Abstract
Class I HLA sample and display peptides from thousands of endogenous proteins at the cell surface. During infection, the influenza virus modifies the host cell proteome by (i) triggering host anti-viral responses, (ii) hijacking host processes, and (iii) inhibiting host mRNA processing. In turn, the catalog of HLA class I peptides that decorate the surface of an infected cell is positioned to reflect an altered host cell proteome. To understand the host-encoded peptides presented by class I molecules following influenza infection, we compared by mass spectrometry (MS) peptides eluted from the HLA of naïve and infected cells. We identified 20 peptide ligands unique to infected cells and 347 peptides with increased presentation following infection. Infection with different influenza strains demonstrated that proteome changes are predominantly strain-specific, with few individual cellular interactions observed for multiple viral strains. Modeling by pathway analysis, however, revealed that strain specific host peptide changes represent different routes to the same destination; host changes mediated by influenza are found predominantly clustered around HLA-B, ACTB, HSP90AB1, CDK2, and ANXA2. The class I HLA proteome scanning of influenza infected cells therefore indicates how divergent strains of influenza pursue alternate routes in order to access the same host cell processes.
Keywords: HLA class I, MHC class I, influenza virus, host, mass spectrometry
Introduction
Class I HLA molecules are nature’s proteome scanning chip. Thousands of peptides derived from proteins in multiple cellular compartments, including mitochondrial, secreted, and nuclear proteins are sampled by the class I HLA molecule and presented at the cell surface (1). Found on all nucleated cells, class I HLA proteome scanning chips sample and reflect the protein contents of a cell.
Traditionally, interest has focused upon the discovery of virus-derived peptides presented by class I HLA; a virus infects a cell, class I HLA binds a viral peptide for display at the cell surface, immune surveillance mechanisms discern the viral peptide/HLA complex, and the virus infected cell is destroyed (2). However, viral peptide presentation by the class I of infected cells represents a small, albeit significant, chapter of a much larger story. Following their entry into a host cell, viruses activate host gene transcription, hijack host proteins for viral replication, and degrade host proteins that impede virion production and assembly (3–5). As class I molecules reflect the proteome and changes therein, and as viruses markedly reorganize the infected host cell, we hypothesize that influenza A virus infection will evoke changes in the host class I HLA peptide repertoire.
Analogous to studies of changes in gene expression following infection (6–11), we posit that changes in the presentation of self epitopes following infection will unveil viral interactions within the host cell and imply points of therapeutic intervention. For example, self peptides unique to unhealthy cells may serve as diagnostic and/or therapeutic immune targets, as is the case for putative cancer cytotoxic T cell (CTL) epitopes (12–14). In addition, self peptides unique to infected cells reveal cellular proteins that have a role in virus replication. For example, increased class I HLA presentation of eIF4G peptide fragments following HIV infection coincides with degradation of eIF4G by the viral protease (15). Immunologically, CTL may target host-encoded class I HLA peptides that are unique to infected cells. During acute measles virus infection in humans, CTL target two HLA-A*0201 host encoded peptides (16). Realization of host ligands unique to infected cells is also positioned to help unravel the autoimmune responses associated with viral infection (17–19). Understanding changes in the host proteome following infection is poised to reveal targets for both small molecules and immunotherapy.
In this study we characterize the host peptides revealed by class I HLA following influenza A virus infection. Class I HLA-peptide complexes were affinity purified from cells expressing the soluble HLA (sHLA) class I molecule A*0201 (sHLA-A*0201). Peptides eluted from the sHLA-A*0201 of naïve cells and from cells infected with three different influenza A virus strains were separated by reverse-phase (RP) HPLC, comparatively mapped by mass spectrometry (MS), and amino acid sequenced by MS/MS fragmentation. Comparative MS analysis of HLA-A*0201 presented peptides identified 20 host-encoded ligands unique to influenza-infected cells and 347 self ligands upregulated during virus infection. Several of the class I HLA-presented host peptides are derived from proteins reported to directly interact with influenza such that numerous points of host-virus interaction, both direct and indirect, are discovered herein by scanning influenza induced peptide changes. The immune targeting and potential therapeutic impact of the changes in the class I HLA peptide repertoire following influenza infection are discussed.
MATERIALS AND METHODS
Cell Lines and Transfectants
HeLa cells (ATCC CCL-2) cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin were electroporated with a pcDNA 3.1- expression vector (Invitrogen) encoding secreted HLA-A*0201. The cytoplasmic and transmembrane domains of HLA-A*0201 were removed by PCR mutagenesis. The production of sHLA-A*0201 was measured by ELISA using anti-W6/32 (20) capture and β2-microglobulin primary antibodies (21). The peptide repertoire of HLA-A*0201 was examined as it is the most common HLA-A allele in North America (22).
Virus Production
Influenza viruses A/Puerto Rico/8/34 (PR8), A/Oklahoma/7485/01 (7485), and A/Oklahoma/309/06 (309) were propagated in Madin-Darby canine kidney (MDCK) cells. Roller bottles were seeded with MDCK cells at a density of 20,000 cells/cm2, cultured for 2 days, washed with PBS, and inoculated with virus in 10 ml CaMg PBS at a MOI of 0.05. Infection medium (DMEM/F12K supplemented with 6% ITS, 1% penicillin/streptomycin, 1% non-essential amino acids, and 1% sodium pyruvate) was added to the cell culture after 2 h incubation at 37°C. Virus production was monitored daily by HA titration on human red blood cells. Cell supernatant containing virus was centrifuged at 3,000 rpm for 30 min to remove cell debris and stored in 200 ml aliquots at −80°C.
Cell Pharm Infection
7.5 × 109 HeLa cells were pelleted, washed 3 times with CaMg PBS, and incubated with virus at a MOI of 200 for 2 h at 37°C. Influenza-infected HeLa cells were inoculated into a cell pharm hollow fiber bioreactor (Biovest International) and cultured in DMEM supplemented with 6% ITS. Cellular glucose consumption, pH, and sHLA production were monitored daily. The percentage of HeLa cells infected with influenza was measured by intracellular staining with an anti-nucleoprotein antibody (Meridian Life Science Inc.) and anti-serum targeting the influenza core (23) followed by flow cytometric analysis.
Peptide Isolation and Purification
Approximately 25 mg of sHLA-A*0201 peptide complex was affinity purified with W6/32 antibody from the supernatant of naïve and influenza infected HeLa cells. Peptide was eluted from the HLA class I binding groove with a 10% acetic acid boil and separated from class I HLA by passage through a 3 kDa membrane (Millipore). Naïve and infected peptide pools were separated by RP-HPLC into 40 peptide containing fractions. Prior to mass spectrometric analysis, 10% of the naïve and infected peptide pools underwent 14 cycles of N-terminal Edman degradation to confirm the HLA-A*0201 origin of the eluted peptide (data not shown) (24).
Mass Spectrometric Analysis
Peptides in naïve and influenza-infected RP-HPLC fractions were sprayed 3 times via nanospray into a Qstar Elite quadrupole time-of-flight (Q-TOF) mass spectrometer to create a MS ion map for each fraction. The MS ion maps for corresponding naïve and infected fractions were aligned at 20 amu increments to identify ions (putative peptides) unique to influenza infected cells. Peptides exhibiting a fold increase of at least 1.5-fold following infection were identified by summing the intensity of each ion in 3 naïve and infected MS ion maps and calculating the normalized fold increase in the intensity of the ion in the infected versus the naïve MS ion maps (24). A threshold of 1.5-fold or more was established after mass spectrometric analysis of the HLA class I peptide pools isolated from repeat naïve cell pharm runs revealed the extent of variability in peptide presentation between cell pharm runs (unpublished). This threshold limits the impact that stochastic or random changes in protein processing have upon the data presented herein. The identity of peptides in infected MS ion maps was determined by MS/MS fragmentation followed by amino acid sequencing de novo and/or with MASCOT (25). MS/MS fragmentation of peptides in pre, post, and corresponding RP-HPLC fractions of three separate uninfected cell pharm runs and affinity purifications confirmed the absence of unique peptides in the naïve class I HLA-A*0201 peptide pool.
Predicted Binding Affinity
The binding affinity of unique and increased peptides (8-, 9-, 10-, and 11-mers) to HLA-A*0201 was predicted with the SMM binding algorithm accessed on the Immune Epitope Database (IEDB) and Analysis Resource website (26).
Pathway Analysis
Functional analysis of host proteins encoding unique or increased HLA-A*0201 peptides following influenza infection was performed with Ingenuity Pathways Analysis software (Ingenuity Systems). Analysis was limited to direct protein interactions.
RESULTS
Influenza Strain Specificity Among Class I HLA-A*0201 Self Peptides
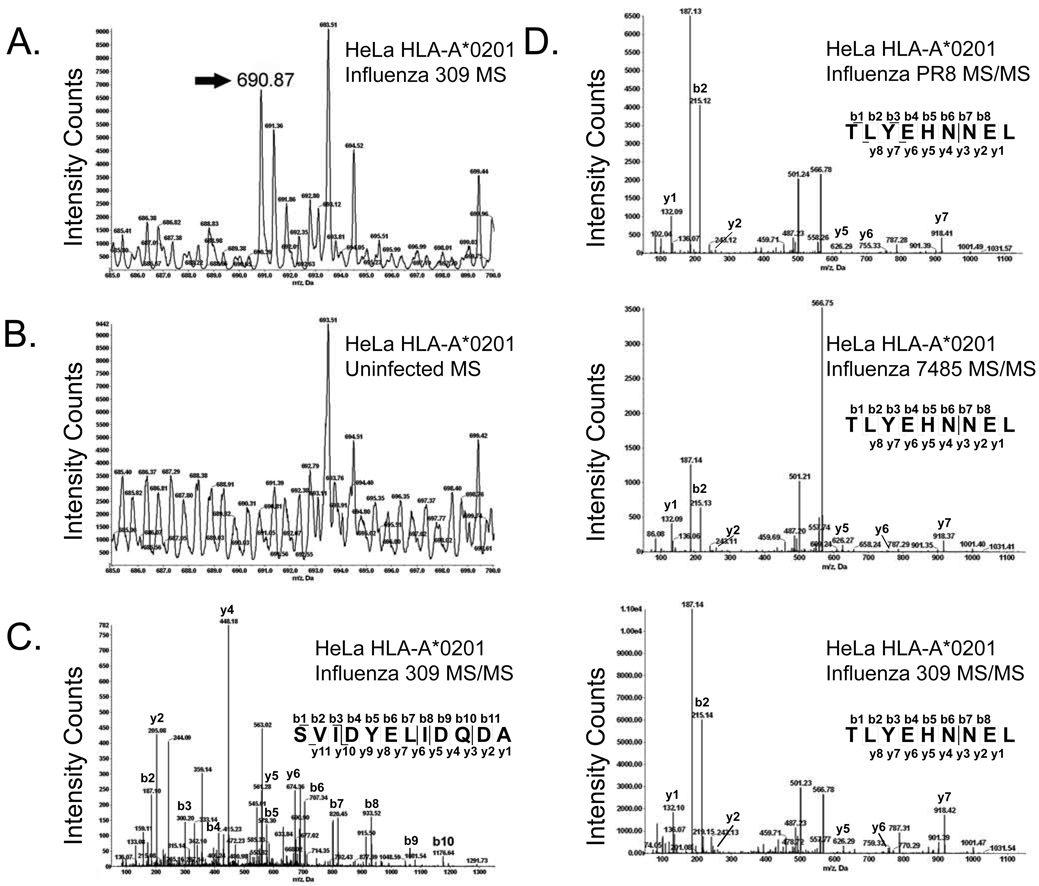
In this study we assessed the impact of influenza A virus infection upon class I HLA presentation of host encoded peptides. We initially gathered HLA/peptide complexes from naïve and influenza infected cells. A catalog of self peptides eluted from naïve cells was utilized for comparison to the class I repertoire of cells infected with multiple strains of influenza: PR8, a well characterized H1N1 influenza A virus laboratory strain, and two recent influenza A (H1N1 7485 and H3N2 309) circulating strains. Comparative mass spectrometry of HLA-A*0201 peptides gathered from naïve and infected cells identified 20 host-encoded peptides unique to infected MS ion maps (Fig. 1A–C) and 347 different self peptides with presentation increased 1.5 fold or greater following infection (Fig. 1D). Upregulated host peptides were increased by 1.5 to 33.09 fold following influenza infection with an average increase of 4-fold.
Fig. 1.
Identification of unique and increased peptides by mass spectrometry following influenza A virus infection. The annexin A2202–213 (SVIDYELIDQDA) ligand was unique to influenza 309 infected cells. An ion with the mass of 690.87 atomic mass units (amu) was identified as unique to infected cells by aligning the MS spectra of corresponding infected (A) and naïve (B) RP-HPLC fractions at 20 amu increments. Amino acid sequencing of the unique ion by MS/MS fragmentation identified a peptide derived from annexin A2 (SVIDYELIDQDA) (C). TLYEHNNEL, a nine amino acid peptide derived from the AAAS protein, was increased following infection with all three influenza A virus strains (fold increase: PR8-2.49, 7485-2.84, and 309-2.2). The amino acid sequence was determined by MS/MS fragmentation (D).
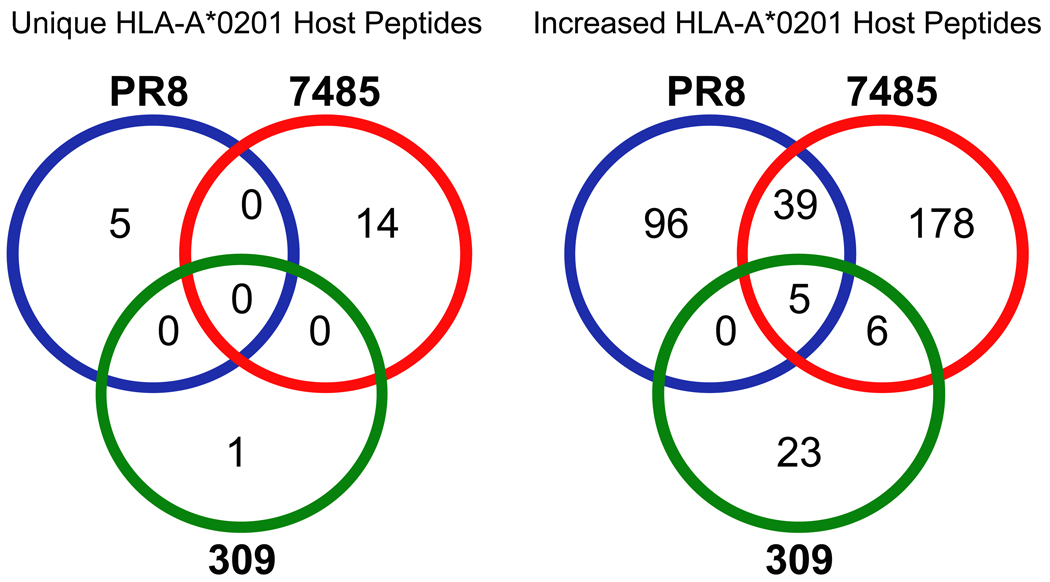
Many host peptides demonstrated increased presentation following infection, yet only a handful were increased by multiple strains of the influenza A virus (Fig. 2). With 347 upregulated host ligands characterized following influenza infection, 39 were increased by both strains of H1N1 influenza A (PR8 and 7485) and 6 were shared by recent H1N1 and H3N2 influenza A virus isolates 7485 and 309 (Fig. 2). Only five host-derived peptides (GVYDGEEHSV-MAGEB2, KLDQDLNEV-SMC3, TLYEHNNEL-AAAS, LLDVVHPA-CCT7, and SLFEGTWYL-HMGCS1) exhibited increased presentation following infection with all three influenza strains studied (Fig. 1D and 2, Table 2). These data show that each viral strain has a unique signature of altered self ligands, that influenza A H1N1 strains overlap by approximately 10% of their upregulated ligands (39/347), and that a neglible number of upregulated ligands (6/347) overlap between the H3N2 and H1N1 isolates circulating in the human population.
Fig. 2.
Host encoded HLA-A*0201 peptides that are unique or increased following influenza A virus infection. Comparative mass spectrometric analysis of class I HLA-A*0201 peptides purified from naïve and influenza infected HeLa cells identified host-encoded peptides with unique or increased presentation following influenza infection. A Venn diagram reveals overlap in the presentation of unique and increased peptides observed between the PR8 (blue), 7485 (red), and 309 (green) influenza strains. The twenty host peptides unique to influenza infection were presented on a strain-by strain basis (PR8-5, 7485-14, and 309-1). While the majority of peptides with upregulated presentation following influenza infection were limited to the infecting strain, 39 host encoded HLA-A*0201 peptides had increased presentation during infection with both H1N1 strains (PR8 and 7485), 6 peptides were upregulated following 7485 and 309 infection, and 5 peptides exhibited increased presentation during infection with all influenza A virus three strains.
Table 2.
Increased HLA-A*0201 host peptides following influenza A infection
| Fractiona | Ionb | Peptide | Protein | AAc | IC50d | PR8 | 7485 | 309 | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 45 | 460.2 | FAGDDAPRA | Actin | 9 | 1083.8 | NA | 5.21 | 7.48 | ACTB |
| 45 | 500.2 | NLAEKPKTV | Proteasome associated protein ECM29 | 9 | 281.2 | NA | 2.84 | 2.66 | ECM29 |
| 46 | 508.2 | ALKDKIEKA | Heat shock protein 94c | 9 | 1392.9 | 2.8 | 3.2 | NA | GRP94C |
| 46 | 800.4 | IIPASTGAA | Glyceraldehyde-3 phosphate dehydrogenase | 9 | 6817.7 | 17.7 | 12.67 | NA | GAPDH |
| 47 | 450.2 | IINEPTAAA | Heat shock protein 70 | 9 | 1060.7 | 21.71 | 4.28 | NA | HSPA2/5/6/8/1A/1B/1L |
| 48 | 546.2 | GVYDGEEHSV | Melanoma antigen, family B | 10 | 289 | 9.92 | 1.97 | 1.66 | MAGEB2 |
| 50 | 464.7 | VVQDGIVKA | Peroxiredoxin 5 | 9 | 880.2 | 7.38 | 10.11 | NA | PRDX5 |
| 53 | 516.2 | GLIKGSGTAEV | Pyruvate kinase | 11 | 1940.7 | NA | 2.23 | 1.69 | PKM2 |
| 53 | 537.2 | KLDQDLNEV | Structural maintenance of chomosome 3 | 9 | 22.5 | 1.98 | 2.76 | 1.57 | SMC3 |
| 53 | 578.8 | FIQTQQLHAA | Pyruvate kinase | 10 | 2280 | 10.17 | 24.26 | NA | PKM2 |
| 53 | 959.5 | SQVPEVTTV | Phosphoinositide-3-kinase, regulatory subunit 4 | 9 | 335.2 | NA | 7.52 | 1.55 | PIK3R4 |
| 54 | 497.2 | NLDPAVHEV | GTPase 1 | 9 | 27.8 | 5.03 | 2.88 | NA | GPN1 |
| 54 | 499.8 | RVPPPPPIA | Heterogeneous nuclear ribonucleoprotein C | 9 | 8604 | 6.54 | 4.96 | NA | HNRNPC |
| 54 | 502.7 | NMVAKVDEV | Ribosomal protein L10a | 9 | 116.9 | 2.93 | 2.12 | NA | RPL10A |
| 54 | 566.7 | TLYEHNNEL | Achalasia, adrenocortical insufficiency, alacrimia | 9 | 81.5 | 2.49 | 2.84 | 2.2 | AAAS |
| 55 | 589.7 | YAYDGKDYIA | MHC-B antigen | 10 | 413.9 | 1.6 | 2.74 | NA | HLA-B |
| 56 | 444.2 | AVSDGVIKV | Cofilin 1 | 9 | 140.4 | 8.98 | 2.02 | NA | CFL1 |
| 56 | 530.2 | ILGYTEHQV | Glyceraldehyde-3 phosphate dehydrogenase | 9 | 171.1 | 23.91 | 4.47 | NA | GAPDH |
| 57 | 495.3 | TLAEVSTRL | Salt-inducible protein kinase-like kinase | 9 | 101.6 | NA | 1.88 | 2.61 | SNF1LK |
| 58 | 432.2 | LLDVVHPA | TCP-1 HIV-1 Nef interacting protein | 8 | 644.4 | 33.09 | 1.83 | 1.7 | CCT7 |
| 58 | 467.7 | LLDVVHPAA | TCP-1 HIV-1 Nef interacting protein | 9 | 265 | 3.43 | 3.82 | NA | CCT7 |
| 60 | 474.2 | ILSGVVTKM | Ribosomal protein S11 | 9 | 384.7 | 1.95 | 1.59 | NA | RPS11 |
| 60 | 567.3 | ILMEHIHKL | Ribosomal protein L19 | 9 | 9.8 | 5.16 | 3.04 | NA | RPL19 |
| 62 | 507.3 | ALNEKLVNL | Eukaryotic translation initiation factor 3, subunit F | 9 | 110.7 | 1.52 | 2.49 | NA | EIF3F |
| 62 | 528.7 | ALNDHFVKL | Glyceraldehyde-3 phosphate dehydrogenase | 9 | 95.4 | 23.02 | 8.32 | NA | GAPDH |
| 64 | 455.8 | GVLPNIQAV | Histone H2A | 9 | 509.1 | NA | 8.69 | 4.16 | HIST3H2A |
| 64 | 466.7 | ALDKATVLL | Programmed cell death 4 | 9 | 100.7 | 2.13 | 3.8 | NA | PDCD4 |
| 64 | 514.8 | GLIEKNIEL | DNA methyltransferase | 9 | 56.7 | 1.58 | 1.54 | NA | DNMT1 |
| 64 | 528.8 | KVFDPVPVGV | Deadbox polypeptide 9 | 10 | 112.8 | 1.74 | 1.97 | NA | DHX9 |
| 64 | 630.3 | TLWDIQKDLK | Lactate dehydrogenase B | 10 | 17930.7 | 1.64 | 4.79 | NA | LDHB |
| 65 | 1014.5 | SMTLAIHEI | Degenerative spermatocyte homolog 1 | 9 | 143.6 | 2.11 | 1.58 | NA | DEGS1 |
| 66 | 471.2 | GLDIPTVQV | Deadbox polypeptide 49 | 9 | 193.4 | 4.78 | 1.78 | NA | DDX49 |
| 66 | 479.2 | GLIDGRLTI | Signal peptidase complex subunit 2 | 9 | 201.1 | 1.67 | 2.21 | NA | SPCS2 |
| 67 | 544.2 | TLWGIQKEL | Lactate dehydrogenase A | 9 | 197.4 | 3.27 | 1.72 | NA | LDHA |
| 67 | 878.5 | AIIGGTFTV | ER-golgi intermediate compartment 1 | 9 | 27 | 4.17 | 2.79 | NA | ERGIC |
| 68 | 476.2 | LLGPRLVLA | Transmembrane trafficking protein 10 | 9 | 276.8 | 2.09 | 5.6 | NA | TMED10 |
| 68 | 480.2 | SLLEKSLGL | Eukaryotic translation elongation factor 1 | 9 | 46 | 1.64 | 3.7 | NA | EEF1E1 |
| 68 | 622.8 | KLIGDPNLEFV | Ras-related nuclear protein | 11 | 245.3 | 2.82 | 7.2 | NA | RAN |
| 69 | 537.2 | IMLEALERV | Small nuclear ribonucleoprotein polypeptide G | 9 | 14.1 | 1.64 | 1.54 | NA | SNRPG |
| 70 | 537.7 | SMPDFDLHL | AHNAK nucleoprotein | 9 | 85.4 | 1.83 | 2.78 | NA | AHNAK |
| 70 | 625.8 | FLAEEGFYKF | Subunit of the oligosaccharyltransferase complex | 10 | 284.3 | 2.98 | 2.86 | NA | STT3A |
| 70 | 1019.5 | GIVEGLMTTV | Glyceraldehyde-3 phosphate dehydrogenase | 10 | 205.5 | 3.1 | 1.57 | NA | GAPDH |
| 73 | 541.3 | TLAKYLMEL | Cyclin B1 | 9 | 16.4 | 6.81 | 3.95 | NA | CCNB1 |
| 73 | 588.3 | GLQDFDLLRV | Protein kinase C, iota | 10 | 43.7 | 2.29 | 2.9 | NA | PRKCI |
| 74 | 485.8 | LLDVTPLSL | Heat shock protein 70 | 9 | 153.6 | 9.68 | 1.92 | NA | HSPA2/8/9 |
| 75 | 488.7 | GIWGFIKGV | Proline-rich coiled-coil 1 | 9 | 116.7 | 6.1 | 2.01 | NA | PRRC1 |
| 75 | 533.8 | FLPSYIIDV | Cleavage and polyadenlyation specificity factor 1 | 9 | 2.8 | 2.57 | 2.97 | NA | CPSF1 |
| 75 | 593.8 | IIMLEALERV | Small nuclear ribonucleoprotein polypeptide G | 10 | 43.2 | 13.68 | 4.89 | NA | SNRPG |
| 76 | 515.3 | SLLDIIEKV | Tuberous sclerosis 2 | 9 | 9.8 | 2.56 | 1.82 | NA | TSC2 |
| 79 | 558.2 | SLFEGTWYL | Hydroxymethylglutaryl coA synthase | 9 | 2.6 | 2.36 | 1.83 | 2.79 | HMGCS1 |
RP-HPLC fraction containing the endogenous peptide
Ion mass in atomic mass units corresponding to the endogenous peptide
Amino acids
IC50=Inhibitory concentration 50 percent
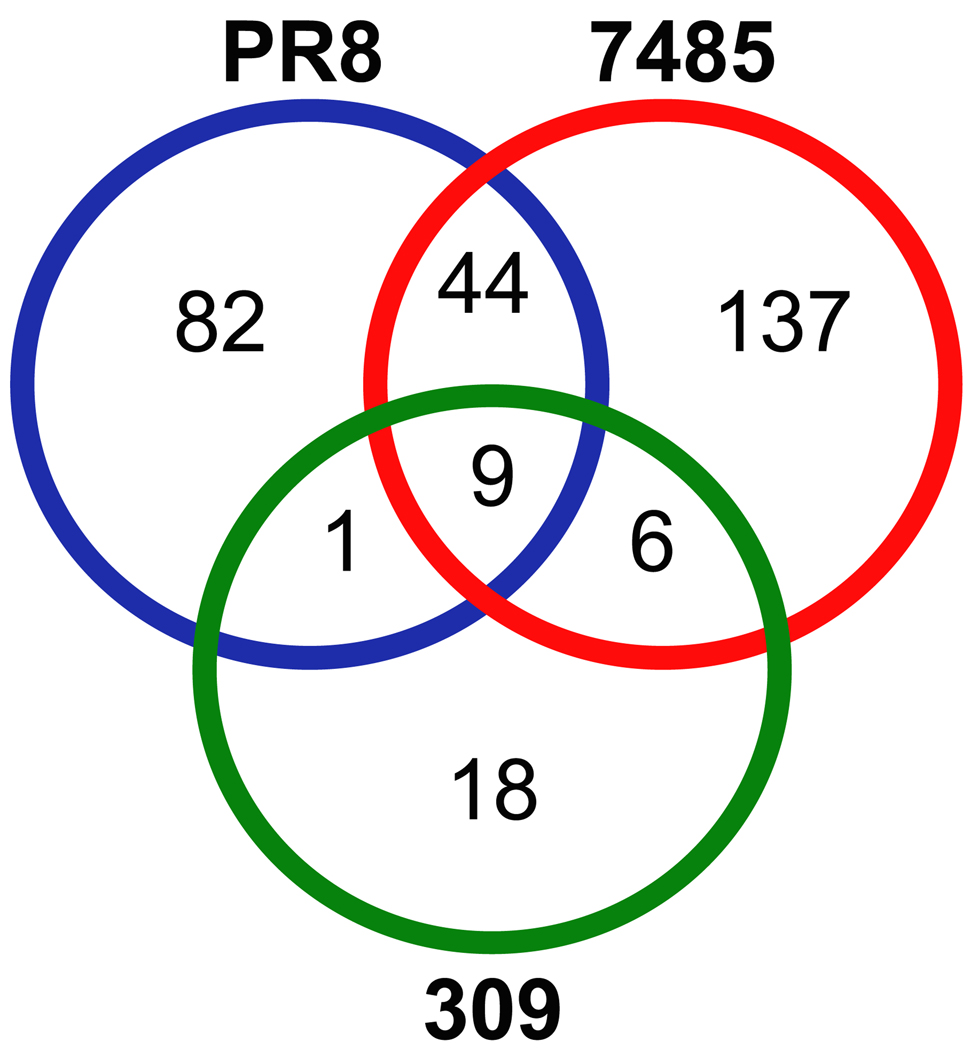
Although overlap in the presentation of unique and increased host ligands between virus strains was minimal, it is possible that the class I HLA of influenza infected cells sample different peptides of the same host protein. Therefore, we determined the overlap in source proteins revealed by HLA-A*0201 following influenza infection. The 20 unique and 347 increased host HLA-A*0201 peptides are encoded by 297 different proteins. Figure 3 confirms the minimal overlap between the three influenza strains in the source proteins sampled by class I HLA, reflecting the strain specificity of unique/increased self HLA-A*0201 peptides following PR8, 7485, and 309 influenza infection. Approximately 3% of the host proteins (9/297) encoding unique/increased ligands were revealed by class I HLA following infection with all three influenza A virus strains (Fig. 3). The two H1N1 influenza virus strains PR8 and 7485 share 15% (44/297) of host proteins, the recent H1N1 and H3N2 influenza isolates (7485 and 309) overlap in 2% (6/297) of the host proteins exposed by HLA-A*0201, and class I HLA sampled only one protein exclusive to PR8 and 309 influenza strains (Fig. 3). As influenza viruses diverge, so does the signature of class I HLA presented self ligands associated with each viral strain: Unique patterns of self-ligand presentation and host proteins sampled distinguish the various strains of influenza.
Fig. 3.
Source proteins encoding HLA-A*0201 peptides with unique or increased presentation following influenza A virus infection. HLA-A*0201 self peptides with unique or increased presentation following influenza A virus infection were encoded by 297 different proteins. A Venn diagram illustrates the overlap between PR8 (blue), 7485 (red), and 309 (green) influenza strains in regard to the source proteins encoding unique and increased peptides identified during influenza infection. Minimal overlap was observed in the host proteins sampled by class I HLA among the three influenza A virus strains. The most extensive overlap was displayed by the two H1N1 influenza virus strains (44/297). Both PR8 and 7485 H1N1 strains exhibited slight overlap with the recent H3N2 isolated 309. Only 9 host proteins were shared by the three influenza strains.
Physical Characteristics Distinguish Upregulated and Unique Ligands
Nineteen of 20 unique self peptides resulting from influenza infection are not nonamers and vary in length from 7 to 13 amino acids (Table 1). In contrast, 78% of host peptides with increased presentation in multiple influenza strains are nine amino acids in length (Table 1 and 2). Unique and upregulated ligands were further distinguished by their HLA-A*0201 binding affinity (Table 1 and 2). The average HLA-A*0201 peptide unique to infected cells has a lower (IC50 7570 nM) binding affinity compared to self peptides with increased presentation (IC50 971 nM) (Table 1 and 2). These data indicate that class I HLA presented self peptides unique to influenza infected cells vary in size and have a substantially lower HLA-A*0201 binding affinity when compared to ligands with increased presentation which are predominantly nonamers and exhibit a much higher predicted HLA-A*0201 binding affinity.
Table 1.
Unique HLA-A*0201 host peptides following influenza A infection
| Fractiona | Ionb | Peptide | Strain | AAc | IC50d | Protein | Gene |
|---|---|---|---|---|---|---|---|
| 61 | 594.3 | KLMELHGEGSS | PR8 | 11 | 1369.7 | Ribosomal protein S3A | RPS3A |
| 66 | 579.8 | LMTTVHAITAT | PR8 | 11 | 693.7 | Glyceraldehyde-3 phosphate dehydrogenase | GAPDH |
| 73 | 614.8 | TLAEVERLKGL | PR8 | 11 | 136.1 | Small nuclear ribonucleoprotein polypeptide A | SNRPA1 |
| 75 | 757.8 | KLFDDDETGKISF | PR8 | 13 | NA | Caltractin | CETN2 |
| 80 | 656.7 | TLQEVFEKATF | PR8 | 11 | 406.6 | Nucleolin | NCL |
| 47 | 618.3 | YVPEDEDLKK | 7485 | 10 | 37536.2 | Coiled-coil domain containing 12 | CCDC12 |
| 53 | 456.2 | GQIGQLPQA | 7485 | 9 | 1029.2 | E1A binding protein p300 | EP300 |
| 53 | 563.8 | VLPTETEVAPA | 7485 | 11 | 566.1 | Microtubule-associated protein 4 | MAP4 |
| 59 | 799.4 | LLDVPTAA | 7485 | 8 | 20959.2 | Interferon gamma inducible protein 30kDa | IFI30 |
| 66 | 823.4 | GLFDTAIS | 7485 | 8 | 1209.9 | Nucleoporin 160 kDa | NUP160 |
| 68 | 588.3 | GLFGGAGVGKTVL | 7485 | 13 | NA | Mitochondrial ATPase H+ beta subunit | ATP5B |
| 71 | 1071.5 | FLDLEPIPGA | 7485 | 10 | 26.5 | 5-3 nucleotidase | NT5C |
| 73 | 1002.5 | SLFEGTWY | 7485 | 8 | 4763.9 | Hydroxymethylglutaryl coA synthase | HMGCS1 |
| 77 | 931.5 | FLLSLLIL | 7485 | 8 | 820.6 | Chemokine-like factor superfamily 6 | CMTM6 |
| 78 | 673.3 | ILAADFEIGHFL | 7485 | 12 | NA | Nucleosome assembly protein | NAP1L1 |
| 78 | 802.4 | SLLDELL | 7485 | 7 | NA | Sterol O-acyltransferase | SOAT1 |
| 79 | 461.2 | FVFPGELL | 7485 | 8 | 33102.8 | Neutral amino acid transporter B | SLC1A5 |
| 82 | 938.4 | YLFDSFF | 7485 | 7 | NA | Carbamoylphosphate synthase I | CPS1 |
| 84 | 895.5 | YLFLGGIL | 7485 | 8 | 3353.5 | Bax inhibitor 1 | TMBIM6 |
| 64 | 690.8 | SVIDYELIDQDA | 309 | 12 | NA | Annexin A2 | ANXA2 |
RP-HPLC fraction containing the endogenous peptide
Ion mass in atomic mass units corresponding to the endogenous peptide
Amino acids
IC50=Inhibitory concentration 50 percent
Different Influenza Strains are Related in Their Host Interaction
The biologic function of the host proteins most frequently sampled by HLA class I molecules following influenza infection is poised to reveal host cell functions disrupted during viral replication. The functions of source proteins encoding multiple unique and/or increased HLA-A*0201 self peptides were compared to identify common cellular functions altered by viral replication. Table 3 illustrates that infection with H1N1 influenza virus strains PR8 and 7485 triggered the class I presentation of multiple peptides encoded by proteins involved in cellular metabolism. In particular, proteins involved in aerobic and anaerobic glycolysis (i.e. GAPDH, LDHA, LDB, and PKM2) were frequently sampled by class I HLA. Numerous peptides derived from several proteins that function in protein translation elongation such as RPL10A and RPLP0 were presented by HLA-A*0201 following 7485 H1N1 and 309 H3N2 influenza infection respectively indicating a disturbance in protein synthesis (Table 3). In addition, PR8 infection displayed several unique/increased class I peptides encoded by proteins that process RNA (CPSF1, IMP3, and SNRNPG) and chaperone proteins (HSPA2 and CCT7) in the host cell nucleus (Table 3).
Table 3.
Most frequently sampled source proteins
| Gene | Peptides | Biologic Function |
|---|---|---|
| PR8 | ||
| GAPDH | 9 | Metabolism: Glycolysis |
| CCT7 | 2 | Protein Binding |
| CPSF1 | 2 | RNA processing: pre-mRNA processing |
| GILT | 2 | Oxidation Reduction |
| HSPA2/8 | 2 | Protein Binding |
| IMP3 | 2 | RNA processing: rRNA processing |
| SNRNPG | 2 | RNA processing: RNA Splicing |
| VIM | 2 | Cell Motility |
| 7485 | ||
| LDHA | 7 | Metabolism: Anaerobic Glycolysis |
| ASS1 | 6 | Metabolism: Arginine Biosynthesis |
| CPS1 | 6 | Metabolism: Glutamine/Nitrogen Metabolism |
| GAPDH | 6 | Metabolism: Glycolysis |
| LDHB | 4 | Metabolism: Anaerobic Glycolysis |
| RPL10A | 4 | Protein Synthesis: Translation Elongation |
| ACTB | 3 | Cell Motility |
| ATP5B | 3 | Proton Transport |
| CCT7 | 3 | Protein Binding |
| ADSL | 2 | Metabolism: Purine Ribonucleotide Biosynthesis |
| AHNAK1 | 2 | Nervous System Development |
| ECHS1 | 2 | Metabolism: Lipid Metabolism |
| EEF2 | 2 | Translation Elongation |
| HMGCS1 | 2 | Metabolism: Lipid Metabolism |
| KRT18 | 2 | Cell Cycle |
| MTAP | 2 | Metabolism: Nucleoside Metabolism |
| NOMO3 | 2 | Nodal Signaling |
| NUP160 | 2 | mRNA Export from Nucleus |
| PKM2 | 2 | Metabolism: Glycolysis |
| RPL12 | 2 | Protein Synthesis: Translation Elongation |
| RPL13a | 2 | Protein Synthesis: Translation Elongation |
| RPL19 | 2 | Protein Synthesis: Translation Elongation |
| RPS11 | 2 | Protein Synthesis: Translation Elongation |
| RPS17 | 2 | Protein Synthesis: Translation Elongation |
| SERPINH1 | 2 | Protein Binding |
| SNRPG | 2 | RNA processing: RNA Splicing |
| VCP | 2 | Vesicle Transport |
| 309 | ||
| RPLP0 | 2 | Protein Synthesis: Translation Elongation |
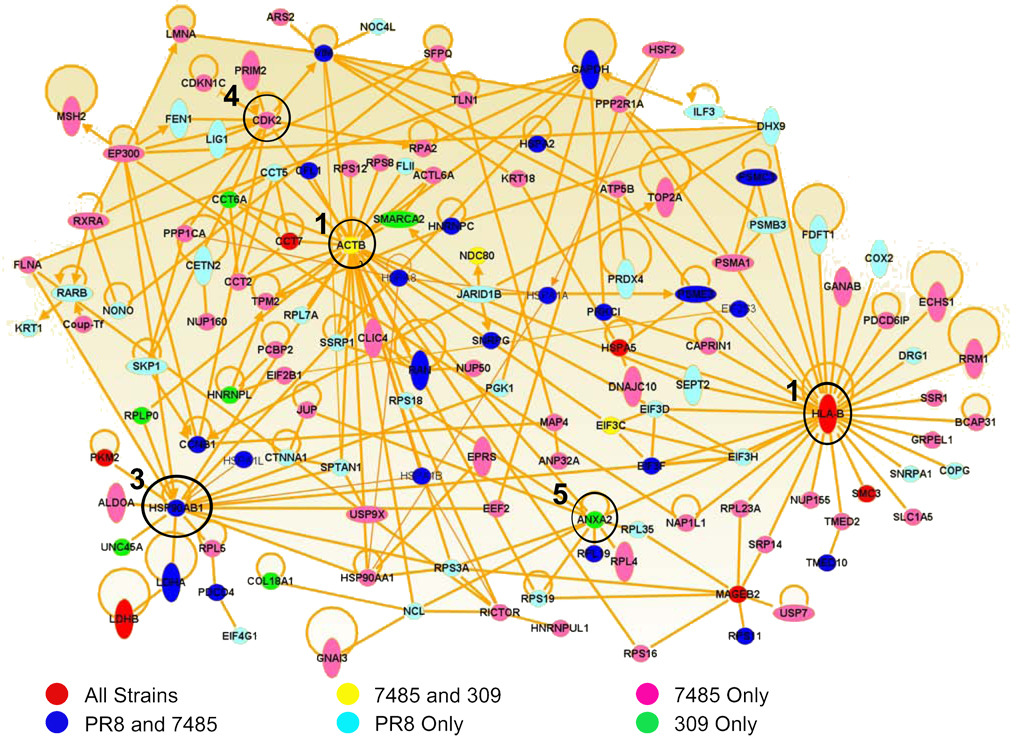
To determine associations among influenza mediated changes in the infected cell, host proteins encoding HLA-A*0201 unique/increased self peptides were mapped by Ingenuity Pathway Analysis software. Pathway analysis revealed that 45% (134/297) of the host proteins encoding unique/increased HLA-A*0201 ligands directly interact in the host cell (Fig. 4). Five prominent hubs of influenza/host protein interaction were revealed by these direct interactions: HLA-B, beta-actin (ACTB), heat shock protein 90 (HSP90AB1), cyclin-dependent kinase 2 (CDK2), and annexin A2 (ANXA2). All three influenza A virus isolates influenced related source proteins in these five hubs as detected by HLA-A*0201 peptide sampling (Fig. 4).
Fig. 4.
Direct interactions of proteins encoding unique/increased HLA-A*0201 following PR8, 7485, and 309 influenza infection. Direct interactions between the source proteins of all unique and increased HLA-A*0201 peptides following PR8, 7485, and 309 infection were mapped by Ingenuity Pathway Analysis. Proteins encoding unique and/or increased peptides that were sampled by HLA-A*0201 during infection with all three influenza strains are shown in red. Host proteins shared by H1N1 influenza A virus strains PR8 and 7485 are illustrated in blue and proteins common to the recent H1N1 and H3N2 isolates 7485 and 309 are colored yellow. The remaining source proteins were unique to either PR8 (light blue), 7485 (pink), or 309 (green) influenza strains. The five most prominent hubs of protein interaction were centered around the HLA-B, ACTB, HSP90AB1, CDK2, and ANXA2 molecules which are circled and labeled in accordance to their dominance in the protein network.
Almost half of the proteins in the network directly interact with HLA-B (31/134) and/or beta-actin (31/134), hinting at a major reorganization in the HLA-B class I presentation pathway and cytoskeleton during influenza virus infection (Fig. 4). Approximately 30% of network source proteins directly interact with HSP90AB1 (18/134), CDK2 (11/134), and/or ANXA2 (10/134) molecules which function in protein folding, cell cycle, and calcium-ion dependent phospholipid binding respectively, indicating additional host pathways likely modified by influenza replication (Fig. 4). Moreover, host changes that did not directly interact with one another in these pathways were nonetheless clustered in the same regions. For example, several ribosomal proteins that do not directly interact with one another in the host cell circle around ACTB (RPS3A, RPS8, RPS12, RPS18, RPS19, RPLP0 and RPL7A) and ANXA2 (RPS3A, RPS18, RPS19, RPL19, and RPL35). Collectively, these data reveal that despite minimal overlap in peptide presentation, the host proteins sampled by class I HLA reveal common cellular functions altered by influenza virus infection. Source proteins encoding host HLA class I peptides with unique or increased presentation following influenza infection form a highly integrated web of functional and physical interaction.
The Influenza A Virus Directly Interacts with Unique and Increased Self Peptide Source Proteins
Literature reveals that seven HLA-A*0201 host peptides with increased or unique presentation following influenza A virus infection are derived from five proteins that are directly involved in viral propagation (Table 4). The presentation of a nine amino acid peptide derived from actin (FAGDDAPRA) was upregulated following infection with 7485 and 309 influenza virus strains (Table 1). The binding of influenza nucleoprotein (NP) molecules to the actin cytoskeleton helps maintain cytoplasmic retention of vRNPs following nuclear export (27). Infection with the two H1N1 strains resulted in increased presentation of two nonamers (IINEPTAAA and LLDVTPLSL) derived from heat shock protein 70 (HSP70) (Table 1). During influenza infection, HSP70 facilitates nuclear export of vRNPs by binding influenza matrix 1 (M1) molecules attached to vRNPs (28, 29). Infection with the H1N1 strains also resulted in the increased presentation of two nonamers (ILDKKVEKV-PR8 and ALLSSGFSL-7485) derived from heat shock protein 90 (HSP90) (Table 4). During influenza infection, the inducible (HSP90AA1) and constitutively (HSP90AB1) expressed forms of HSP90 promote viral replication by facilitating the nuclear import and assembly of influenza polymerase proteins (30–32). PR8 infection revealed the unique presentation of an undecamer peptide derived from nucleolin (TLQEVFEKATF) (Table 2). During influenza infection the non-structural protein 1 (NS1) protein colocalizes with nucleolin in the nucleoli of the host cell, although the significance of this interaction is yet unknown (33). Influenza 309 infection identified a 12 amino acid peptide encoded by annexin A2 (SVIDYELIDQDA) unique to HLA-A*0201 of infected cells (Fig. 1, Table 2), and annexin A2 is positioned to impact viral maturation by activating plasmin which cleaves the HA0 molecule into the HA1 and HA2 subunits (34–36). Thus, within the host cell the influenza virus hijacks actin, HSP70, HSP90, and annexin A2 to carry various stages of viral replication. Collectively, these data indicate that five proteins encoding HLA-A*0201 ligands unique (nucleolin and annexin A2) or increased (actin, HSP70, and HSP90) following influenza infection directly interact with the influenza A virus during various stages of viral replication.
Table 4.
Host source proteins encoding HLA-A*0201 unique and increased peptides with direct interaction with the influenza A virus following infection.
| Source Protein | Peptide Sequence | Function | Influenza Interaction |
|---|---|---|---|
| Cytoskeleton Component | Influenza virus NP binds the actin cytoskeleton to promote cytoplasmic retention of vRNPs |
||
| Actin | FAGDDAPRA | Cell Structure, Integrity, and Mobility | Integrity of cytoskeleton is necessary for virus internalization into epithelial cells |
| Maintains organization of lipid rafts during virus budding |
|||
| IINEPTAAA | Correct folding of nascent polypeptides | Colocalizes with nuclear M1 to promote nuclear export of vRNPs |
|
| HSP70 | |||
| LLDVTPLSL | Disassembly of clathrin-coated vesicles | Inhibits nuclear export of vRNPs at 41°C | |
| ILDKKVEKV | Correct folding of nascent polypeptides | Nuclear import of influenza polymerase proteins |
|
| HSP90 | |||
| ALLSSGFSL | Refolding denatured proteins | Assembly of influenza polymerase complex | |
| Transportation of ribosomes and ribosomal proteins between nucleus and cytoplasm |
|||
| Nucleolin | TLQEVFEKATF | Influenza virus NS1 localizes with nucleolin in host cell nucleoli |
|
| During Cell Stress: Inhibits DNA replication, Stimulates DNA repair, and Down regulates c-myb |
|||
| Calcium dependent phospholipid binding protein | Incorporated into influenza virions | ||
| Annexin A2 | SVIDYELIDQDA | Regulates cell growth | |
| Converts plasminogen into plasmin. Plasmin can cleave HA into HA1 and HA2 |
|||
| Involved in signal transduction pathways | |||
Discussion
Systematic characterization of the HLA class I peptide repertoire is positioned to provide a unique view into the influenza infected cell. Here, we found that infection with 3 different influenza A virus strains identified 20 unique and over 345 increased class I peptides. We were initially surprised at the minimal overlap in peptide presentation and source protein origin between the three strains of influenza A virus. Overlap was limited to upregulated class I peptides; 5 ligands were increased during infection with all strains, 39 upregulated peptides were shared amongst both H1N1 influenza strains, and 6 class I ligands were increased during infection with the recent H1N1 and H3N2 strains. Substantial variation was also observed between the three influenza strains in regards to the source proteins sampled by HLA-A*0201 following infection. There was little overlap in specific peptide presentation or in source protein origin between strains of influenza.
This minimal overlap among individual peptide ligands and source proteins sampled led to the rather surprising preliminary interpretation that these three strains were markedly different in their viral-host interactions. However, a picture of host-influenza interactions began to emerge when the cell biology of these individual ligands was considered. For example, source proteins for the ligands frequently sampled following influenza infection revealed proteins involved in RNA processing, cell metabolism, and protein synthesis; functions required for host cell survival and virus propagation. In corroboration of these results, microarray studies following in vivo and in vitro influenza infection indicate an increase in the expression of genes encoding proteins involved in growth and proliferation (7, 9). The dominance of source proteins associated with cell metabolism (especially glycolysis following 7485 infection) may reflect the rapid production of viral proteins within the host cell. Most significant was that pathway modeling of source proteins for the unique and increased peptides revealed that the three different strains of influenza substantially overlap in their interaction with the infected host cell. Each of the three viruses mediated changes in the same five prominent hubs: HLA-B, ACTB, HSP90AB1, CDK2, and ANXA2 molecules were at the core of these virus-host interaction hubs. Thus, differences in peptide presentation indicate alternate routes taken by each strain of influenza A strain towards the same five cellular destinations.
It is intriguing to consider the five host destinations at which influenza arrives within the infected cell. Protein interactions revolving around HLA-B may reflect an alteration in the HLA class I presentation pathway. We have recently reported that class I molecules encoded at the HLA-B locus present immune dominant influenza peptide epitopes to CTL, and these data are collaborated by others (37–39). Moreover, it has been reported that HLA-B molecules are vital to the targeting of viruses such as HIV and EBV (40–42). Furthermore, while it is clear that influenza peptide epitopes are presented by class I HLA to CTL, the level of presentation often falls at or below the peptide presentation of low-abundance self peptide ligands (37). It is therefore possible that influenza moderates (but does not abrogate) the presentation of viral peptide ligands to immune surveillance mechanisms, especially by class I molecules encoded at the HLA-B gene locus.
Understanding how the influenza virus interacts with the host cell following infection is positioned to aid the development of anti-viral drugs and therapeutics. Realization of virus-host interactions has led to the development of the anti-viral drugs amantadine and rimantadine that target the influenza matrix protein 2 (M2) ion channel. The anti-viral drugs Tamiflu and Relenza target neuraminidase (NA) molecules at early and late stages of viral replication. Although these current influenza anti-viral drugs inhibit host-virus interactions by targeting viral proteins, the identification of host proteins that interact with viral proteins during viral replication may also serve as potent anti-viral targets. In our study we identified several host HLA class I peptides with increased or unique presentation that are derived from proteins that directly interact with influenza viral proteins in the host cell, including actin and HSP90. Other laboratories have demonstrated that drug inhibitors of actin and HSP90 reduce influenza replication in vitro. An intact actin cytoskeleton is required for early (virus internalization) and late (nuclear export of vRNA and budding of progeny virions) stages of influenza replication (27, 43, 44). Disruption of the actin cytoskeleton can drastically reduce clathrin-mediated internalization of influenza virus particles to near undetectable levels (44). Treatment of polarized epithelial cells with cytochalasin D, an actin inhibitor, drastically impedes the internalization of influenza virions to near undetectable levels (44) but increases viral infectivity of LLC-MK2 cells (45). Inhibition of HSP90 activity in vitro with HSP90 specific inhibitors results in decreased influenza replication (31, 32). Thus, virus-host dynamics can be disrupted during influenza infection by targeting viral proteins and host molecules necessary for viral replication.
Host HLA class I peptides with unique or increased presentation following influenza infection may also serve as primary targets for CTL eliciting vaccines and T cell receptor mimic (TCRm) antibodies which recognize specific HLA class I-peptide complexes. CTL reactive host HLA class I peptides have been demonstrated during HIV-1 and measles virus infection and in the case of HIV-1 these virus-associated host ligands are being explored as vaccine candidates (16, 46). Targeting host HLA class I peptides is not a novel concept as several tumor vaccines have been generated against endogenous class I peptides that mark the surface of tumor tissue (47, 48). Recently, antibodies that mimic the T Cell Receptor (TCRm) and recognize tumor associated HLA class I peptides have also been developed, and these TCRm drastically reduce tumor burden in vivo in a tumor mouse model (14). TCRm antibodies specific for host class I HLA peptides that distinguish influenza infected cells from healthy cells may also serve as novel therapeutics, especially against anti-viral resistant and recombinant influenza virus strains.
As nature’s proteome scanning chip, HLA class I molecules are poised to reflect alterations in the proteome of virus infected cells. Characterization of the class I HLA peptide repertoire of infected cells acts as a compliment to gene and protein expression arrays, highlighting virus-induced changes in the protein composition of host cells. The host class I peptide profile presented at the cell surface not only reflects alterations in host gene and protein expression, but it also reveals changes in the post-translational modification of proteins, the subcellular location of proteins, protein degradation, and protein-protein interactions. Host proteins that are disregulated following viral infection appear to function in viral propagation and in the anti-viral host cell response, and the identification of such virus-associated changes is positioned to provide targets for therapeutics and diagnostics. How CTL target unique/increased host HLA class I peptides on the surface of infected cells, as is the case during HIV-1 and measles virus infection, must also be explored to determine the immunogenic nature of unique/upregulated host ligands. Discovery of host ligands with altered class I HLA presentation following infection provides potential immune targets at the cell surface as well as possible points of intervention within the influenza infected cell.
ACKNOWLEDGEMENTS
We thank Dr. Gillian Air for the human influenza A virus isolates and her insight on the influenza virus. Also, we thank Dr. Ken Jackson of the University of Oklahoma Health Sciences Center Molecular Biology Proteomics Facility for technical assistance. We thank Dr. Igor Dozmorov for his assistance with the Ingenuity Pathway Analysis software. This work was supported by the National Institutes of Health contract HHSN266200400027C (W.H.H.) and the National Institute of Allergy and Infectious Disease Institutional Training Grant A1007633-006 (A.W.)
Abbreviations
- HLA
human leukocyte antigen
- RNA
ribonucleic acid
- DNA
deoxyribonucleic acid
- MS
mass spectrometry
- CTL
cytotoxic T lymphocyte
- HIV
human immunodeficiency virus
- sHLA
soluble human leukocyte antigen
- HPLC
high pressure liquid chromatography
- RP-HPLC
reverse phase high pressure liquid chromatography
- DMEM
Dulbecco’s modification of Eagle’s medium
- FBS
fetal bovine serum
- PCR
polymerase chain reaction
- ELISA
enzyme-linked immunosorbent assay
- MDCK
Madin-Darby canine kidney cell
- MOI
multiplicity of infection
- PBS
phosphate buffered saline
- ITS
insulin-transferrin-selenium
- SMM
stabilized matrix method
- IEDB
Immune Epitope Database
- PR8
Influenza A/Puerto Rico/8/34
- 7485
Influenza A/Oklahoma/7485/01
- 309
Influenza A/Oklahoma/309/06
- IC50
inhibitory concentration 50 percent
- HA
hemagglutinin
- vRNP
viral ribonucleoprotein
- NP
nucleoprotein
- M1
matrix protein 1
- M2
matrix protein 2
- NS1
non-structural protein 1
- NA
neuraminidase
- EBV
Epstein-Barr Virus
- HSP70
heat shock protein 70
- HSP90
heat shock protein 90
- TCRm
T cell receptor mimic
- Q-TOF
quadrupole-time of flight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hickman HD, Luis AD, Buchli R, Few SR, Sathiamurthy M, VanGundy RS, et al. Toward a definition of self: proteomic evaluation of the class I Peptide repertoire. J Immunol. 2004 Mar 1;172(5):2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell J, Bennink JR, Hosaka Y. Cells process exogenous proteins for recognition by cytotoxic T lymphocytes. Science. 1988;239:637–640. doi: 10.1126/science.3257585. [DOI] [PubMed] [Google Scholar]
- 3.Ptak RG, Fu W, Sanders-Beer BE, Dickerson JE, Pinney JW, Robertson DL, et al. Cataloguing the HIV type 1 human protein interaction network. AIDS Res Hum Retroviruses. 2008 Dec;24(12):1497–1502. doi: 10.1089/aid.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kash JC, Goodman AG, Korth MJ, Katze MG. Hijacking of the host-cell response and translational control during influenza virus infection. Virus Res. 2006 Jul;119(1):111–120. doi: 10.1016/j.virusres.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003 Oct;110(2):163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001 Oct 26;294(5543):870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 7.Geiss GK, An MC, Bumgarner RE, Hammersmark E, Cunningham D, Katze MG. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J Virol. 2001 May;75(9):4321–4331. doi: 10.1128/JVI.75.9.4321-4331.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu XF, Liu QZ, Li C, Dong J, Zhou JF, Wang M, et al. The differential expression of the human lung carcinoma cells infected with high pathogenic avian influenza virus A/Anhui/1/2005 (H5N1) Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2008 Jun;22(3):180–182. [PubMed] [Google Scholar]
- 9.Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, et al. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008 Nov;82(22):11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Su YA, Hu P, Yang J, Zheng B, Wu P, et al. Signature patterns revealed by microarray analyses of mice infected with influenza virus A and Streptococcus pneumoniae. Microbes Infect. 2006 Jul;8(8):2172–2185. doi: 10.1016/j.micinf.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawada J, Kimura H, Kamachi Y, Nishikawa K, Taniguchi M, Nagaoka K, et al. Analysis of gene-expression profiles by oligonucleotide microarray in children with influenza. J Gen Virol. 2006 Jun;87(Pt 6):1677–1683. doi: 10.1099/vir.0.81670-0. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins OE, Vangundy RS, Eckerd AM, Bardet W, Buchli R, Weidanz JA, et al. Identification of breast cancer peptide epitopes presented by HLA-A*0201. J Proteome Res. 2008 Apr;7(4):1445–1457. doi: 10.1021/pr700761w. [DOI] [PubMed] [Google Scholar]
- 13.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994 Apr 29;264(5159):716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 14.Wittman VP, Woodburn D, Nguyen T, Neethling FA, Wright S, Weidanz JA. Antibody targeting to a class I MHC-peptide epitope promotes tumor cell death. J Immunol. 2006 Sep 15;177(6):4187–4195. doi: 10.4049/jimmunol.177.6.4187. [DOI] [PubMed] [Google Scholar]
- 15.Hickman HD, Luis AD, Bardet W, Buchli R, Battson CL, Shearer MH, et al. Cutting edge: class I presentation of host peptides following HIV infection. J Immunol. 2003 Jul 1;171(1):22–26. doi: 10.4049/jimmunol.171.1.22. [DOI] [PubMed] [Google Scholar]
- 16.Herberts CA, van Gaans-van den Brink J, van der Heeft E, van Wijk M, Hoekman J, Jaye A, et al. Autoreactivity against induced or upregulated abundant self-peptides in HLA-A*0201 following measles virus infection. Hum Immunol. 2003;64(1):44–55. doi: 10.1016/s0198-8859(02)00707-3. [DOI] [PubMed] [Google Scholar]
- 17.van der Werf N, Kroese FG, Rozing J, Hillebrands JL. Viral infections as potential triggers of type 1 diabetes. Diabetes Metab Res Rev. 2007 Mar;23(3):169–183. doi: 10.1002/dmrr.695. [DOI] [PubMed] [Google Scholar]
- 18.Toussirot E, Roudier J. Epstein-Barr virus in autoimmune diseases. Best Pract Res Clin Rheumatol. 2008 Oct;22(5):883–896. doi: 10.1016/j.berh.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Christensen T. Human herpesviruses in MS. Int MS J. 2007 Jun;14(2):41–47. [PubMed] [Google Scholar]
- 20.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. Journal of Immunology. 1979;123:342–249. [PubMed] [Google Scholar]
- 21.Prilliman KR, Lindsey M, Zuo Y, Jackson K, Zhang Y, Hildebrand WH. Large-Scale Production of Class I Bound Peptides: Assigning a Peptide Signature To HLA-B*1501. Immunogenetics. 1997;45:379–385. doi: 10.1007/s002510050219. [DOI] [PubMed] [Google Scholar]
- 22.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 2003 May;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Air GM. Expression of functional influenza virus A polymerase proteins and template from cloned cDNAS in recombinant vaccinia virus infected cells. Biochem Biophys Res Commun. 1994 Apr 15;200(1):95–101. doi: 10.1006/bbrc.1994.1419. [DOI] [PubMed] [Google Scholar]
- 24.Wahl A, Weidanz J, Hildebrand W. Direct class I HLA antigen discovery to distinguish virus-infected and cancerous cells. Expert Rev Proteomics. 2006 Dec;3(6):641–652. doi: 10.1586/14789450.3.6.641. [DOI] [PubMed] [Google Scholar]
- 25.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [pii] [DOI] [PubMed] [Google Scholar]
- 26.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005 Mar;3(3):e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Digard P, Elton D, Bishop K, Medcalf E, Weeds A, Pope B. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J Virol. 1999 Mar;73(3):2222–2231. doi: 10.1128/jvi.73.3.2222-2231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K, Fuse T, Asano I, Tsukahara F, Maru Y, Nagata K, et al. Identification of Hsc70 as an influenza virus matrix protein (M1) binding factor involved in the virus life cycle. FEBS Lett. 2006 Oct 16;580(24):5785–5790. doi: 10.1016/j.febslet.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 29.Hirayama E, Atagi H, Hiraki A, Kim J. Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J Virol. 2004 Feb;78(3):1263–1270. doi: 10.1128/JVI.78.3.1263-1270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007 Feb;81(3):1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, et al. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008 Aug 1;377(2):431–439. doi: 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 32.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002 Nov 22;277(47):45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 33.Murayama R, Harada Y, Shibata T, Kuroda K, Hayakawa S, Shimizu K, et al. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem Biophys Res Commun. 2007 Nov 3;362(4):880–885. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- 34.LeBouder F, Morello E, Rimmelzwaan GF, Bosse F, Pechoux C, Delmas B, et al. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J Virol. 2008 Jul;82(14):6820–6828. doi: 10.1128/JVI.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, et al. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem. 2001 May;268(10):2847–2855. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- 36.Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975 Dec;68(2):440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 37.Wahl A, Schafer F, Bardet W, Buchli R, Air GM, Hildebrand WH. HLA class I molecules consistently present internal influenza epitopes. Proc Natl Acad Sci U S A. 2009 Jan 2; doi: 10.1073/pnas.0811271106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boon AC, De Mutsert G, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. Preferential HLA usage in the influenza virus-specific CTL response. J Immunol. 2004 Apr 1;172(7):4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- 39.Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, et al. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J Virol. 2002 Jan;76(2):582–590. doi: 10.1128/JVI.76.2.582-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004 Dec 9;432(7018):769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 41.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006 Apr 1;176(7):4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 42.Hollsberg P. Contribution of HLA class I allele expression to CD8+ T-cell responses against Epstein-Barr virus. Scand J Immunol. 2002 Feb;55(2):189–195. doi: 10.1046/j.0300-9475.2001.01043.x. [DOI] [PubMed] [Google Scholar]
- 43.Simpson-Holley M, Ellis D, Fisher D, Elton D, McCauley J, Digard P. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology. 2002 Sep 30;301(2):212–225. doi: 10.1006/viro.2002.1595. [DOI] [PubMed] [Google Scholar]
- 44.Sun X, Whittaker GR. Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells. Cell Microbiol. 2007 Jul;9(7):1672–1682. doi: 10.1111/j.1462-5822.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 45.Arcangeletti MC, De Conto F, Ferraglia F, Pinardi F, Gatti R, Orlandini G, et al. Host-cell-dependent role of actin cytoskeleton during the replication of a human strain of influenza A virus. Arch Virol. 2008;153(7):1209–1221. doi: 10.1007/s00705-008-0103-0. [DOI] [PubMed] [Google Scholar]
- 46.Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007 Nov;3(11):e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N, Hirohashi Y, Tsukahara T, Kikuchi T, Sahara H, Kamiguchi K, et al. Molecular pathological approaches to human tumor immunology. Pathol Int. 2009 Apr;59(4):205–217. doi: 10.1111/j.1440-1827.2009.02353.x. [DOI] [PubMed] [Google Scholar]
- 48.Kessler JH, Melief CJ. Identification of T-cell epitopes for cancer immunotherapy. Leukemia. 2007 Sep;21(9):1859–1874. doi: 10.1038/sj.leu.2404787. [DOI] [PubMed] [Google Scholar]