabolism was also investigated following the same approach as for DCF. The data in Figure 7 show similar trends as DCF for inhibiting and activating the cyt P450 3A enzymes. TGZ is metabolized by both cyt P450 3A and 2C8 enzymes and KET only inhibits the catalytic activity of the cyt P450 3A enzyme. Cyt P450 2C8 activity is not influenced. Hence, TGZQ is still being generated. As discussed above, in the presence of QIN the activity of Cyt P450 3A enzymes is elevated, thus more metabolite is generated when compared to TGZ metabolism in the absence of QIN.
Figure 7.
Inhibition and acceleration of enzymes: (a) The effect of KET and QIN on the TGZ metabolism by Cyt P450 3A present in microsome-bioreactors. Data collected after 10 min. incubation in the presence of NADPH and QIN or KET. (b) CapLC chromatograms illustrating the metabolites formed with exposure of TGZ to HLM-bioreactors in the presence of NAPDH with and without KET.
DISCUSSION
Results above demonstrate the feasibility of microsome-coated bioreactors integrated into a high throughout array system coupled with automated LC-MS (Scheme 1) for rapid multi-enzyme metabolic profiling of drugs. Silica nanoparticles coated with human or rat liver microsomes, or bicistronic cyt P450 3A4 provide high local enzyme concentrations to ensure rapid metabolite formation via relevant oxidation and conjugation pathways. The use of a well plate platform enabled 96 reactions to be conducted simultaneously with fast filtration and transfer to autosampler vials for automated LC-MS/MS. The approach significantly decreases the sample complexity and work up, providing cleaner samples by retaining the microsomes on the particles. These samples facilitated faster and better chromatographic separations and more sensitive MS analysis, eliminating the concern for suppressed ionization. LC-MS enhances the bioreactor approach by providing high sensitivity while requiring a small sample size, and provides metabolite structure elucidation and determination of unknown entities. In addition, the methodology reported here is amenable to further automation via robotics, e.g. to handle deposition of microsome bioreactors and reaction solutions into the wells.
The ability of this approach to concurrently develop drug metabolite profiles, identify enzymes responsible for metabolism, measure relative metabolite formation rates and screen for potential drug-drug interactions was illustrated by investigating the metabolism of 3 drugs using 3 microsomal enzyme sources. The 96-well plate format significantly shortens the time-consuming and labor-intensive sample workup while generating higher metabolite turnover rates (Supporting information, Table S1) when compared to microsome dispersions where microsomes were not fixed on silica nanoparticles (Figure 4). When the enzyme-rich microsomes are coated onto a solid support, the available enzyme concentration in the reaction volume is increased, with the net result of higher enzyme turnover rates in shorter incubation times.31
Multiple metabolic pathways such as 5’-hydroxylation and 4’-hydroxylation were confirmed for DCF (Figure 1a), in agreement with previous reports.27 Subsequent degradation of 5’-OH-DCF into diclofenac-2,5-quinone was also detected (Scheme 2a), illustrating the ability of this method to detect major as well as minor metabolites. TGZQ metabolite was detected using both RLM and HLM-bioreactors (Figures 2 and 3). Capacities for mass measurements of parent and fragment ions and signature fragmentation patterns greatly facilitate the successful metabolic profiling.
The capability of the method to elucidate bioconjugation pathways was demonstrated by confirming glucuronidation and glutathionylation pathways of raloxifene. The two major raloxifene glucuronides, 4-G-R and 6-G-R (Scheme 2c), 25 were detected when raloxifene was catalyzed by UGT enzymes present in HLM-bioreactors. The formation of raloxifene-glutathione conjugates (Figure 5c and 5d) illustrated the capability of microsome-bioreactors in catalyzing glutathionylation pathway, and shed light on activation by cyt P450s as well, because of the function of glutathione as a trapping reagent.26
The exploitation of bicistronic cyt P450 3A4 and UGT1A10 supersome bioreactors assists enzyme-specific metabolic pathway studies. Thus, contributions of cyt P450 3A4 to DCF and TGZ metabolism, and of UGT1A10 to raloxifene metabolism were assessed by generating the single metabolites 5’-OH-DCF, TGZQ and 4-R-G. These approaches also aided in identifying metabolites with same m/z and similar fragmentation patterns in the absence of authentic standards, i.e. 5’-OH-DCF and 4-R-G.
As illustrated in CID spectra for 5’-OH-DCF (Figure 1c), 5’-OH-DCF affords good ionization efficiencies and yielded characteristic spectra rich in fragment ions. The fragmentation pattern proves the presence of the metabolite. Similar CID experiments were conducted to confirm the presence of 4’-OH-DCF (Supporting information, Figure S1).
In many instances more than one enzyme can be involved in the metabolism of a single compound and many times these multiple enzymatic pathways generate the same metabolite. It has been reported that cyt P450 3A enzymes play a major role in the bioactivation of troglitazone,32 however, cyt P450 2C8 enzymes are also involved in the hydroxylation of troglitazone to troglitazone quinone (Scheme 2b).33 In drug-drug interaction studies it is important to know the exact contribution of each enzyme in the metabolism of each drug. The formation of TGZQ was detected in reactions catalyzed by all three types of bioreactors (Figures 2 and 3). The ESI-MS spectra, illustrated in Fig. 2b, with m/z 458 [M+H]+ and m/z 475 [M+NH4]+ is representative of the TGZQ metabolite structure shown in Scheme 2b. The SRM chromatogram illustrated in Figure 3 was generated by monitoring the transition from m/z 458 to m/z 222, corresponding to a loss of m/z 235 (-C14H19O3). The loss of m/z of 235, assigned to 1,4-benzoquinone-ring is a characteristic TGZQ fragment generated from CID.34
KET and QIN, cyt P450 3A specific substrates, were utilized to demonstrate the ability of this approach to study drug-drug interactions. KET a potent specific cyt P450 3A inhibitor, exhibited a significant inhibitory effect on the metabolism of both TGZ to TGZQ and DCF to 5’-OH-DCF (Figures 6 and 7). KET did not inhibit the activity of cyt P450 2C8 or cyt P450 2C9 (Figures 6b and 7). As illustrated in Figure 6b the 4’-OH-DCF metabolite was still produced. Additionally, TGZ was metabolized to some extent by cyt P450 2C8, thus the peak corresponding to TGZQ was still present in the capLC chromatogram (Figure 7b). Many reports suggest that QIN stimulates the metabolism of several cyt P450 3A4-dependent substrates, including 5’-OH-DCF.35 The results presented in Figures 6 and 7, show that TGZ and DCF metabolites are elevated in the presence of QIN compared to reactions without QIN. These results clearly indicate that our semi-automated bioreactor approach can be used to identify and evaluate potential drug-drug interactions and their effect (inhibitory or activating) on cyt P450 3A4 enzymes when more then one drug is administered.
Additionally, as illustrated in Figure 4 and Table S1, this proposed approach can be used to set up time dependent studies to determine the relative enzyme turnover rates, an important factor in drug metabolism and pharmacokinetic studies. Microsome-bioreactors fabricated on silica nanoparticles provided ~2-3 larger metabolite turnover rates when compared microsome dispersions alone.
In summary, we have demonstrated a novel, cost-effective, high throughput approach featuring multi-well plates and nanoparticle-based microsomal bioreactors followed by automated LC-MS/MS that provides a bioanalytical tool for rapid metabolite profiling. The integration of microsome-bioreactors in a multi-well high throughput format eliminates complex mixtures while providing significantly shorter sample preparation steps and cleaner samples for LC-MS. The applicability and versatility of this approach was illustrated by concurrently studying in vitro oxidation and conjugation reactions, enzyme specific metabolism and drug-drug interactions, identifying reactive metabolites and elucidating metabolic pathways. We envision that this approach can be combined with current bioanalytical and microbiological assays to provide a more comprehensive assessment of drug metabolism. The system provides a significant step towards the full automation of the enzyme-bioreactor approach for in vitro metabolic profiling.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported financially by US PHS grant No. ES03154 from the National Institute of Environmental Health Sciences (NIEHS), NIH, USA. The authors thank Dr. Eli G. Hvastkovs for providing the bisistronic-CYP3A4 microsomes. JFR is grateful to Science Foundation Ireland for a Walton Research Fellowship.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Experimental LC separation gradients and mass spectrometer operating parameters are described in detail. Two additional figures for UGT1A10 catalyzed reaction, diclofenac metabolism and relative metabolite formation rates. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Kramer JA, Sagartz JE, Morris DL. Nat. Rev. Drug Disc. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 2.Wienkers LC, Heath TG. Nat. Rev. Drug Disc. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 3.Williams AJ, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup RJ, Ball SE. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Samuel K, Subramanian R, Braun PM, Stearns AR, Chiu S-HL, Evans CD, Baillie AT. J. Pharmacol. Exp. Ther. 2002;303:969–978. doi: 10.1124/jpet.102.038992. [DOI] [PubMed] [Google Scholar]
- 5.Yu S, Crawford E, Tice J, Musselman B, Wu J-T. Anal. Chem. 2009;81:193–202. doi: 10.1021/ac801734t. [DOI] [PubMed] [Google Scholar]
- 6.Carlson TJ, Fisher MB. Comb. Chem. High Throughput Screening. 2008;11:258–264. doi: 10.2174/138620708783877717. [DOI] [PubMed] [Google Scholar]
- 7.Chang M, Kim EJ, El-Shourbagy TA. Rapid Commun. Mass Spectrom. 2007;21:64–72. doi: 10.1002/rcm.2808. [DOI] [PubMed] [Google Scholar]
- 8.a Chauret N, Tremblay N, Lackman RL, Gauthier J-Y, Silva JM, Marois J, Yergey JA, Nicoll-Griffith DA. Anal. Biochem. 1999;276:215–226. doi: 10.1006/abio.1999.4348. [DOI] [PubMed] [Google Scholar]; b Jenkins KM, Angeles R, Quintos MT, Xu R, Kassel DB. J. Pharm. Biomed. Anal. 2004;34:989–1004. doi: 10.1016/j.jpba.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Nicoli R, Curcio R, Rudaz S, Veuthey J-V. J. Med. Chem. 2009;52:2192–2195. doi: 10.1021/jm900201b. [DOI] [PubMed] [Google Scholar]
- 10.Rendic S. Drug Metab. Rev. 2002;34:83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- 11.a Zhang NY, Fountain ST, Bi HG, Rossi DT. Anal. Chem. 2000;72:800–806. doi: 10.1021/ac9911701. [DOI] [PubMed] [Google Scholar]; b Trunzer M, Faller B, Zimmerlin A. J. Med. Chem. 2009;52:329–335. doi: 10.1021/jm8008663. [DOI] [PubMed] [Google Scholar]
- 12.Bajrami B, Hvastkovs EG, Jensen GC, Schenkman JB, Rusling JF. Anal. Chem. 2008;80:922–932. doi: 10.1021/ac702025f. [DOI] [PubMed] [Google Scholar]
- 13.a Hvastkovs EG, So M, Krishnan S, Bajrami B, Tarun M, Jansson I, Schenkman JB, Rusling JF. Anal. Chem. 2007;79:1897–1906. doi: 10.1021/ac061975q. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Krishnan S, Bajrami B, Hvastkovs EG, Choudhary D, Schenkman JB, Rusling JF. Anal. Chem. 2008;80:5279–5285. doi: 10.1021/ac800763r. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Zhao L, Krishnan S, Zhang Y, Schenkman JB, Rusling JF. Chem. Res. Toxicol. 2009;22:341–347. doi: 10.1021/tx8004295. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Rusling JF, Hvastkovs EJ, Schenkman JB. In: Drug Metabolism Handbook. Nassar A, Hollenburg PF, Scatina J, editors. J. Wiley; N. J.: 2009. pp. 307–340. [Google Scholar]
- 14.Bajrami B, Krishnan S, Rusling JF. Drug Met. Let. 2008;2:158–162. doi: 10.2174/187231208785425854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull D, Bajrami B, Jansson I, Schenkman J, Rulsing JF. Anal. Chem. 2009;81:716–724. doi: 10.1021/ac802179s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillam EMJ, Guo ZY, Guengerich FP. Arch. Biochem. Biophys. 1994;312:59–66. doi: 10.1006/abbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 17.Tarun M, Bajrami B, Rusling JF. Anal. Chem. 2006;78:624–627. doi: 10.1021/ac0517996. [DOI] [PubMed] [Google Scholar]
- 18.Small RE. Clin. Pharm. 1989;8:545–558. [PubMed] [Google Scholar]
- 19.Tang W, Stearns RA, Bandiera SM, Zhang Y, Raab C, Braun MP, Dean DC, Pang J, Leung KH, Doss GA. Drug Metab. Dispos. 1999;27:365–372. [PubMed] [Google Scholar]
- 20.Shen S, Marchick RM, Davis RM, Doss AG, Pohl RL. Chem. Res. Toxicol. 1999;12:214–222. doi: 10.1021/tx9802365. [DOI] [PubMed] [Google Scholar]
- 21.Saltiel AR, Olefsky JM. Diabetes. 1996;45:1661–1669. doi: 10.2337/diab.45.12.1661. [DOI] [PubMed] [Google Scholar]
- 22.Henry RP. Curr. Ther. Diabetes. 1997;26:553–573. [Google Scholar]
- 23.Sahi J, Hamilton G, Sinz M, Barros S, Huang S-M, Lesko LJ, LeCluyse EL. Xenobiotica. 2000;30:273–284. doi: 10.1080/004982500237668. [DOI] [PubMed] [Google Scholar]
- 24.Heringa M. Int. J. Clin. Pharmacol. Ther. 2003;41:331–345. doi: 10.5414/cpp41331. [DOI] [PubMed] [Google Scholar]
- 25.Kemp DC, Fan PW, Stevens JC. Drug Metab. and Dispos. 2002;30:694–700. doi: 10.1124/dmd.30.6.694. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q, Ngui JS, Doss GA, Wang RW, Cai XX, DiNinno FP, Blizzard TA, Hammond ML, Stearns RA, Evans DC, Baillie TA, Tang W. Chem. Res. Toxicol. 2002;15:907–914. doi: 10.1021/tx0200109. [DOI] [PubMed] [Google Scholar]
- 27.Tang W. Cur. Drug. Metab. 2003;4:319–329. doi: 10.2174/1389200033489398. [DOI] [PubMed] [Google Scholar]
- 28.Leemann T, Transon C, Dayer P. Life. Sci. 1993;52:29–34. doi: 10.1016/0024-3205(93)90285-b. [DOI] [PubMed] [Google Scholar]
- 29.Masubuchi Y. Drug Metab. Pharmacokinet. 2006;21:347–356. doi: 10.2133/dmpk.21.347. [DOI] [PubMed] [Google Scholar]
- 30.Tettey NJ, Maggs LJ, Rapeport GW, Pirmohamed M, Park KB. Chem. Res. Toxicol. 2001;14:965–974. doi: 10.1021/tx0001981. [DOI] [PubMed] [Google Scholar]
- 31.Iglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat. Chem. Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 32.Kassahun K, Pearson P, Tang W, McIntosh I, Leung K, Elmore C, Dean D, Wang R, Doss G, Baillie AT. Chem. Res. Toxicol. 2000;14:62–70. doi: 10.1021/tx000180q. [DOI] [PubMed] [Google Scholar]
- 33.Smith M. Chem. Res. Toxicol. 2003;16:679–687. doi: 10.1021/tx034033e. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto Y, Yamazaki H, Ikeda T, Watanabe T, Iwabuchi H, Nakajima M, Yokoi T. Drug. Metab. Dispos. 2002;30:155–160. doi: 10.1124/dmd.30.2.155. [DOI] [PubMed] [Google Scholar]
- 35.Ngui SJ, Tang W, Stearns AR, Shou M, Miller RR, Zhang Y, Lin HJ, Baillie T. Drug. Metabol. Disp. 2000;28:1043–1050. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.