Abstract
Histone methylation plays an important role in regulating chromatin-mediated gene control and epigenetic-based memory systems that direct cell fate. Enzymes termed histone demethylases directly remove the methyl marks from histones, thus contributing to a dynamically regulated histone methylated genome, however the biological functions of these newly identified enzymes remains unclear. The JMJD2A-D family belongs to the JmjC domain-containing family of histone demethylases (JHDMs). Here, we report the cloning and functional characterization of the Drosophila HDM gene Dmel\Kdm4A that is a homolog of the human JMJD2 family. We show that homologs for three human JHDM families, JHDM1, JHDM2 and JMJD2 are present in Drosophila and that are each expressed during the Drosophila lifecycle. Disruption of Dmel\Kdm4A results in a reduction of the male lifespan and a male-specific wing extension/twitching phenotype that occurs in response to other males, and is reminiscent of an inter-male courtship phenotype involving the courtship song. Remarkably, certain genes associated with each of these phenotypes are significantly downregulated in response to Dmel\Kdm4A loss, most notably the longevity associated Hsp22 gene and the male sex-determination fruitless gene. Our results have implications for the role of the epigenetic regulator Dmel\Kdm4A in the control of genes involved in lifespan and male-specific sex-determination in the fly.
INTRODUCTION
Histones are chromatin proteins that play an important role in DNA packaging and gene regulation. The initial level of chromatin packaging consists of the nucleosome, made up of DNA wrapped around two copies each of core histone proteins H2A, H2B, H3 and H4. Histones are subjected to a wide variety of covalent modifications that include acetylation, phosphorylation and methylation (Berger, 2002; Felsenfeld and Groudine, 2003; Fischle et al., 2003; Martin, 2005; Luger, 2006). Distinct combinatorial patterns of such modifications are believed to serve as epigenetic marks that control chromatin packaging and subsequent gene expression by providing recognition sites for downstream chromatin regulatory factors (Nowak and Corces, 2000; Rice and Allis, 2001; Fischle et al., 2003; Bottomley, 2004).
Histone methylation plays an important role in many biological processes including heterochromatin formation, homeotic gene silencing, X-chromosome inactivation, genomic imprinting and transcriptional regulation (Lachner et al., 2001; Feinberg et al., 2002; Santos-Rosa et al., 2002; Margueron et al., 2005; Martin and Zhang, 2005; Martin, 2005) and may exist on both the lysine (K) and arginine (R) residues of histones. Lysine methylation can occur on a variety of specific sites on histone H3 (K4, K9, K27, K36, and K79) and histone H4 (K20), thus allowing for the generation of distinct histone methylation patterns that directly influence chromatin regulated cellular processes. Importantly, lysine residues can also be mono-, di-, or trimethylated, and such differential methylation states serve to diversify the docking sites for effector chromatin proteins and modifiers, thus underscoring the complexity of histone methylation in regulating biological processes (Zhang and Reinberg, 2001; Santos-Rosa et al., 2002; Wang et al., 2004). Histone methylation had long been considered an irreversible epigenetic mark, however this viewpoint was challenged with the discovery of the first H3-K4 and K9 specific histone demethylase (HDM) LSD1 (Metzger et al., 2005; Shi et al., 2005). Since then, numerous different HDMs have been discovered that display distinct substrate and methylation conversion state specificity, supporting the concept that histone methylation, like acetylation, is a reversible and dynamically regulated process (Chang et al., 2007; Shi, 2007). Investigation of the specific biological roles of these newly identified HDMs will undoubtedly contribute to our understanding of histone methylation regulated cellular processes in development and disease.
The JmjC-domain-containing histone demthylases (JHDMs) represent the largest class of HDMs (Wang et al., 2004; Tsukada et al., 2006; Shin and Janknecht, 2007b). These HDM enzymes are characterized by containing a conserved JmjC domain that catalyzes lysine demethylation of histones via an oxidative reduction reaction that requires iron Fe(II) and alpha-ketoglutarate (aKG) cofactors. Unlike LSD1, that reduces only mono- and dimethyl lysine modifications (Shi et al., 2005), certain JHDM family members can also reduce tri- histone lysine-methylation states (Tsukada et al., 2006; Whetstine et al., 2006; Shi, 2007). Additionally, different JHDM families display distinct substrate specificity. For instance, the JHDM1 family reduces H3K36, the JHDM2 family reduces H3K9 while certain members of the JHDM3/JMJD2 family can reduce both H3K9 and H3K36. There are four JHDM3/JMJD2 genes within the human genome, designated JHDM3/JMJD2A-D (here, they will be referred to as JMJD2A-D for simplicity) (Tsukada et al., 2006; Whetstine et al., 2006). It has been suggested that JMJD2D gave rise to two additional human genes, JMJD2E and JMJD2F via local retrotransposition. Genes JMJD2A-C encode for proteins containing N-terminal JmjC and JmjN domains, followed by two C-terminus PhD domains and two Tudor domains (Klose et al., 2006; Whetstine et al., 2006). In contrast, the JMJD2D family member encodes for a shorter protein product that lacks the C-terminal PHD and Tudor domains (Klose et al., 2006).
Prior in vitro analysis of the catalytic activity of the four human JMJD2A-D protein family members reveals differences in both their substrate specificity and their ability to mediate different degrees of demethylation, supporting distinct biological functions for these family members. For example, while all JMJD2 family members can reduce H3-K9Me1, only JMJD2A and C have the capacity to also reduce H3-K36Me3. Additionally, while JMJD2A-D can all convert H3-K9Me3 to H3-K9Me2, only JMJD2D can reduce H3-K9Me3 to H3-K9Me2 and H3-K9Me1 (Whetstine et al., 2006; Shin and Janknecht, 2007a; Shin and Janknecht, 2007b). However, although these newly identified HDMs are now well characterized in terms of their enzymatic specificity and activity, the biological role of these proteins during multicellular development remains to be elucidated.
Here, we report the cloning and functional characterization of the Drosophila HDM gene Dmel\Kdm4A that is a homolog of the human JMJD2 family. We show that homologs for the three main human JHDM families, JHDM1, JHDM2 and JMJD2 (Klose et al., 2006; Tsukada et al., 2006), are each present in Drosophila and that each is expressed during the Drosophila lifecycle. Disruption of Dmel\Kdm4A in the fly results in a reduction in the male lifespan and a male-specific wing extension/twitching phenotype that occurs in response to the presence of other males, and is reminiscent of an inter male courtship phenotype involving the courtship song (Certel et al., 2007). Remarkably, certain genes associated with each of these phenotypes are significantly downregulated in response to Dmel\Kdm4A loss, most notably the longevity associated heat shock protein 22 (Hsp22) gene (Morrow et al., 2004) and the male sex-determination fruitless gene (Dickson, 2002; Dickson, 2008), in which mRNA levels in male flies is almost undetectable. Taken together, our results support an essential role for epigenetic regulator Dmel\Kdm4A in the transcriptional activation of genes required for lifespan control and male-specific sex-determination and courtship behavior.
MATERIALS AND METHODS
Identification of D. melanogaster histone demethylases, isolation of cDNA clones and DNA sequencing
BLAST searches were carried out using the BLAST algorithm at both FLYBASE (1999) and NCBI with sequences corresponding to either JHDM1B, JHDM2B and JHDM3/JMJD2A. Drosophila sequences were identified that displayed high homology to each of these sequences (CG11033 for Dmel\JHDM1, CG8165 for Dmel\JHDM2 and CG15835 for Dmel\Kdm4A). As we failed to identify a cDNA clone corresponding to Dmel\Kdm4A in the expressed sequence tag (EST) database at the time we began this work, we cloned a cDNA using RT-PCR. Total RNA was isolated from Canton S. D. melanogaster pupae or adult flies using TRIzol (Invitrogen) and treated twice with DNA-free™ (Ambion) to remove DNA. First strand cDNA was prepared using the SuperScript™ II reverse transcriptase kit (Invitrogen) according to the manufacture’s instructions with 1μg total RNA and 15 ng/μL of random hexamer primers (Roche). The full ORF for Dmel\Kdm4A was amplified by PCR using the forward primer, 5′-GAT ATAAAGCGGCCGCGCCATCATGTCCACGAGATCTTCATTCGCC3′ containing eight additional base pairs to aid in restriction enzyme digest (underlined), a NotI site (bold), followed by a KOZAC sequence (in italics), and sequence corresponding to the first 7 codons of Dmel\Kdm4A. The reverse strand primer, 5′-GCTCTAGATCATCATCAATCCTCGTCGTCAAGTGTGAG-3′ contained eight additional base pairs to aid in restriction digest, a XbaI site (bold), followed by two in frame stop codons (italics), and the last five codons of Dmel\Kdm4A. PCR reactions were carried out using Expand™ High Fidelity PCR System (Roche) according to the manufactures instructions using 400 nM of each forward and reverse primers. The cycling parameters were 30 cycles of 95° for 2 min, 55° for 1 min, and 72° for 3 min, using Mastercycler (Eppendorf). The correct sized PCR amplification products were cloned into the TOPO pCR2.1 vector (Invitrogen) according to the manufacture’s instructions. The entire insert DNA sequence for each of these constructs was determined by the University of Pennsylvania DNA Core Sequencing Facility, Philadelphia, PA.
Semi-Quantitative RT-PCR of staged Drosophila RNA
Total RNA was isolated from staged Canton S. D. melanogaster (12-24h embryo, 1st instar larvae, 2nd instar larvae, 3rd instar larvae, pupae, and adult fly) using TRIzol (Invitrogen) and treated twice with DNA-free™ (Ambion) to remove DNA. First strand cDNA was prepared using the SuperScript™ II reverse transcriptase kit (Invitrogen) according to the manufacture’s instructions with 1μg total RNA and 15 ng/μL of random hexamer primers (Roche). Primer sets for Dmel\JHDM1 (forward primer: 5′-CGCGTGAAACAGGAGATAAAG3′, reverse primer: GCTGGTGGCAATCACACTAATAG3′) amplified a 464-bp fragment, Dmel\JHDM2 (forward primer: 5′-GTTTTCAGTGCATGACCAAG-3′, reverse primer: 5′GGCAACGAGCTCTAGTGATG-3′) amplified a 417-bp fragment and Dmel\Kdm4A (forward primer: 5′-GTTTCCAGCCAGAGCGATAC-3′, reverse primer: GACAGGGCAGTTCATTCCATAG3) amplified a 401-bp fragment and RP49 (forward primer: 5′-5′GCCCAGCATACAGGCCCAAG3′3′, reverse primer: 5′CGTTCTCTTGAGAACGCAGG3 3′) amplified a 402-bp fragment. All PCR reactions were carried out in triplicate in 40 μl total reaction volumes containing: 0.5 U Taq (Qiagen), 1 μl cDNA (from the RT reaction described above), 250 μM dNTPs (Amersham Pharmacia Biotech), and 10 μM each of forward and reverse primer. The PCR cycling conditions were: 34 cycles at 95° for 3 min , 55° for 1 min, and 72° for 1 min with a 7 min. extension after each cycle.
q PCR analysis
Total RNA was isolated from 21 day old male Dmel\Kdm4A P-supp and Dmel\Kdm4AREV flies using TRIzol (Invitrogen) and treated twice with DNA-free (Ambion) to remove DNA. cDNA was prepared using the SuperScript II reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions with 1 μg total RNA and 0.2μg/mL random hexamer primers (Roche Applied Science). PCR reactions were performed in a 20 μL reaction volume containing cDNA, 1× Power SYBR® Green PCR Master Mix (Applied Biosystems), and 10μM both forward and reverse primers (primer pairs available upon request). PCR was performed using an ABI 7500 Real-Time PCR system (Applied Biosystems) following the manufacturer’s instructions. Fold change in mRNA expression were determined by the ΔΔCt method (Livak and Schmittgen, 2001; Yuan et al., 2006).
Immunohistochemical staining of embryos
The antibodies used in immunohistochemical staining of embryos were as follows: mouse anti-ELAV (Developmental Studies Hybridoma Bank, University of Iowa); mouse anti-REPO (Developmental Studies Hybridoma Bank, University of Iowa); mouse 22C10 (Developmental Studies Hybridoma Bank, University of Iowa); biotin-conjugated anti-mouse secondary antibody (Vectastain ABC Elite kit; Vector Laboratories); biotin-conjugated anti-rat secondary antibody (Vector Laboratories). Embryos collected from grape-agar plates (Flystuff) were dechorionated with 50% Clorox bleach, rinsed with 0.1% Triton-X solution in water, then transferred to eppendorf tubes. To fix eggs, the Triton-X solution was removed and equal volumes of 4% paraformaldehyde in PBS and heptane (1ml) were added. The tubes were gently and continuously shaken for 2 minutes by hand before removing first the bottom paraformaldehyde phase and then the top heptane phase. Eggs were devitellinized in a 1:1 heptane/methanol mixture (1ml), rinsed once with methanol, and then washed twice with PT (PBS, 0.1% Tween-20).
Antibody staining was performed by first washing the embryos in phosphate-buffered saline (PBS) every 30 minutes with 0.1% Tween (PBT) over a 3 hour period at room temperature. Embryos were incubated with primary antibody (diluted 1:500 in PBT) overnight at 4°C in 1.5 ml microcentrifuge tubes with constant rotation. Embryos were washed with PBT every 30 min for 3 h at room temperature. Biotinylated anti-mouse secondary antibody (Vectastain ABC Elite kit; Vector Laboratories) diluted 1:400 in PBT was added to the embryos and incubated overnight at 4°C. Embryos were then washed with PBT every 30 min over a 3 hour period at room temperature and incubated in biotin—streptavidin—horseradish peroxidase complex (Vectastain ABC Elite kit; Vector Laboratories) at room temperature for 1.5 h. Embryos were then washed eight times in PBT for 2 h. The signal was developed by incubation with 500 μl ImmPACT DAB (Vector Laboratories) in the presence of 1 μl of 10% H2O2. The reaction was terminated by washing the embryos with PBT and then with ethanol. The embryos were mounted in methyl salicylate and viewed with Zeiss Axioplan2 optics.
Identification of Dmel\Kdm4AP-supp and revertant fly lines
The P-element suppressor of Dmel\Kdm4A was identified by searching for stocks under the gene accession number for the gene: CG15835 at Flybase.org. The Bloomington fly stock number is 13828 and the genotype is: y1 w67c23; P{SUPor-P}CG15835KG04636. These flies were designated as line Dmel\Kdm4AP-Supp. To create Dmel\Kdm4A revertant fly lines, the P-element was remobilized and excised as described (Palladina et al., 2002). Such excision flies were examined for precise excision of the P-element by single fly genomic PCR. Briefly, single male and female flies from the 56 potential P-element excision lines were collected. Genomic DNA was extracted by homogenizing flies in SB buffer (10 mM Tris-Hcl pH 8.0, 1mM EDTA, 25 mM NaCl, 200 g/ml Proteinase K) and the crude DNA extract was directly used for PCR reactions. To verify the presence of a single P-element insertion in Dmel\Kdm4AP-Supp, sequencing was carried out on genomic DNA using primers P2 and P3, corresponding to the 3′ and 5′ends of the P-element, respectively. Precise excision was verified using PCR with primer sets P1 and P2, corresponding to the 5′ genomic insertion site and 3′ end of the P-element, respectively and primer sets P3 and P4 corresponding to the 5′ end of the P-element and 3′ end of the genomic insertion site sequence. The primers were: P1 (5′-GAGATTCGTTTCGCTTGCTT-3′), P2(5′-GGCAAGAAAGTAGGTTGATAAAGC-3′), P3(5′- GTCTGACCTTTTGCAGGTGC-3′), and P4 (5′-GCTGGATGTTGATTTGCTGG-3′). All PCR fragments were sequenced to confirm their correct identity.
Climbing assay
The climbing assay was performed as follows. Twenty female flies and twenty male flies (forty flies total) were placed in plastic vials. The number of flies at the top of the vial were counted after either 7s or 18s of climbing over a period of 14 days. Each time point was repeated a minimum of five times and a maximum of ten times. The experiment was repeated three independent times with similar results obtained from each experiment.
Longevity assay
Staged male and female flies were collected at 0-24 hours and grown separately after eclosion to eliminate the affect of mating on longevity. 113 Canton S males, 112 Canton females, 222 Dmel\Kdm4AREV A male, 196 Dmel\Kdm4AREV A females, 226 Dmel\Kdm4AP-supp males and 194 Dmel\Kdm4AP-supp females were maintained in embryo collection chambers capped with grape juice plates. The plates were applied with fresh yeast paste and changed every day and the number of dead flies was recorded. The data was analyzed using 2-way ANOVA with SAS programming and Microsoft Excel.
Twitching Assay
Staged 0-24 hour Dmel\Kdm4AP-Supp and Dmel\Kdm4AREV A male and female flies were collected in separate vials and allowed to acclimate for 4 days. 10 vials containing 3 male flies and 10 vials containing 3 female flies (60 flies total) were observed and the number of times the flies twitched was counted over 5 minutes. The number obtained was divided by 3 to calculate average number of twitches per fly.
RESULTS
Identification and cloning of the Drosophila Dmel\Kdm4A gene that is a homolog of the human JMJD2 gene family
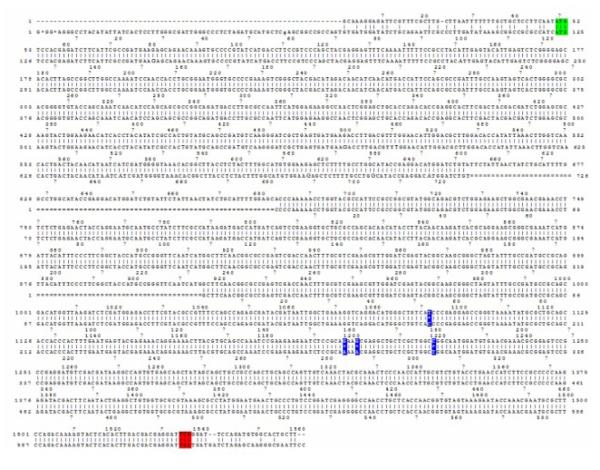
To identify human homologs of the JMJD2 gene family in Drosophila, conserved sequences within the JMJD2 genes were used to query the Drosophila Genome database for genomic DNA encoding homologous sequences. Two homologous genes (accession numbers: CG15835 and CG33182) were identified that were both located on chromosome 2, arm 2R: 3810274 to 3812488 and 9073721 to 9035781 respectively. Both proteins displayed the greatest structural similarity to family member JMJD2D in their protein structure, as both of them lacked the C-terminal PHD and Tudor domains. As the conceptual protein product encoded by gene CG15835 displayed a greater homology to the JMJD2 family due to its longer N-terminus, it was chosen for further analysis (Figure 2). Because no EST cDNA clones were available for CG15838 at the time we began this work, we cloned the gene using an RT-PCR based strategy on RNA isolated from Canton S pupae. The expected 1487 bp PCR product was cloned into the TOPO vector and the full sequence was determined and aligned to the CG15835 gene sequence at FlyBase. Four nucleotide differences were identified and these same base pair changes were also found to be present in cDNA clones prepared from RNA isolated from adult Canton S flies (Figure 1). Importantly, these changes did not alter the amino acid sequence of the CG15835 conceptual translation product posted at FlyBase.
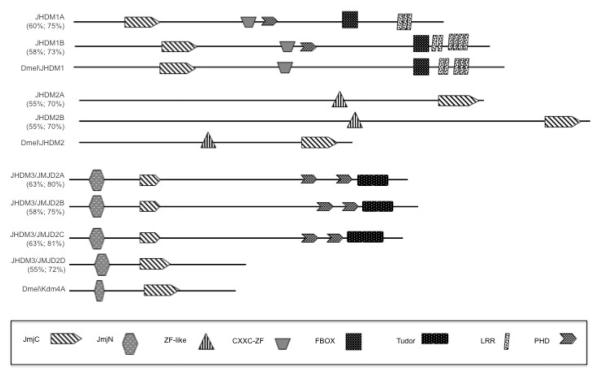
Figure 2. Schematic representation of histone demethylase families.
Dmel\JHMD1, Dmel\JHMD2 and Dmel\Kdm4A are each highly conserved with their human homolog counterparts. Shown is a schematic representation (drawn to scale) of the conserved domains and their location within each of the JHMD1, JHMD2 and Dmel\Kdm4A family members. Structural domains and locations were obtained at CDD/NCBI. Numbers represent percentage identity and similarity with respect to the corresponding Drosophila homolog. The positions of the JumonjiC (JmjC) and JumonjiN (JmjN) domains are indicated. Zinc-like finger, CXXC-zinc finger, PHD and tudor domains and their locations are also shown.
Figure 1. Dmel\Kdm4A DNA sequence.
Alignment of the Dmel\Kdm4A cDNA and sequence found at NCBI. Mismatches are indicated in blue, start codon in green and stop codon in red.
Analysis of the conceptual translation product for the CG15835 gene (designated Dmel\Kdm4A) indicated that this isolated Drosophila gene is the homolog of the human JMJD2 family. First, an alignment between the Dmel\Kdm4A and each of the human JMJD2D proteins demonstrated significant homology over their entire coding sequences (Figure 2). Structural protein data obtained using the conserved domain architecture retrieval tool (CDART) at NCBI demonstrated that the predicted protein domains specific for Dmel\Kdm4A and their locations within the protein are highly conserved between the human JMJD2D protein and Dmel\Kdm4A; both human family member JMJD2 and Dmel\Kdm4A each contain JmjN and JmjC domains within their N-termini and do not contain the C-terminal PhD and Tudor domains that JMJD2A-C contain. However, despite the strong structural similarity between human JMJD2D and Dmel\Kdm4A, Dmel\Kdm4A displays the highest amino acid conservation with JMJD2A: 63% identity and 80% similarity and JMJD2C: 63% identity and 81% similarity (Figure 2). Taken together, our data strongly indicate that the Dmel\Kdm4A gene is homologous to the human JMJD2 family.
Drosophila homologs of the three main JHDM families are each expressed during Drosophila development
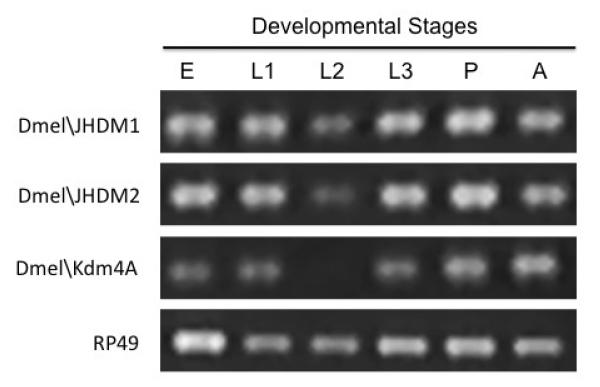
To determine whether additional JHDM family members are also present in Drosophila, we carried out data base searches which revealed the presence of Drosophila homologous sequences corresponding to JHDM1 and JHDM2. Analysis of the conceptual protein sequence of each gene indicated the presence of the distinct conserved domains specific for their classification (Figure 2). To determine whether these genes were expressed during the Drosophila lifecycle, RNA was isolated from staged Drosophila melanogaster (12-24 h staged embryos, first, second and third instar larvae, pupae, adult flies) and DNaseI treated. cDNAs were generated for each developmental stage by RT priming with random hexamers and the RT products were amplified using PCR with primer pairs specific for each HDM. Importantly, primers amplifying the gene for ribosomal protein RP49 were used as an internal control. In general, we found that each of the three HDM transcript levels were present during the Drosophila lifecycle (Figure 3). These data demonstrate that Drosophila contains actively transcribed homologous genes for each of the human Jmc family member homologs analyzed, and that each of these JHDM genes are expressed during the Drosophila life cycle.
Figure 3. Dmel\JMHD1, Dmel\JMHD2 and Dmel\Kdm4A are each differentially expressed during Drosophila development.
Semi-quantitative real-time PCR analysis of Dmel\JMHD1, Dmel\JMHD2 and Dmel\Kdm4A transcript levels using stage specific Drosophila melanogaster cDNAs (12-24 h staged embryos (E), first (L1), second (L2) and third (L3) instar larvae, pupae (P), adult (A) flies) prepared by RT priming of equal amounts of DNase treated RNA with random hexamers and PCR primer sets amplifying 400 bp regions specific for each HDMs. −RT controls were used for each sample. All experiments were repeated at least 3 independent times with consistent results.
Disruption of the Dmel\Kdm4A gene causes a male-specific wing extension/twitching phenotype
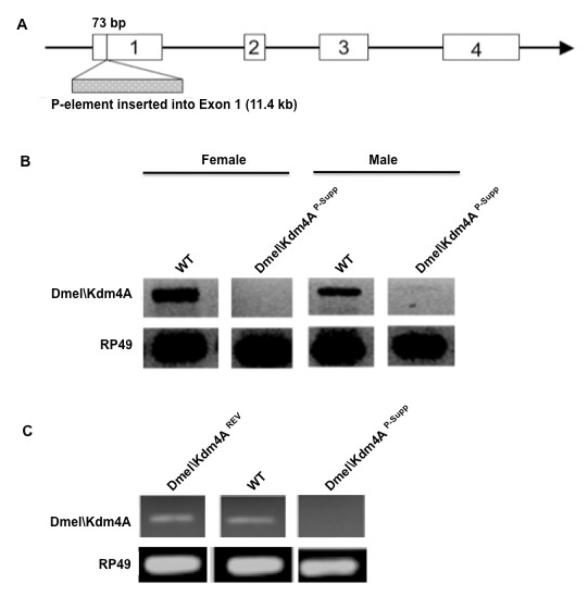
To decipher the cellular function of Dmel\Kdm4A during Drosophila development, we identified a P-element insertion fly line from the Flybase collection that contained a single 11.4 kb P-element inserted 76 bp downstream of the Dmel\Kdm4A start codon (Figure 4A). Sequence analysis confirmed that this was a single P-element insertion that disrupted only the Dmel\Kdm4A ORF and that no other genes were located in close proximity of the insertion site (Figure 4B). RT-PCR analysis using RNA isolated from male and female adult flies with primers spanning the entire Dmel\Kdm4A ORF demonstrated that Dmel\Kdm4A transcripts were completely absent in female flies and were significantly reduced in males (Figure 4B). These results confirmed that this P-element insertion disrupted Dmel\Kdm4A gene transcription and we designated this fly line Dmel\Kdm4AP-Supp. The characterization of this fly line provided us with the opportunity to study the biological function of Dmel\Kdm4A in the Drosophila multicellular developmental model setting.
Figure 4. Characterization of Dmel\Kdm4AP-supp and Dmel\Kdm4AREV fly lines.
(A) Schematic representation of P-element location within the Dmel\Kdm4A locus. (B) Semi-quantitative RT-PCR analysis of transcript levels in Dmel\Kdm4AP-Supp and Canton S flies. RNA was isolated from either male or female adult flies and equal amounts of RNA for each sample was subjected to cDNA preparation using RT priming with random hexamers and PCR using primer sets spanning the Dmel\Kdm4A open reading frame (ORF).
(C) Semi-quantitative RT-PCR analysis of transcript levels in Dmel\Kdm4AP-supp and Dmel\Kdm4AREV A flies. All experiments were repeated at least 3 independent times with consistent results and similar results were obtained for Dmel\Kdm4AREV B.
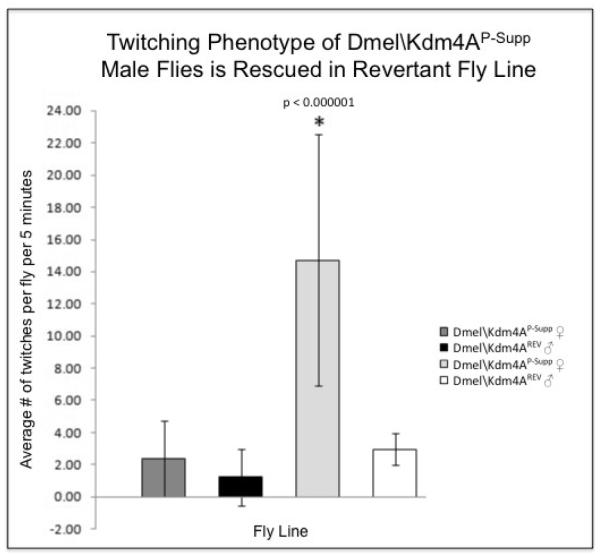
Initial characterization of the Dmel\Kdm4AP-Supp flies revealed that the flies displayed a twitching of their wings when compared to wild-type Canton S flies. The Dmel\Kdm4AP-Supp flies extended and shook their wings in quick succession, making it appear as if their wings were twitching. This phenotype appeared at approximately five days of age and appeared to become more apparent as the flies aged. To confirm that the twitching phenotype we observed was specifically due to lack of Dmel\Kdm4A transcript, we re-mobilized the P-element to excise it from the Dmel\Kdm4A gene. Two independent lines of flies, shown by sequence analysis to carry precise excisions (designated Dmel\Kdm4AREV A and Dmel\Kdm4AREV B ) restored Dmel\Kdm4A transcript levels back to those comparable to wild-type flies (Figure 4C). Initial observation revealed that both the precise excision independent fly lines showed no evidence of the twitching phenotype. Importantly, a quantitative twitching assay of staged 5 day old flies (the stage when the phenotype is first readily observable) revealed that the average number of wing extension/twitches per Dmel\Kdm4AP-Supp male flies was significantly higher than that of female Dmel\Kdm4AP-Supp flies and both male and female Dmel\Kdm4AREV A control flies (Figure 5). Remarkably, this careful behavioral analysis revealed that the male-specific Dmel\Kdm4AP-Supp wing extension/twitching we observed was identical in appearance to the previously described unilateral wing extension and vibration used to produce the ritualistic courtship song (Billeter et al., 2006; Certel et al., 2007; Dickson, 2008) and did not occur randomly, but almost exclusively as a result of male specific interaction. This behavior did not appear to be aggressive in nature, as no fencing, indicative of aggressive behavior, was observed (Certel, 2007 PNAS). Importantly, when 10 males and 10 females were observed in a vial, the male wing extension/twitching behavior was almost exclusively directed in response to male and not female flies, indicating male preference in this behavior. These results indicated that the male-specific reciprocal wing extension/ twitching phenotype we observed was similar to an inter-male wing extension/singing courtship behavior, and resulted directly from the P-element disruption of the Dmel\Kdm4A gene as the phenotype could be rescued by precisely excising the P-element to restore wild-type Dmel\Kdm4A transcript levels (Figure 5).
Figure 5. Disruption of Dmel\Kdm4A gene results in a twitching phenotype.
Staged 0-24 hour Dmel\Kdm4AP-Supp and Dmel\Kdm4AREV A flies males and females were collected in separate vials and allowed to acclimate for 4 days. 10 vials containing 3 male flies and 10 vials containing 3 female flies were observed and the number of times the flies twitched was counted over 5 minutes. The number obtained in each vial was divided by 3 to calculate average number of twitches per fly.
Normal Drosophila displays a negative geotactic response in that when they are tapped to the bottom of a vial, they rapidly climb to the top and remain there (Feany and Bender, 2000). This natural response is compromised due to aging and defects in neurological and muscle processes. The climbing assay is widely used to quantitate the severity of defects in such processes as well as to monitor the progression of severity in a number of Drosophila neurological disease models (Chan and Bonini, 2000; Chen and Feany, 2005) including Alzheimer’s(Crowther et al., 2004), Parkinson’s(Feany and Bender, 2000) and Huntington’s(Agrawal et al., 2005) disease. To test the climbing ability of the Dmel\Kdm4AP-Supp flies, 40 male or female Dmel\Kdm4AP-Supp flies and 40 male or female Dmel\Kdm4AREV A control flies were placed in separate plastic vials and gently tapped to the bottom of each vial. The number of flies at the top of the vial was counted after 18 seconds of climbing. This climbing assay was performed at 3, 5, 8, 10 and 14 days of age. The results revealed that Dmel\Kdm4AP-Supp flies showed no significant loss of climbing ability for male or female flies when compared to the revertant flies at all time points (our unpublished results). Immunostaining of mutant and wild-type embryos with antibodies specific for either the differentiated neuronal marker protein elav or the glial cell marker protein repo revealed a normal neuronal staining pattern for all embryonic stages observed, indicating normal embryonic neuronal formation in the Dmel\Kdm4AP-Supp flies (our unpublished results).
Disruption of the Dmel\Kdm4A gene leads to a reduction in the male lifespan
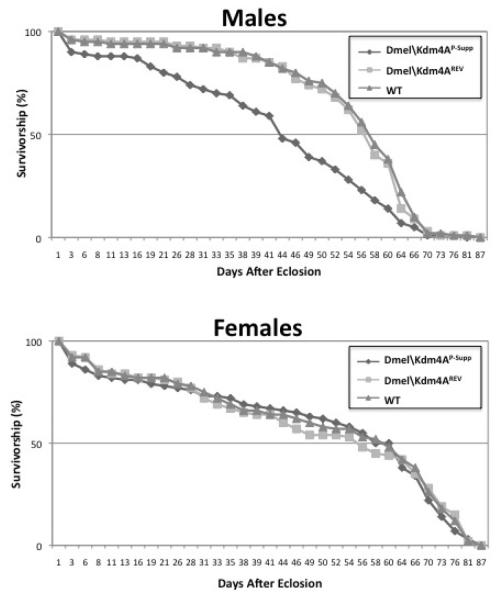
We did not observe a significant decrease in Dmel\Kdm4A P-supp viability when compared to Dmel\Kdm4AREV flies. Therefore, we assessed the effects of the Dmel\Kdm4A mutation on fly lifespan. An equal number of staged 0-24 hour Dmel\Kdm4AP-Supp and control Dmel\Kdm4A REV A flies were transferred to grape juice agar plates in a collection chamber. Importantly, male and female flies used in this experiment were grown separately directly after their eclosion to eliminate the affect of mating on longevity. The plate was changed daily with fresh yeast paste over a period of 87 days with the number of dead flies per day recorded. The results of this assay were graphed as survival curves for each of the fly lines (Figure 6). We observed a significant reduction in the lifespan for male flies and no significant reduction for female flies, indicating that disruption of the Dmel\Kdm4A gene reduces the fly life-span in a male specific fashion.
Figure 6. Disruption of Dmel\Kdm4A results in a reduction in male specific longevity.
Survival curves of male (A) and female (B) flies that were separately reared after eclosion at 25°C. Mt is Dmel\Kdm4AP-Supp and WT is Dmel\Kdm4AREV A control flies. Flies were maintained in embryo collection chambers at 25°C. The flies were changed each day and the number of dead flies was recorded per day. The data was analyzed using 2-way ANOVA with SAS programming and Microsoft Excel.
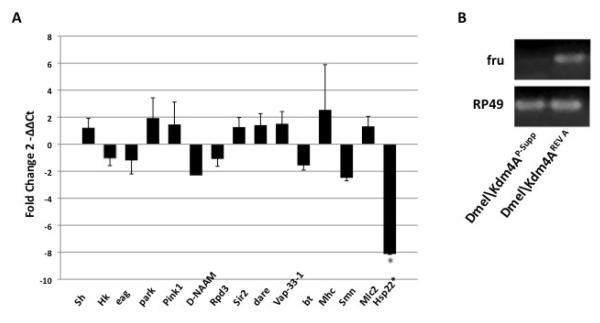
Specific genes associated with mutant Dmel\Kdm4AP-Supp phenotypes are significantly downregulated in response to Dmel\Kdm4A loss
Histone methylation patterns within the genome play an important role in establishing and maintaining specific gene expression profiles required for proper cell function (Lachner et al., 2001; Fischle et al., 2003). To investigate a potential molecular basis underlying the twitching and longevity defects we observed in the Dmel\Kdm4AP-Supp flies, we asked whether loss of Dmel\Kdm4A resulted in misexpression of genes known to be associated with such phenotypes. The mRNA levels of 16 specific genes from Dmel\Kdm4AP-Supp and Dmel\Kdm4AREV A A flies was assessed by quantitative real time PCR and the fold change in gene expression levels between the two fly lines was determined (Figure 7). As the twitching and longevity phenotypes we observed were male specific and intensified with age, we chose to use staged 21 day old adult males for RNA analysis to enhance our opportunity to detect potential changes in gene expression associated with these defects. The putative Dmel\Kdm4A target genes chosen to be assessed were: Shaker (Sh) (Wang et al., 2000; Cirelli et al., 2005), Hyperkinetic (Hk) (Ueda and Wu, 2008), and ether a go-go (Zhong and Wu, 1993) chosen for their involvement in K+ channel function and shown to display a shaking leg phenotype when mutated; park and pink1, involved in Parkinson disease (Greene et al., 2003; Tan and Dawson, 2006); Drosophila Nicotinamidase (D-NAAM), Silent information regulator 2 (Sir2), and rpd3, selected for their involvement in a deacetylase-mediated longevity pathway (Rogina and Helfand, 2004); bent (bt) and myosin heavy chain (Mhc), each involved in muscle function (Redowicz, 2002); defective in the avoidance of repellents (dare) (Freeman et al., 1999), Vap-33-1 (DVAP-33A) (Chai et al., 2008)}, and survival motor neuron (SMN) (Chan et al., 2003) each involved in appropriate neuromuscular junction (NMJ) function; Heat shock protein 22 (Hsp22), the mitochondrial small heat shock protein involved in stress and aging (Morrow et al., 2004); and fruitless (fru) involved in male-specific neuron formation that promotes masculinization (Dickson, 2002; Dickson, 2008; Yamamoto, 2008) . The results of our analysis demonstrated that out of the 16 genes assessed, two of the genes (Hsp22 and fru) were significantly affected, with a marked decrease in mRNA levels for each of them. Of note, the fru gene was the most significantly downregulated (Figure 7A), with fru mRNA levels so low in the Dmel\Kdm4AP-Supp flies, that they were undetectable in our qPCR assay and thus their downregulation was confirmed using RT-PCR and gel electrophoresis analysis (Figure 7B). That only certain genes were affected indicate that the gene changes we observed were specific. Additionally, Dmel\Kdm4A target gene downregulation in response to Dmel\Kdm4A loss is consistent with the function of Dmel\Kdm4A as an enzyme with potent histone demethylase activity for the removal of specific methyl groups from chromatin environments marked for repression (Shi, 2007; Lin CH, 2008; Wallrath and Elgin, 2008). Significantly, each of the affected genes was associated with the wing twitching and longevity phenotypes we observed in our Dmel\Kdm4AP-Supp mutant fly line. For example, Hsp22 has been proposed to be involved in the aging process (Morrow et al., 2004). Quite notably, the most downregulated gene, fru, functions in male specific neuronal processing involved in masculinization, with fru fly mutants displaying inter-male courtship behaviors consistent with the inter-male courtship song behaviors we observed in our Dmel\Kdm4AP-Supp mutant flies (Billeter et al., 2006) . Taken together, our results support an essential role for Dmel\Kdm4A in the transcriptional activation of genes involved in the aging process and male- specific neuronal formation and courtship behavior.
Figure 7. Specific genes associated with Dmel\Kdm4AP-Supp male-specific twitching and longevity pheonotypes are significantly downregulated in response to Dmel\Kdm4A loss.
(A) Shown is a histogram depicting qPCR analysis of the expressionof the indicated genes in staged 21 day old male Dmel\Kdm4AP-Supp and Dmel\Kdm4AREV flies. The relative fold change in mRNA expression levels were measured using the comparative Ct method with rp49 as the internal control gene. Error bars represent standard deviation. Astrics (*) indicates significant fold changes between Dmel\Kdm4A mutant and revertant flies with values of p < 0.05.
(B) Semi-quantitative RT-PCR analysis of fruitless transcript levels in Dmel\Kdm4AP-supp and Dmel\Kdm4AREV A flies. All experiments were repeated at least 3 independent times with consistent results.
DISCUSSION
Using Drosophila, we describe the consequences of eliminating Dmel\Kdm4A function in an animal model. Our results help to place the previously described biochemical activities and certain functional activities of JMJD2 into a developmental context. To investigate the role of Dmel\Kdm4A during development, we identified a P-element insertion fly line in the FlyBase collection (designated Dmel\Kdm4AP-Supp), and confirmed that it disrupted Dmel\Kdm4A expression (Figure 4A). Our creation of two precise P-element excision lines (designated Dmel\Kdm4AREVA and Dmel\Kdm4AREVB ) restored transcripts to wild-type levels, making our Dmel\Kdm4AP-supp and Dmel\Kdm4AREV fly lines a powerful multicellular model system to explore developmental Dmel\Kdm4A function. Importantly, while this work was in progress, other groups also identified the Dmel\Kdm4A gene and demonstrated by overexpression assays the ability of this enzyme to specifically demethylate H3-K36 in vivo in flies and in Drosophila S2 cell lines (Lin CH, 2008; Lloret-Llinares et al., 2008) and specifically demethylate H3K36me2 and H3K36me3 both in vitro and in vivo (Lin CH, 2008). These studies confirm the demethylation activity of Dmel\Kdm4A in Drosophila and strongly indicate that the cause of the phenotypes we describe here for the Dmel\Kdm4AP-supp fly line is due to an imbalance of histone methylation in tissues and developmental stages where Dmel\Kdm4A transcripts are lacking.
When first characterizing the Dmel\Kdm4AP-supp fly line, we observed that the Dmel\Kdm4AP-Supp flies exhibited a wing extension/twitching phenotype. Quantitative analysis of this phenotype revealed that the average number of twitches per Dmel\Kdm4AP-Supp male fly was significantly higher than that of female Dmel\Kdm4AP-Supp flies and both male and female Dmel\Kdm4AREV A control flies, indicating that the twitching phenotype was male specific and caused by disruption of the Dmel\Kdm4A gene. Previous studies had demonstrated that mutations in genes that encode voltage gated ion channels and are associated with electrical signal transmission, display a twitching phenotype (Wang et al., 2000) and thus we reasoned that the Dmel\Kdm4AP-supp twitching phenotype may have originated from similar neurological defects. However, unlike these mutant fly lines, we observed no significant defects in the ability of either Dmel\Kdm4AP-supp male and female flies to perform the climbing assay, a test used to monitor neurological defects (Chan and Bonini, 2000; Chen and Feany, 2005). We also did not detect any gross abnormalities in embryonic CNS and PNS development as assessed by immunohistochemical staining of embryonic glial cells, neuronal cells and embryonic axonal cytoskeleton formation, consistent with the defect being observed in the adult fly. Moreover, qPCR analysis revealed that expression levels of the major genes involved in voltage-gated ion-channel formation were unaffected in the Dmel\Kdm4AP-Supp mutant flies when compared to revertants (Figure 7). Taken together, these results indicated that the wing extension/twitching phenotype in Dmel\Kdm4AP-Supp flies was reminiscent of another biological pathway.
A detailed behavioral analysis of the mutant flies revealed that the male-specific Dmel\Kdm4AP-Supp wing extension/twitching we observed was not random, but occurred almost exclusively in response to the presence of other males and not females, demonstrating an inter-male preference for this behavior. Further observation revealed that the behavior was identical in nature to a central component of the courtship ritual, the courtship song, which is produced by a visible unilateral wing extension and vibration and is commonly used as a measurable readout of the male’s decision to court (Dickson, 2008). This behavior was often reciprocal in nature between the males, did not appear to be aggressive, as absolutely no fencing, indicative of aggressive behavior, was observed (Certel et al., 2007) and was identical in nature to previous studies describing inter-male wing extension courtship behaviors (Certel et al., 2007; Clyne and Miesenbock, 2008). Phenotypes involving male-male courtship preference have been well characterized in the fly and predominantly result from disruption of the fruitless (fru) gene, shown to play a prominent role in the development of appropriate male sexual behavior. The transcriptional regulation of the fru gene is complex, in that the single fru gene contains four different promoters, P1, P2, P3, and P4 that each encode closely related BTB/POZ (Broad complex, Tramtrack, and Bric-a-brac/Poxvirus and Zinc finger)-Zn finger (ZnF) proteins, that likely act as transcription factors (Song et al., 2002). The function of fru in directing male-specific sex determination depends on transcripts initiated from the P1 promoter (Song et al., 2002). These transcripts are sex-specifically spliced, and subsequently translated into male-specific FruM proteins that directs the formation of the masculinized P1 neuronal cluster in male flies (Kimura et al., 2008; Yamamoto, 2008) . Transcripts produced from promoters P2-P4 function in sex-nonspecific roles in axonal pathfinding (Song et al., 2002). Remarkably, qPCR analysis using primers designed to detect fru transcripts revealed almost a complete absence of these transcripts in the male Dmel\Kdm4AP-Supp flies (Figure 7 B). Our finding that fru transcripts are almost absent in male Dmel\Kdm4AP-Supp flies, and that these flies exhibit inter-male courtship behavior is consistent with previous studies demonstrating that mutations at the fru locus that lead to inter-male courtship behavior are always associated with a global reduction in the levels of fru gene expression (Billeter et al., 2006). Interestingly, the inter-male courtship behavior we observed was confined to the wing extension/courtship song stage of the well characterized courtship repertoire (Billeter et al., 2006). Moreover, although the flies distinctly exhibit male preference in performing this step of the courtship sequence, Dmel\Kdm4AP-Supp flies do produce offspring, indicating that male-female mating does take place. This observation is consistent with previous studies demonstrating that different fru mutant flies exhibit courtship abnormalities to different degrees and at separate stages of the courtship sequence depending on the mutant allele (Villella et al., 1997). Importantly, although the molecular steps leading to the production of male specific FruM proteins via sex-specific differential splicing of fru P1 transcripts is well characterized, the molecular mechanism(s) underlying how different P1-P4 initiated fru isoforms are spatially and temporally regulated remain unclear. Our findings have important implications for Dmel\Kdm4A in the control of fru gene expression, possibly by controlling certain regulators of the fru gene, or by directly modulating histone methylation levels at P1-P4 gene regulatory regions in certain cell types that results in the initiation and/or maintenance of the differential production and levels of different fru transcripts, a model we can now explore with the use of our characterized Dmel\Kdm4A fly lines.
Our longevity assays revealed that the lifespan of the male and not female Dmel\Kdm4AP-supp flies was significantly reduced. Intriguingly, this male-specific reduction in lifespan is consistent with studies of the HDM LSD1 in Drosophila demonstrating that reduction of LSD1 leads to a reduction in fly viability that is more severe in male flies (Di Stefano et al., 2007). It is known that lifespan in Drosophila is influenced by a number of factors including temperature, starvation and caloric restriction (Rogina et al., 2002), oxidative stress (Mourikis et al., 2006), mating (Aigaki and Ohba, 1984), and certain gene mutations (Rogina et al., 2000). Moreover, a number of studies support a role for the epigenetic regulators Sir2 and Rpd3 in controlling longevity. These histone deacetylases (HDACs) influence longevity through a pathway related to calorie restriction (Rogina et al., 2002; Rogina and Helfand, 2004). Calorie restriction triggers downregulation of Rpd3 and upregulation of Sir2 activity, leading to the extension of lifespan in Drosophila, presumably via changes in HDAC production that influences gene expression profiles that control longevity. Here, we show that although levels of Rpd3 and Sir2 gene expression are unaffected in response to Dmel\Kdm4A loss, there is a significant reduction in mitochondrial Hsp22 transcript levels. Notably, disruption of the mitochondrial Hsp22 gene in flies results in a decrease in longevity while overexpression of the gene in all cells or motoneurons increases fly lifespan. Thus, our results suggest that JMJD2 is directly or indirectly involved in regulating the aging process via Hsp22 controlled pathways (Morrow et al., 2004).
Several studies have demonstrated connections between regulation of histone methylation and neurological disorders. Specifically, SMCX, a member of the H3K4me3-specific demethylase family, has been linked to X-linked mental retardation (XLMR) (Tzschach et al., 2006). Morevover, neuron specific genes are misexpressed due to histone demethylase LSD1 knockdown (Di Stefano et al., 2007). Intriguingly, the fru gene, also significantly down-regulated in response to Dmel\Kdm4A loss (Figure 7), plays an essential role in neurogenesis by directing the correct formation and positioning of a male-specific neuronal cluster termed P1, that is located in the dorsal posterior brain and directs typical male fly behavior and courtship (Yamamoto, 2008) . It has been recently postulated that fru determines the level of masculinization of these neurons by regulating the transcription of a set of downstream target genes. Thus, it is tempting to speculate that in our Dmel\Kdm4AP-Supp flies there are additional neuronal genes, particularly those involved in P1 neurite formation, that are also misexpressed, resulting in disruption of the masculinized P1 cluster (Yamamoto, 2008). Our development and characterization of the Dmel\Kdm4AP-supp and Dmel\Kdm4AREV fly lines now provide a powerful multicellular model system to further explore the biological function of JMJD2 in controlling such gender-specific behavioral and neuronal processes.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Daniel Marenda and Dr. Xianmin Zhu for their invaluable advice and technical support. This work was supported by a National Institute of Health grant 1R01HD057939 to F.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- The FlyBase database of the Drosophila Genome Projects and community literature The FlyBase Consortium. Nucleic Acids Res. 1999;27:85–8. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Pallos J, Slepko N, Apostol BL, Bodai L, Chang LW, Chiang AS, Thompson LM, Marsh JL. Identification of combinatorial drug regimens for treatment of Huntington’s disease using Drosophila. Proc Natl Acad Sci U S A. 2005;102:3777–81. doi: 10.1073/pnas.0500055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigaki T, Ohba S. Effect of mating status on Drosophila virilis lifespan. Exp Gerontol. 1984;19:267–78. doi: 10.1016/0531-5565(84)90022-6. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–8. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Billeter JC, Rideout EJ, Dornan AJ, Goodwin SF. Control of male sexual behavior in Drosophila by the sex determination pathway. Curr Biol. 2006;16:R766–76. doi: 10.1016/j.cub.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–9. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, Savella MG, Schlegel DC, Kravitz EA. Modulation of Drosophila male behavioral choice. Proc Natl Acad Sci U S A. 2007;104:4706–11. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai A, Withers J, Koh YH, Parry K, Bao H, Zhang B, Budnik V, Pennetta G. hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum Mol Genet. 2008;17:266–80. doi: 10.1093/hmg/ddm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HY, Bonini NM. Drosophila models of human neurodegenerative disease. Cell Death Differ. 2000;7:1075–80. doi: 10.1038/sj.cdd.4400757. [DOI] [PubMed] [Google Scholar]
- Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trulzsch B, Sattelle DB, Davies KE, van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–76. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chen L, Feany MB. Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci. 2005;8:657–63. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Clyne JD, Miesenbock G. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell. 2008;133:354–63. doi: 10.1016/j.cell.2008.01.050. [DOI] [PubMed] [Google Scholar]
- Crowther DC, Kinghorn KJ, Page R, Lomas DA. Therapeutic targets from a Drosophila model of Alzheimer’s disease. Curr Opin Pharmacol. 2004;4:513–6. doi: 10.1016/j.coph.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol. 2007;17:808–12. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–64. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Wired for sex: the neurobiology of Drosophila mating decisions. Science. 2008;322:904–9. doi: 10.1126/science.1159276. [DOI] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–8. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol. 2002;12:389–98. doi: 10.1016/s1044-579x(02)00059-7. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–53. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Dobritsa A, Gaines P, Segraves WA, Carlson JR. The dare gene: steroid hormone production, olfactory behavior, and neural degeneration in Drosophila. Development. 1999;126:4591–602. doi: 10.1242/dev.126.20.4591. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–69. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lin CH LB, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Molecular Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res. 2008;36:2852–63. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–76. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Martin C.a.Z. Y The diverse functions of histone lysine methylation. Nature Review Molecular Cell Biology. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004;279:43382–5. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Hurlbut GD, Artavanis-Tsakonas S. Enigma, a mitochondrial protein affecting lifespan and oxidative stress response in Drosophila. Proc Natl Acad Sci U S A. 2006;103:1307–12. doi: 10.1073/pnas.0510564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 2000;14:3003–13. doi: 10.1101/gad.848800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redowicz MJ. Myosins and pathology: genetics and biology. Acta Biochim Pol. 2002;49:789–804. [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–73. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- Rogina B, Reenan RA, Nilsen SP, Helfand SL. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–40. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–33. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun. 2007a;359:742–6. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007b;353:973–7. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- Song HJ, Billeter JC, Reynaud E, Carlo T, Spana EP, Perrimon N, Goodwin SF, Baker BS, Taylor BJ. The fruitless gene is required for the proper formation of axonal tracts in the embryonic central nervous system of Drosophila. Genetics. 2002;162:1703–24. doi: 10.1093/genetics/162.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JM, Dawson TM. Parkin blushed by PINK1. Neuron. 2006;50:527–9. doi: 10.1016/j.neuron.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tzschach A, Lenzner S, Moser B, Reinhardt R, Chelly J, Fryns JP, Kleefstra T, Raynaud M, Turner G, Ropers HH, Kuss A, Jensen LR. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat. 2006;27:389. doi: 10.1002/humu.9420. [DOI] [PubMed] [Google Scholar]
- Ueda A, Wu CF. Effects of hyperkinetic, a beta subunit of Shaker voltage-dependent K+ channels, on the oxidation state of presynaptic nerve terminals. J Neurogenet. 2008;22:1–13. doi: 10.1080/01677060701807954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Gailey DA, Berwald B, Ohshima S, Barnes PT, Hall JC. Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics. 1997;147:1107–30. doi: 10.1093/genetics/147.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC. Stimulating conversations between HP1a and histone demethylase dKDM4A. Mol Cell. 2008;32:601–2. doi: 10.1016/j.molcel.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Humphreys JM, Phillips JP, Hilliker AJ, Wu CF. A novel leg-shaking Drosophila mutant defective in a voltage-gated K(+)current and hypersensitive to reactive oxygen species. J Neurosci. 2000;20:5958–64. doi: 10.1523/JNEUROSCI.20-16-05958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Yamamoto D. Brain sex differences and function of the fruitless gene in Drosophila. Journal of Neurogenetics. 2008;15:1–24. doi: 10.1080/01677060802298491. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN., Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wu CF. Modulation of different K+ currents in Drosophila: a hypothetical role for the Eag subunit in multimeric K+ channels. J Neurosci. 1993;13:4669–79. doi: 10.1523/JNEUROSCI.13-11-04669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]