Summary
Vectors based on the adeno-associated virus are attractive and versatile vehicles for in vivo gene transfer. The virus capsid is the primary interface with the cell that defines many pharmacological, immunological and molecular properties. Determinants of these interactions are often restricted to a limited number of capsid amino acids. In this study, a portfolio of novel AAV vectors was developed following a structure-function analysis of naturally occurring AAV capsid isolates. Singletons, which are particular residues on the AAV capsid that were variable in otherwise conserved amino acid positions were found to impact on vector's ability to be manufactured or to transduce. Data for those residues that mapped to monomer-monomer interface regions on the particle structure suggested a role in particle assembly. The change of singleton residues to the conserved amino acid resulted in the rescue of many isolates that were defective upon initial isolation. This led to the development of an AAV vector portfolio that encompasses 6 different clades and 3 other distinct AAV niches. Evaluation of the in vivo gene transfer efficiency of this portfolio following intravenous and intramuscular administration highlighted a clade-specific tropism. These studies further the design and selection of AAV capsids for gene therapy applications.
Keywords: gene transfer, gene therapy, adeno-associated viruses, AAV, vector, capsid, muscle, liver
Introduction
Adeno-associated virus (AAV) is a ssDNA virus from the Parvoviridae family.1 Initially AAV serotype 2 was considered as a vehicle for gene transfer in various in vitro and in vivo experimental and therapeutic settings.2 More recently, several alternative serotypes have been used as vectors for gene therapy.3 One impetus for exploring the diversity of AAV biology is the remarkable differences in gene transfer and vector tropism observed between the AAV serotypes. Enhanced gene transfer efficiency of alternative capsids in small and large animal models may overcome hurdles towards successful therapeutic gene transfer.4–6 Mechanisms underlying the distinct biological performance of different AAV serotypes have been attributed to differences in viral entry, trafficking, uncoating and genome processing.7–9
By definition, serotypes distinguish themselves in their susceptibility to serological neutralization and the lack of cross-neutralization, a property that is determined by the absence of shared antigenic determinants on the exposed viral surface.10 In humans, a common host for AAV natural infection, neutralizing antibodies (NAB) are highly prevalent to several commonly used serotypes.11 Vector administration in the presence of circulating neutralizing antibodies can significantly impact the efficiency of gene transfer, an important hurdle that alternative serotypes can possibly circumvent.11–13
The control of cellular and humoral responses against the transgene product is pivotal for any safe and efficacious gene therapy. The capsid serotype may play a determining role in generating a tolerogenic environment in order to achieve persistent expression of transgene product.14 AAV serotype vectors also differentiate themselves in their propensity to activate T cells against their capsid through processes of cross presentation.15
Characterization of the diversity in capsid sequences from latent AAV genomes identified over 100 AAV isolates from humans and other nonhuman primates (NHPs) that clustered into phylogenetically related clades.10,16–21 The majority of the sequence variation is located on the external surface regions of the capsid in select hypervariable domains.20,22–24 These regionscomprise approximately 19% of the viral capsid structure leaving the majority of viral architecture largely conserved.22,23 Even minimal variation on the AAV capsid can phenotypically alter vector biology. For example, limited amino acid substitutions due to natural variation confer heparin affinity and liver tropism onto AAV6 25 and AAV2.26 Within a single clade, capsid variants have also been shown to demonstrate dramatically different in vivo gene transfer following intrapleural administration.27
AAV vector biology is largely determined by the capsid, its primary point of interaction with the host. Indeed, the use of alternative or modified capsids has been a valuable tool in increasing vector efficiency, altering tropism, evading serological neutralization or minimizing immunological responses.3,6,15 We and others have previously used the natural diversity of AAV as a source to identify vectors with novel biology.10,21,28–31 This approach led to the identification of 3 novel AAV serotypes and yielded over 100 isolates from various sources of primate tissues for vector development.
In this report, the available sequence dataset was analyzed in relation to the functional aspects of the respective isolated viral reagents. Subsequent mutational studies led to define the direct and/or indirect impact on AAV vector biology of minor modifications in the primary and tertiary structure. These data were used to design and optimize an AAV vector portfolio with a broad representation of structural and phylogenetic distinct AAVs. This expanded set of AAV vectors was tested for in vitro transduction of primary neuronal cultures32, was used to identify the optimal vector for gene transfer to conducting airway33 and was evaluated for capsid-specific variation in in vivo tropism in the CNS34 and the eye.35 Here, in addition to the development of the AAV portfolio, we present the evaluation of these viral vectors in an in vivo model for both muscle- and liver-directed gene transfer.
Material and Methods
Sequences and cloning
All novel and modified AAV sequences were deposited in Genbank. Most wild type isolate sequences were described previously.10,20,21 Site-directed mutagenesis was done with Quickchange from Stratagene for all but one of the modified capsid sequences and confirmed by sequencing. The non-conservative modifications and the nomenclature of the subsequent clones are described in Table 1. Rh.32.33 was generated by cloning a BsiWI – BbvCI restricted rh.33 fragment into AAV2/rh.32. Novel capsid sequences were transferred from a PCR isolate into TOPO (Invitrogen) and then subsequently into an AAV packaging construct by partial restriction digestion at the first internal cap XhoI site on the 5' end and, depending on the orientation in the TOPO plasmid, a PmeI or EcoRV blunt end digestion.
Table 1.
Structure-function analysis of clade E vector subset.
| Isolate | Clade | Species of Isolation | Δhu.37 | No. of Singleton | Yield (× 103 GC/cell) | Gene Transfer (μg hAAT/ml) |
|---|---|---|---|---|---|---|
|
| ||||||
| hu.37 | E | Human | 0 | 0 | 50 | 57 |
| hu.41 | E | Human | 3 | 2 | 33 | 1.4 |
| rh.40 | E | Rhesus | 4 | 1 | 1 | n/a |
| AAV8 | E | Rhesus | 48 | 0 | 80 | 72 |
Δhu.37 identifies the number of residues different from hu.37. Yields represent numbers of GC produced per HEK293 cell following transfection. Gene transfer is measured following intraportal administration of 1 × 1011 GC AAV.CB.hAAT particles to NCR nude mice in the serum at peak expression.
AAV Vector Preparations
AAV vector preparations were done by triple transfection of 40 15 cm dishes of HEK293 cells as previously described.6 For small scale production, the transfection protocol was downscaled to a single 15 cm petridish using 13 μg AAV2/X packaging plasmid (X referring to the particular cap isolate), 13 μg cis transgene plasmid pZac2.1, and 26 μg pAdΔF6 Adenoviral helper construct. Small scale vector cultures were kept in DMEM with 10 % FBS for 24 h when media was replaced. 2 days later viral vector was harvested by scraping all cellular material into the medium supernatant. Following 3 subsequent freeze-thaw cycles (−80 °C to 37 °C), cellular debris was precipitated by low speed centrifugation. Benzonase resistant vector genomes were titered by TaqMan PCR amplification, using primer probes for the polyadenylation signal encoded in the transgene constructs used.46 The number of particles per producer cell is the ratio of the measured yields per preparation and number of transfected cells (2 × 107 per 15 cm petri dish). Different experiments were internally controlled with a parallel AAV2/8 vector preparation. For large scale production absolute yields were established subsequent to purification whereas small scale preparations were titered without further purification.
Sequence analysis
In frame nucleotide sequence alignment was done with the ClustalX algorithm in the BioEdit software.47 Singleton, phylogenetic and additional sequence analysis was performed in MEGA 3.1.48 Singleton sites were considered as per definition of the software manufacturer i.e. containing “at least two types of amino acids with, at most, one occurring multiple times. MEGA identifies a site as a singleton site if at least three sequences contain unambiguous amino acids”. Phylogenies were calculated with the Neighbor-Joining algorithm following pairwise deletion of gaps and making use of the Poisson corrected model for substitution. Percent differences in the capsid were calculated at a protein level for full VP1 Cap proteins after pairwise deletion of gaps and using 735 amino-acids as average VP1 length across all sequences.
Structural analysis
The coordinates for the crystal structure of AAV8 VP3 (23 PDB accession number 2QA0) were used to visualize the locations of mutated residues on the capsid using the program O 49 on a Silicon graphics Octane workstation. The icosahedral 2-, 3-, and 5-fold symmetry related VP3 monomers were generated in the program O by applying symmetry operators to a reference VP3 monomer. These symmetry related VP3 monomers enabled the positions of the mutated residues to be analyzed within the context of an assembled capsid. The locations of the mutated residues were then graphically presented using the program PyMOL.23
Animals
The structure-function analysis data were obtained following intraportal injection of male NCR Nude mice at a dose of 1011 genome containing (GC) particles. The comparison studies of singleton modified and novel vectors were done in male C57Bl/6 mice. Animals were injected with 1011 GC by intramuscular injection (i.m.) in the hind limb at two injection sites with a total injection volume of 50 μl. Intravenous tail vein injections (i.v.) were done with 1011 GC in a volume of 100 μl. Serum samples were obtained by retro-orbital venipuncture. Experimental protocols were approved by the Institutional Animal Care and Use Committee, and use of the vectors in the protocols was approved by the Environmental Health and Radiation Safety Office and the Institutional Biosafety Committees of the University of Pennsylvania.
Evaluation of gene transfer efficiency
Vectors expressing GFP driven by a CMV promoter were used to examine in vitro transduction efficiency in 293 cells. For in vivo studies, human α-antitrypsin (hAAT) was used as a quantitative reporter to evaluate vector performance. The expression of hAAT in the AAV vector is under the control of CMV-enhanced chicken β-actin promoter (CB). hAAT concentrations in serum were determined by an ELISA-based assay at various time points following vector administration.10,15
Results
Structure-function analysis
Over 100 AAV Cap sequences were isolated previously from primate tissue sources.10 Whereas highly functional vectors were identified in this effort, preliminary attempts to derive vectors from these isolates were often unsuccessful due poor vector production or gene transfer efficiency. In this study, 26 capsid isolates from primate tissue that were previously described10,20 were tested in an assay for vector production. In this analysis, 11 out of 26 (42%) yielded less vector than AAV2, the lowest yielding serotype in terms of production.
To identify the structural capsid determinants that impact on the apparent defect in vector production, AAV subsets were studied with highly homologous yet phenotypically divergent capsids. The vector yield from large scale vector preparations, in vitro gene transfer and in vivo gene transfer in NCR nude mice following intravenous injection (i.v.) were considered as indicators of function. As an example, three natural variants from the same clade were analyzed (Table 1). The isolate hu.37 was produced at similar titers as AAV8 which belongs to the same clade E yet diverges in 48 amino acids. In comparison to the other isolates (hu.41 and rh.40), hu.37 diverges in a maximum of only 4 amino acids and the vector was produced at much higher yields (50 × 103 GC/cell). Specifically, rh.40, distinct in 4 capsid amino acid residues from hu.37, is severely impaired in vector production. In addition, the in vivo gene transfer of hu.37 was comparable to AAV8 but dramatically superior to the other isolates (Table 1). The isolate, hu.41 has yields comparable to hu.37 but achieves only 2% of its in vivo transduction (Table 1).
To distinguish between amino acids that are tolerated in the capsid without loss in functionality versus those that are detrimental to vector function, we analyzed the degree of conservation of individual residues within the broader primate AAV family. The residues divergent between two homologous yet phenotypically distinct isolates were of particular interest. This analysis identified two types of divergent amino-acid residues. A first type is a variable residue at a position where other isolates also demonstrate divergence. Not surprisingly, many of these residues are located in previously described hypervariable regions.20,23 A second type of divergent residue is variable at otherwise conserved amino-acid positions. These residues are referred to as singletons and are located at what is called a singleton site on the capsid. For example, the aspartic acid in rh.40 at position 329 is not present in most other AAV variants of this clade that carry an asparagine at this location and was therefore defined as a singleton residue. Singleton sites were found throughout Cap and surprisingly located both within hypervariable domains as well as within structurally conserved domains of the capsid.
We hypothesized that the presence of one or more singleton residues on the AAV capsid can impact negatively on vector production and/or gene transfer efficiency. A meta-analysis of available sequence data provided additional support for this hypothesis. For example, in the AAV subset of Clade E vectors that was discussed earlier no singletons were identified for either of the fully functional hu.37 or AAV8. The isolates rh.40 and hu.41 contain 1 and 2 singletons respectively and also have a reduced ability to produce vector or transduce in comparison to hu.37 or AAV8 (Table 1). This analysis was further expanded to a dataset of 115 primate AAV sequences. Singletons were defined by comparison to sequences of a minimum homology of 80% (Supplement S1). AAVs isolated from NHPs contained an average of 1.51±1.10 (average ± standard deviation) singleton residues per VP1 while those from humans averaged to 1.72±1.33. Additional support for this hypothesis can be found in the fact that all but one serotype is free of singleton residues (data not shown). The exception is AAV6 which contained 2 singletons, F129L and K531E (Table 3). Remarkably, in position 129 of AAV6 all known primate AAVs have a leucine where AAV6 has a phenylalanine, with the exception of AAV5, the most distantly related primate AAV serotype (Supplement S1), that also encodes for phenylalanine at this location.
Table 3.
Impact of singleton residues on vector yields.
| Clade | Isolate | Parental | R1/R | R2 | R3 | R4 | R5 | .1 | .2 |
|---|---|---|---|---|---|---|---|---|---|
| A | AAV1 | 141 | |||||||
| hu.44 | 0.42 | 0.49 | 4 | 87 | |||||
| hu.46 | <0.04 | <0.04 | <0.04 | <0.04 | 0.11 | ||||
| hu.48 | 0.30 | 0.38 | 99 | 148 | |||||
| AAV6 | 38 | 110 | 118 | 42 | |||||
| B | AAV2 | 25 | |||||||
| hu.29 | 0.07 | 110 | |||||||
| hu.51 | 27 | ||||||||
| hu.13 | 118 | ||||||||
| C | hu.11 | 68 | |||||||
| hu.53 | 33 | ||||||||
| D | AAV7 | 141 | |||||||
| rh.48 | 1.86 | 76 | 13 | 19 | |||||
| rh.67 | 1.75 | 2.01 | |||||||
| rh.37 | 1.03 | 84 | 57 | 72 | |||||
| cy.5 | 103 | 106 | 103 | 99 | 87 | ||||
| E | AAV8 | 129 | |||||||
| rh.39 | 125 | ||||||||
| rh.10 | 80 | ||||||||
| hu.37 | 144 | ||||||||
| rh.43 | 99 | ||||||||
| rh.64 | 0.19 | 72 | 125 | ||||||
| rh.46 | 28 | ||||||||
| rh.58 | 3.57 | 0.42 | |||||||
| rh.20 | 61 | ||||||||
| rh.2 | 19 | 29 | |||||||
| pi.2 | 22 | ||||||||
| F | AAV9 | 133 | |||||||
| hu.32 | 182 | ||||||||
| Other | rh.8 | 80 | 120 | ||||||
| rh.32.33 | 0.49 | 210 | |||||||
| ch.5 | 57 | 33 |
Numbers represent average GC particles per producer cell (gc/cell) × 103) quantified by Taqman PCR (limit of detection: 0.04 gc/cell) for all of the wild type and capsid modified AAV vectors. Natural AAV isolates segregated per clade A–E or as other when no clade affiliation is available10,20 presented with singleton changed capsid variants. The suffix R refers to the number of sequentially changed singleton residues. For selected isolates, singleton changes were done individually (as opposed to sequentially in a cumulative fashion) and denoted with ".1" and ".2".
Ablation of singleton residues to generate capsid-modified AAV vectors
In order to refine the identification of singleton residues, two subsets of sequences within the AAV phylogeny were segregated based on overall amino acid identity. Cap sequences within each subset had more than 80% identity in primary amino acid sequence. Subset I comprises most serotypes and primate isolates with the exception of AAV4, AAV5 and some AAV isolates from non-primate sources (1, 18, 28) (Fig. 1). AAV4, the bovine AAVs (28) and an isolate similar to the rh.32 isolated from cynomolgus macaque (21) make up singleton subgroup II (Fig. 1). For other monophylic groups such as those containing AAV5, insufficient sequence information within its homology subgroup was available to perform this analysis. These subgroups based on homology are similar to subgroups that were structurally defined in transcapsidation studies by Rabinowitz, et al. making use of serotypes 1, 2, 3, 4 and 5.36
Figure 1. Dendrogram of primate vector portfolio with clade indication.
AAV VP1 Cap sequence Neighbor-Joining phylogeny of primate vector portfolio including all members of primate AAV portfolio in combination with selected mammalian AAV capsid sequences28–30. Isolates and serotypes are grouped according to phylogenetic clade taxonomy and singleton subsets for which members share 80% capsid homology. Bootstrap values (n=1000) are indicated at the branching nodes.
A total of 15 natural isolates with a number of singleton residues ranging from 1 to 5 were selected for further analysis. Through non-conservative site-directed mutagenesis and domain-swapping, singleton residues were altered to the conserved residue to evaluate the impact on vector performance. An overview of the specific mutations per isolate is presented in Table 2. Most capsid isolates underwent a sequential mutagenesis protocol with each vector named after the parental wild type followed by an R and the number of cumulative modified singletons. For example, human clade E isolate, hu.48 contains three singleton residues for which a first is mutated in hu48R1 (G277S). Subsequently hu48R1 was mutated in E322K and resulted in hu48R2. The third singleton was removed in hu48R3. Rh.32 was modified by the introduction of a homologous fragment from an isolate, rh.33, from the same rhesus animal in order to ablate all 5 singleton residues simultaneously. This hybrid capsid is referred to as rh.32.33. Three capsids (AAV6, rh.48 and rh.37), containing 2 singletons, underwent mutagenesis for each singleton residue independently and were combined resulting in 4 capsid variants for evaluation (wild type, two with a single reversion and one with two reversions). These capsid modifications are referred to by the name of the parental isolate followed by “.1” or “.2” depending on the particular residue change that took place. Vectors with the combined two singleton changes are named according to their initial isolate name followed by “R2”.
Table 2.
Singleton modifications and vector nomenclature per clade.
| Clade | Parental | R1/R | R2 | R3 | R4 | R5 | .1 | .2 |
|---|---|---|---|---|---|---|---|---|
| A | hu.44 | G609D | P446L | E137K | ||||
| hu.46 | P156S | R362C | S393F | A676 | ||||
| hu.48 | G277S | E322K | S552N | |||||
| AAV6 | K531E | F129L | ||||||
| B | hu.29 | G396E | ||||||
| D | rh.48 | S304N | K217E | |||||
| rh.67 | L119F | |||||||
| rh.37 | E634K | T207M | ||||||
| cy.5 | G13D | D403N | R51K | N158K | P161Q | |||
| E | rh.64 | R697W | V686E | |||||
| rh.58 | S653N | C696R | ||||||
| rh.2 | V651I | |||||||
| Other | ch.5 | T611I | ||||||
| rh.8 | D531E | |||||||
| rh.32 | S354P | G409E | Y419H | P423L | L482F |
Natural AAV isolates identified per clade A–E or without clade affiliation in 'Other'10,20 side-by-side with singleton changed capsid variants. The suffix R refers to the number of singleton residues that were changed. For these changes, the last sequential reversion is described by the location (wild type sequence VP1 numbering) and residue change. For selected isolates, singleton changes were done individually (as opposed to sequentially) and denoted with ".1" and ".2". When both those individual non-conservative mutations were combined, they yielded an R2 clone.
Improvement of vector yields following singleton-to-conserved residue change
Capsids expressed from homologous vector packaging constructs were used in an assay for vector production. Briefly, Cap expression was driven from the AAV2 p40 promoter and expressed in cis with AAV2 Rep and cotransfected in trans with an adenoviral helper construct and an ITR flanked vector genome construct encoding CMV-eGFP-WPRE. Total vector produced from this culture was measured by quantitative PCR for DNase-resistant vector genomes following repetitive freeze-thaw lysis. Yields are presented in GC particles per producer cell (Table 3).
Six wild type capsid isolates (i.e..hu.44, hu.46, hu.48, hu.29, rh.64 and rh.32) produced less than 1000 GC/cell. For reference, the known AAV serotypes produced 25 to 133 times more vector particles than this threshold. In total, 11 naturally occurring, singleton containing capsid isolates generated yields lower than those for AAV2, the lowest producing vector serotype (Table 3).
The impact of singleton residues on vector yield became apparent in the 16 constructs that underwent site-directed mutagenesis. Of the 11 vectors with yields lower than AAV2, seven demonstrate increased titers when the singleton was reverted, often resulting in total vector yields that were comparable to the known reference serotype of the clade. One isolate (rh.2) demonstrates only marginal production improvement and for 3 isolates, hu.46, rh.67, rh.58, vector production was not rescued following one or more singleton changes. In some vectors, a single amino-acid change (hu.29, rh.64, rh.37) was sufficient to overcome the defect in vector manufacturing. Other vectors demonstrated an increase in vector yield following two or more singleton residue changes (hu.44, hu.48, rh.32.33). Five singleton containing capsids (AAV6, cy.5, rh.2, rh.8 and ch.5) produce vector titers already equivalent to or higher than AAV2 which did not dramatically increase following singleton conversion (Table 3).
For 3 capsids with multiple singletons, AAV6, rh.48 and rh.37, each residue was mutated independently to study its impact on yields (Table 2). In terms of the link between singleton residues and vector yields, three distinct scenarios emerged (Table 3). Vector yields for the Clade D vector rh.48 increased moderately (i.e., 5–10 fold) with each singleton change. Only when both singletons were corrected simultaneously, did the vectors achieve yields in the range of AAV7, the reference serotype of that clade, were obtained, indicating an additive effect of the contribution of each individual singleton residue. Another clade D vector, rh.37, behaved differently. Each residue change independently resulted in a total correction of vector yield. Their combined effect on vector yields resulted in only marginal improvement over what was achieved with a single correction. The wild type AAV6, a clade A vector, results in only moderate yields (AAV2/6: 7.5 ± 3.2 × 1012 GC (n=10)). AAV1, also a clade A member and only 6 residues different from AAV6, was produced 10-fold more efficiently (AAV2/1: 7.0 ± 3.6 × 1013 GC (n=10)). Upon investigation of the potential contribution of the two identified singleton residues to the reduced vector production, we identified a moderate increase of vector titers when a K531E change was introduced into AAV2/6 thereby generating AAV2/6.1 (Table 3). No significant increase in titer was observed for the other singleton residue change (F129L) in AAV2/6.2. The average yields (± standard deviation) of the AAV6 variants based on large scale preparations are as follows: AAV2/6.1: 4.5 ± 3.7 × 1013 GC (n=4), AAV2/6.2: 6.4 ± 2.3 × 1012 GC (n=8), AAV2/6R2: 5.4 ± 3.8 × 1013 GC (n=4).
Impact of singleton reversion on gene transfer efficiency
We speculated that singleton changes may have an effect on gene transfer efficiency in vivo and identified several complete sets amenable to this analysis; all members of the set required a minimum production yield of 25 × 103 GC/cell and detectable infectivity in vitro on HEK293 cells at a multiplicity of infection of 104. All vectors that produced over 104 particles per cell demonstrated GFP expression upon in vitro transduction of 293HEK cells at a MOI of 104 (data not shown). Next, we injected vectors expressing human α-1 antitrypsin (hAAT) intravenously into mice and measured hAAT protein in the serum as a quantitative marker for gene transfer efficiency. The serum levels of hAAT from cy.5 and derivatives increased moderately compared to results with the vector based on the wild type isolate. Rh.64R1, rh.2, rh.8 performed comparable to their singleton ablated homologues (data not shown). Two isolates for which the singleton residues were corrected individually presented distinct phenotypical scenarios. Rh.48R2, a clade D member, outperformed both rh.48.1 and rh.48.2 and yielded a reporter gene expression level comparable to the levels of AAV7 after systemic administration (Fig. 2). AAV6, AAV1, AAV6.1, AAV6.2 and AAV6R2 were administered i.v. and monitored for hAAT expression. Gene transfer efficiency for AAV6.2 was approximately 2-fold higher than AAV6 (Fig. 2). The AAV6 K531 singleton residue was found to be pivotal for its superior liver tropism as previously reported25; the non-singleton glutamic acid at position 531 in AAV1, AAV6.1 and AAV6.1.2 correlated with a significant drop in transgene expression (Fig. 2). We confirmed that the lysine at this position on AAV6 and AAV6.2 confers heparin binding to this vector (data not shown). For vectors based on rh.37 we were unable to generate sufficient yields of the singleton containing parental rh.37 to evaluate the impact of each individual singleton residue on gene transfer. Therefore we evaluated this family of vector for in vitro transduction in a quantitative assay. Here we demonstrated that rh.37.1 demonstrated only marginal improvement of gene transfer compared to rh.37 (although vector production was improved) whereas the other singleton reversion in rh.37.2 had a more substantial effect on improving in vitro transduction (Supplement S2).
Figure 2. Impact of singleton modification on gene transfer efficiency.
AAV vectors encoding hA1AT were administered i.v. at a dose of 1 × 1011 GC per animal Stable levels of hA1AT gene expression are represented as averages per group (n=5) with their standard deviation. Statistical comparison is performed by ANOVA (Dunnett Multiple Comparisons Test) (* p<0.05, ** p<0.01, *** p<0.001)
Structural analysis of the impact of singleton residues on vector function
To understand the structural basis of the impact of the singleton reversion on vector biology, we categorized the mutants into distinct phenotypical classes. For each residue for which data were available, we aimed to attribute the functional impact of a singleton change to the structural change it generated. We classified the singleton residues and locations based on their effect on either vector production or gene transfer efficiency. Table 4 summarizes the residues in 7 distinct categories that were based on these criteria along with their homologous position on AAV8 VP1 as an arbitrary reference to allow their localization on the available high resolution structural model. The structural mapping of these residues per category is presented in Figure 3. The noted effects on vector production and gene transfer summarized in Table 4 are based on data presented in Table 3, Figure 2, Figure 4 and Supplement S2.
Table 4.
Singleton categorization according to phenotypical impact.
| Category | Yield | Gene Transfer | Isolates (Mapped AAV8 VP1 location) |
|---|---|---|---|
| I | + | n/a | rh.64R1(697), hu.44R3 (137, 448, 611), hu.48R2 (324, 279), hu.29R (399) |
| II | + | + | hu.48R3 (554), rh.48.1 (305), rh.48.2 (217) rh.48R2 (217, 305), rh.37R2 (212) |
| III | = | + | AAV6.2 (129), cy.5R1(13), cy.5R5 (159+166) |
| IV | + | = | rh.64R2(686), rh.37R1(643) |
| V | = | = | rh.2R (651), cy.5R3(51), rh.8R(533) |
| VI | = | − | cy.5R2 (410) |
| VII | + | − | AAV6.1 (533), AAV6.1.2 (533) |
| VIII | no rescue | n/a | hu.46R1–R4, rh.67R, rh.58R2 |
The effect of a particular singleton reversion on the capsid is categorized according to the effect on vector yield in production and in vivo gene transfer (upon i.v. administration) (+, increase; = no effect; −, decrease). Residues that upon the change demonstrated the effect are mapped onto the homologous AAV8 sequence (VP1 numbering). Residue locations in italics refer to preceding modifications made in the sequential mutagenesis strategy but for which the phenotypical impact could not be attributed directly. n/a: not available
Figure 3. Structure-function correlates of singleton modification on vector phenotype.
Singleton residues locations from categories I, II, and IV–VII are mapped onto the X-ray crystal structure model of AAV8. Numbers and typesetting refer to AAV8 VP1 residue 220 – 738 of singleton locations of the categories defined in Table 4. For I, II, V and VII, the panels on the left show the outer surfaces of the viral capsid (external) and those on the right show 180° rotated views of the same representation showing the inner surface (internal). An internal view only is provided for IV and VI. The surface representation of the reference VP monomer is shown in yellow, and the backbones of symmetry related VP monomers are shown in cyan, orange, green and aqua for VP monomers that are 5-fold related (labeled 5f), magenta and violet for 3-fold related (3f), and pink for the 2-fold related (2f), respectively. The mutated residues are colored blue (for residues that show a phenotype) or cyan (residues in parenthesis in Fig. 2A with no phenotype). The location of a 5-fold axis is shown as a black pentagon.
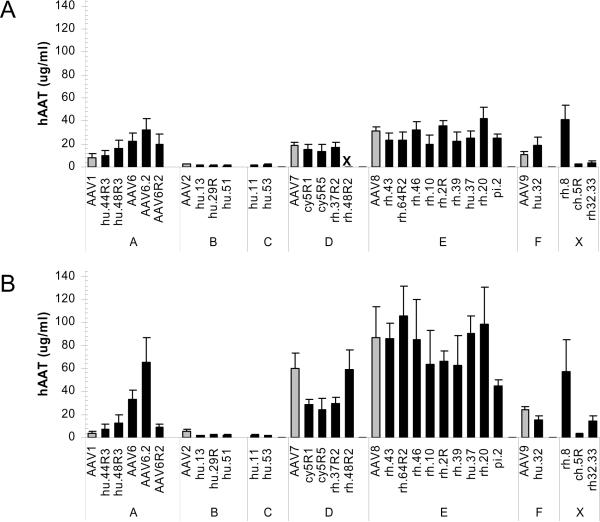
Figure 4. Evaluation of vector portfolio performance upon i.m. and i.v. administration.
Average serum levels of hA1AT at time of stable expression are represented with standard deviation (n=5). (A) Levels following i.m. administration of 1011 GC and (B) following i.v. injection. Gene transfer data for rh.48R2 via the i.m. route is not available.
Category I had a positive effect on yield but the effect on gene transfer efficiency could not be investigated due to insufficient vector production titers of the parental vectors. Category II had a positive effect on both vector yield and gene transfer efficiency. Category III had improved transfer efficiency but no effect on yield, while category IV vectors had the opposite effect. Category V had no effect on either phenotype. Category VI had no effect on yield and a negative effect on gene transfer, and VII had positive effect on production and a negative effect on gene transfer. For residues in category VIII, there was no rescue of vector production and it was therefore non-informative in terms of how the variants affected either vector production or transduction. No residues or classes were identified in this survey for which a singleton reversion had a negative effect on vector yield (Table 4).
The residues in the different singleton categories, with the exception of category III, were mapped onto the available homologous AAV8 VP 3D structure determined by X-ray crystallography37 in order to evaluate the structural impact of singleton reversions at a tertiary and quaternary protein level (Table 4 and Fig. 3). Category III residues are located in a region of the AAV VP that is disordered in all the AAV structures determined to date and thus could not be structurally mapped.
In category I, three residues (E324, G399 and R697) that were critical for improving vector production, when reverted to the conserved amino acid types, lysine, glutamic acid and tryptophan, respectively, are located at icosahedral 5-fold symmetry related VP monomer-monomer interfaces (Fig. 3.I). On the AAV8 capsid structure, residues K324 and E399, located in core beta strands βD and βF, respectively, are involved in 5-fold related interactions. The side chain of K324 forms a hydrogen bond with N338 of a neighboring monomer. A glutamic acid at this position, as observed in isolate hu.48, would not be able to accommodate this interaction due to shorter side-chain length. Significantly, this residue is located at the base of the 5-fold pore which is postulated as the site for the PLA2 externalization in parvoviruses which is required for infection 38. The 5-fold VP-VP interaction observed for residue 399 in the AAV8 capsid involves a backbone carbonyl. However, a glycine residue at this position in hu.29 removes the acidic side-chain of the glutamic acid observed in AAV8 that plays a neutralizing role by pointing towards a basic cavity within the same monomer. W697 is located at the 2-fold interface engaged in multiple van der Waals interactions with the 2-fold related monomer, acting as a hydrophobic anchor into the 2-fold molecule (Fig. 3.I). An arginine side-chain at this position, as observed in the rh.64, would not satisfy these hydrophobic interactions. A fourth category I residue, 137, exhibits a positive phenotype when reverted from a glutamic acid to a lysine as in hu.44R3 and is located outside the VP region that has been resolved by the X-ray crystallography for AAV8 37. Residues G609 and P446 in hu.44 (Table 2), positionally equivalent to AAV8 residues 611 and 448 (Fig. 3.I), respectively, were also found to be located at VP monomer interaction sites at the 3-fold interfaces (Fig. 3.I). For these residues, we were unable to attribute directly their impact on vector yield due to the dominant defect of the last mutated singleton residue in the sequential mutational strategy (Table 3). Category I residue G277 in hu.48 was also observed to have no measurable and attributable effect on vector production when mutated to the conserved serine (hu.48R1, Table 3). This residue, structurally equivalent to AAV8 S279, is located close to beta strand βC37 on the outer surface of the capsid and has a side-chain that is involved in intramolecular VP interactions that may contribute to capsid stabilization (Fig. 3.I).
Two of the four category II residues, S304 and S552 (Fig. 3.II), are located within the ordered region of the AAV8 VP structure (equivalent to N305 and N554), while the remaining two, 212 and 217, are not ordered in the AAV8 crystal structure37. Similar to the observation with the category I residues, residues 305 and 554 are also located at VP-VP monomer interfaces (Fig. 3.II). AAV8 N305 is located within an α-helix, α1, which is conserved at the icosahedral 2-fold axis of all parvovirus structures determined to date. This residue is located on the inside surface of the capsid and has a side-chain that is involved in a bi-dendate hydrogen interaction with the N305 residue from the icosahedral 2-fold related VP monomer. This interaction would not be possible with the shorter side-chain of serine at this position in the rh.48.1 isolate. AAV8 N554 is involved in a hydrogen bond interaction with a 3-fold related VP monomer on the surface of the capsid (Fig. 3.II). The 3-fold interface interactions are the most extensive in the AAV capsids and likely confer significant stability to the capsid.
Category IV residues, V634 and V686, positionally equivalent to K643 and E686, respectively, in AAV8, are located in beta strands βH (643) and βI (686) which form the core AAV capsid. AAV8 K643 is located on the inside surface of the capsid with a side-chain that is proximal to the single nucleotide base observed in the AAV8 crystal structure. A single base is similarly ordered inside the AAV4 crystal structure22 suggesting that its binding site is a conserved DNA interaction pocket inside the AAV capsid. It is possible that K643 plays a role in genomic DNA stabilization. AAV8 E686 is part of a highly conserved stretch of amino acids in beta strand βI, with a side-chain that points into the capsid interior positioned to stabilize the positive charge of the R239 and R313 side-chains.
Mutation of the category V residues 51, 533, 624 and 651 (Table 2), from arginine to lysine (R51K in cy.5R3), aspartic acid to glutamic acid (D531E in rh.8R1), threonine to isoleucine (T624I in ch.5R1) and valine to isoleucine (V651I in rh.2R1) had no significant effect on vector yield or gene transfer, consistent with the conserved nature of the residue changes. Amino acid 51 is not ordered in the AAV8 capsid structure. Residue 531 (positionally equivalent to AAV8 533) is located in variable region VI22 on the capsid surface on the wall of the 2- fold/base of the 3-fold protrusions (Fig. 3.IV & 3.VI). This residue is involved in an intramolecular ionic interaction with K530. Interestingly, the acidic nature of this residue is conserved in most of the AAVs except AAV6 which contains a lysine (K531) at this position. A reversion of this residue to glutamic acid, as in the category VII AAV6.1 virus, results in an increased virus yield suggesting a requirement for K530 neutralization for improved virus production. AAV8 I624 is located on the inner surface of the capsid near the icosahedral 3-fold axis within van der Waals contact distance to P633. It is not involved in any other interactions. AAV8 I651 is buried within the capsid inside a hydrophobic pocket and as such the change from valine to isoleucine in rh.2R1 will only increase the hydrophobic interactions in this capsid region.
Residue D403 of category VI is equivalent to AAV8 N410. This residue is located on the inside of the capsid at the interface between 5-fold related monomers (Fig. 3.VI). Its side-chain makes a hydrogen bond contact with the backbone amide of S225 of a 5-fold symmetry related VP monomer. Interestingly, the aspartic acid renders cy.5 a better gene transfer virus than the same vector with asparagine in that position (Table 4).
Construction of vector portfolio representative of primate AAV biodiversity
To avoid the potential of structural incongruence and reduced functionality, we set out to select and develop novel tools for gene transfer from capsid isolates free of singleton residues, unless functional data dictates otherwise. Isolates were selected in order to maximally represent the primate AAV biodiversity.10,18 Figure 1 summarizes the selected members in their phylogenetic niches in a dendrogram. Differences on a protein level with the known AAV serotypes range from less than 1% (e.g., hu.48R3 versus AAV1) to 9% (pi.2 versus AAV8) within the different clades (Table S1). The rh32.33 clone was most distinct from the established serotypes with AAV4 (18% difference) being the closest relative. Few AAV isolates devoid of singleton residues [hu.36 (clade B) and hu.61 (clade C)] did not meet the minimal requirement for vector production (data not shown).
Characterization of gene transfer potential of expanded AAV portfolio
AAV vectors were produced expressing CB driven hAAT cross-packaged from all singleton corrected and singleton-free capsids represented in Figure 1. All large scale vector preparations reached absolute titers of 2 × 1012 to 1 × 1014 GC. In one set of experiments, 1 × 1011 GC were injected intramuscularly (i.m.) in C57Bl/6 mice and transgene expression was monitored in serum at regular time points following vector administration, and stable gene expression levels are represented in Fig. 4A. In another set of experiments, an identical dose was administered i.v. to in C57Bl/6 mice (Fig. 4B). All vectors demonstrated detectable levels of transgene expression. A clear clade-specific segregation of gene transfer efficiency became apparent following both i.m. and i.v. administration.
Clade E and rh.8-based AAV vectors achieved the highest levels of systemic hAAT following i.m. administration (Fig. 4A). All vectors within clade E performed similarly, indicating that the substantial structural diversity within this clade does not affect its inherent muscle tropism. Clade D vectors also achieved high levels of gene transfer although no vector performed better than the least efficient clade E vector, pi.2. Within clade A, AAV6 and the singleton variant AAV6.2 and AAV6.1.2 appeared to outperform AAV1. Clade B and clade C members, isolate ch.5 and rh.32.33, has a similar level of transduction that was overall inferior to all other vectors tested (Fig. 4).
The gene transfer profile of vectors administered i.v. was similar to that achieved in muscle with clade E members outperforming all other clades. None of the clade E vectors were found to be substantially more efficient than AAV8. Clade F vectors established stable levels of gene transfer that were 2 to 5-fold lower than clade E members. Within clade A, AAV6 and AAV6.2 both distinctly outperformed other clade members including AAV1. The levels of AAV6.2 even reached those obtained by some clade E vectors (Fig. 4B).
When comparing the liver versus muscle-directed gene transfer routes of administration per vector, clade D and E vectors demonstrated a relatively higher performance in liver. Vectors from other clades demonstrated similar levels of vector-mediated gene expression independently of the route of administration. Notable exceptions again were AAV6 and AAV6.2 for which the levels of hAAT gene expression were significantly higher following i.v. administration, further supporting the enhanced liver tropism previously reported for this vector.25
Discussion
Remarkable differences in vector biology, mainly in vector production yield, were noted in our initial characterization of vectors made from AAV capsids isolated from primate tissue by PCR. We were able to attribute many of the apparent defects in production to individual amino acids defined as singleton residues. Reversing these singleton residues to the residue conserved among homologous AAV capsids, was shown to increase vector production and/or gene transfer efficiency in most instances. Given the lack of reagents to study these novel AAVs, these studies did not address whether capsid assembly, genome packaging or capsid stability. Only in the case of AAV6 and cy.5 did one of the singleton changes, K531E, confer a detrimental effect on gene transfer.
How the singleton defects arose and persisted in AAV virus populations to levels allowed for their isolation remains unclear. It is impossible to exclude the contribution of PCR-mediated errors in the methodology of isolation to some of these changes, however, the overall frequency of singleton residues observed exceeds the predictions of the frequency of non-synonymous introduced by the polymerase used for the initial isolation39. It is possible that these viruses are capable of robust replication in the natural host with the reduced vector production being caused by the artificial context of vector production. A homologous rep sequence (whereas in a vector setting heterologous AAV2 rep is provided) or a critical cellular cofactor, lacking in 293 producer cells, may be required. However, our data and structural modeling indicate a structural incompatibility as the basis of some of these defects, suggesting that these sequenced genomes are indeed defective in that they lead to the production of unfit viruses. It is important to highlight that the methodology in previous studies20,21 identified and characterized AAV sequences and not viruses. Therefore, no evidence is available that in vivo these sequences would lead to fit and infectious viral particles. Although it is possible that these defective genomes can be cross-packaged in the setting of a viable co-infection with a non-defective AAV, it is unlikely that these would be propagated in vivo since they are not fit. The high frequency of detection of singleton variants in vivo suggests a low fidelity of DNA replication. Although these findings are inconsistent with previous reports on the error rate of Dependovirus40, they are consistent with the rapid evolution of the virus in vivo.10,20,41 This high frequency of mutations is not only observed in the context of AAV but also for other Parvoviridae where this property was shown to facilitate host switching and immune evasion42,43, characteristics generally thought to be restricted to RNA viruses.44
Irrespective of the singleton origin, the singleton methodology has proven to be an effective method for enhancing the performance of an AAV-based vector. The power of this technique lies in the ability to differentiate potential structural defects from other variation. In addition, this “reverse mutagenesis” approach highlighted the importance of conserved amino acids at VP monomer interfaces or regions predicted to interact with genomic DNA for vector production. Significantly, previous mutagenesis studies in AAV2, the best characterized AAV serotype, have suggested roles for some of the singleton residues in transduction properties and capsid assembly. The mutation of a stretch of amino acids encompassing the acidic 533 residue in the category V and VII viruses has been shown to produce DNA containing capsids that are non-infections (mut37 in Wu, et al.45). In the same study, a charge to alanine mutation of AAV2 681-EIE-683, equivalent to 684-EIE-686 in AAV8, which includes residue 686 of the category IV viruses, resulted in a non-infectious no-capsid phenotype.45 In addition, the mutation of a stretch of conserved amino acids 689-ENSKR-693 in AAV2 equivalent to AAV8 692-ENSKR-696, which is immediately adjacent to W697 singleton change in category I and also resulted in a non-infectious no-capsid phenotype. Thus the involvement of the majority of the singleton residues in monomer interactions suggests functions in capsid assembly and/or stability, properties that would play roles in vector yield and gene transfer. These findings are consistent with previous observations that minimal differences in capsid composition can have dramatic effects on its biology and performance as a vector.15,25,27 Aside from structural changes that may lead to problems with assembly and/or packaging, singletons can have an effect on gene transfer efficiency due to electrostatic changes (e.g., AAV6 K531E) and/or structural rearrangements on the viral surface.
These structure-function correlates were used to design a portfolio of AAV vectors. Vectors were selected or optimized to be free of singleton residues as well as representative for all niches of the known primate AAV biodiversity. Vectors were selected based on production yield and efficiency of in vitro and in vivo gene transfer in muscle and liver directed gene transfer. With few exceptions, the clade classification determined the relative production yield and the vector's performance in vivo, suggesting that a domain conserved within, but unique to each clade, determines its gene transfer performance for liver and muscle. This is of interest for the clade D and E members especially since the internal diversity within the clade can be up to 9% (Table S1).
The expanded portfolio of AAV vectors provides a powerful tool to map structural determinants of vector biology. In one application, we previously made use of these vectors to define a structural determinant on the AAV2 capsid that activates T cell populations towards the capsid.15 Many variables determine the profile of an optimal gene transfer vehicle, most of which are known to be controlled by the AAV capsid structure. Much like candidate selection strategies in small drug development, one can consider AAV as a lead for further optimization specific for the therapeutic aim. In several studies, this AAV portfolio has led to the identification of vectors with novel biology relevant to applications in lung33 and central nervous system-directed gene transfer.32,34 In addition, characterization of the most structurally distinct AAV from this portfolio, identified it as a serotype with a uniquely low serological prevalence.11 In summary, these vectors based on the natural AAV diversity provide a portfolio that can serve as a resource in the determination and selection of the safest and most efficient vector for a particular application and target organ.
Supplementary Material
Acknowledgements
Supported by R01-GM082946 (MA-M), P30-DK-47757, P01-HL-059407, P01-HL-051746 and grants from GlaxoSmithKline Pharmaceuticals, Inc. and the Cystic Fibrosis Foundation (JMW). LV and GPG hold patents on technology described in this manuscript. JMW is an inventor on patents licensed to various biopharmaceutical companies including ReGenX for which he has equity in, consults for and receives a grant from.
This research was supported by R01-GM082946 (MA-M), P30-DK-47757 (JMW), P01-HL-059407 (JMW), P01-HL-051746 (JMW) and grants from GlaxoSmithKline Pharmaceuticals, Inc. (JMW) and the Cystic Fibrosis Foundation (JMW).
Footnotes
Supplementary information is available at Gene Therapy's website.
References
- 1.Fields BN, Knipe DM, Howley PM. Fields' virology. 5th edn. Wolters kluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 2.Flotte TR, Carter BJ. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 3.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Gao GP, Lu Y, Sun X, Johnston J, Calcedo R, Grant R, et al. High-level transgene expression in nonhuman primate liver with novel adeno-associated virus serotypes containing self-complementary genomes. J Virol. 2006;80:6192–6194. doi: 10.1128/JVI.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathwani AC, Gray JT, McIntosh J, Ng CY, Zhou J, Spence Y, et al. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109:1414–1421. doi: 10.1182/blood-2006-03-010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Calcedo R, Nichols TC, Bellinger DA, Dillow A, Verma IM, et al. Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8- mediated, liver-directed gene therapy. Blood. 2005;105:3079–3086. doi: 10.1182/blood-2004-10-3867. [DOI] [PubMed] [Google Scholar]
- 7.Sanlioglu S, Monick MM, Luleci G, Hunninghake GW, Engelhardt JF. Rate limiting steps of AAV transduction and implications for human gene therapy. Curr Gene Ther. 2001;1:137–147. doi: 10.2174/1566523013348788. [DOI] [PubMed] [Google Scholar]
- 8.Thomas CE, Storm TA, Huang Z, Kay MA. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Xie J, Lu H, Chen L, Hauck B, Samulski RJ, et al. Existence of transient functional double-stranded DNA intermediates during recombinant AAV transduction. Proc Natl Acad Sci U S A. 2007;104:13104–13109. doi: 10.1073/pnas.0702778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scallan CD, Jiang H, Liu T, Patarroyo-White S, Sommer JM, Zhou S, et al. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood. 2006;107:1810–1817. doi: 10.1182/blood-2005-08-3229. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Calcedo R, Vandenberghe LH, Figueredo JM, Wilson JM. Impact of preexisting vector immunity on the efficacy of adeno-associated virus-based HIV-1 Gag vaccines. Hum Gene Ther. 2008;19:663–669. doi: 10.1089/hum.2008.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang TP, Jin DY, Wardrop RM, 3rd, Gui T, Maile R, Frelinger JA, et al. Transgene expression levels and kinetics determine risk of humoral immune response modeled in factor IX knockout and missense mutant mice. Gene Ther. 2007;14:429–440. doi: 10.1038/sj.gt.3302881. [DOI] [PubMed] [Google Scholar]
- 15.Vandenberghe LH, Wang L, Somanathan S, Zhi Y, Figueredo J, Calcedo R, et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt M, Voutetakis A, Afione S, Zheng C, Mandikian D, Chiorini JA. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J Virol. 2008;82:1399–1406. doi: 10.1128/JVI.02012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt M, Grot E, Cervenka P, Wainer S, Buck C, Chiorini JA. Identification and characterization of novel adeno-associated virus isolates in ATCC virus stocks. J Virol. 2006;80:5082–5085. doi: 10.1128/JVI.80.10.5082-5085.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnepp BC, Jensen RL, Chen CL, Johnson PR, Clark KR. Characterization of adeno-associated virus genomes isolated from human tissues. J Virol. 2005;79:14793–14803. doi: 10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnepp BC, Jensen RL, Clark KR, Johnson PR. Infectious molecular clones of adeno-associated virus isolated directly from human tissues. J Virol. 2009;83:1456–1464. doi: 10.1128/JVI.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A. 2003;100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, et al. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, et al. Structure of Adeno-Associated virus serotype 8, a gene therapy vector. J Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, et al. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De BP, Heguy A, Hackett NR, Ferris B, Leopold PL, Lee J, et al. High levels of persistent expression of alpha1-antitrypsin mediated by the nonhuman primate serotype rh.10 adeno-associated virus despite preexisting immunity to common human adeno-associated viruses. Mol Ther. 2006;13:67–76. doi: 10.1016/j.ymthe.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Arbetman AE, Lochrie M, Zhou S, Wellman J, Scallan C, Doroudchi MM, et al. Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol. 2005;79:15238–15245. doi: 10.1128/JVI.79.24.15238-15245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lochrie MA, Tatsuno GP, Arbetman AE, Jones K, Pater C, Smith PH, et al. Adeno-associated virus (AAV) capsid genes isolated from rat and mouse liver genomic DNA define two new AAV species distantly related to AAV-5. Virology. 2006;353:68–82. doi: 10.1016/j.virol.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt M, Katano H, Bossis I, Chiorini JA. Cloning and characterization of a bovine adeno-associated virus. J Virol. 2004;78:6509–6516. doi: 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bossis I, Chiorini JA. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J Virol. 2003;77:6799–6810. doi: 10.1128/JVI.77.12.6799-6810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Royo NC, Vandenberghe LH, Ma JY, Hauspurg A, Yu L, Maronski M, et al. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain Res. 2008;1190:15–22. doi: 10.1016/j.brainres.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limberis MP, Vandenberghe LH, Zhang L, Pickles RJ, Wilson JM. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol Ther. 2009;17:294–301. doi: 10.1038/mt.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cearley CN, Vandenberghe LH, Parente MK, Carnish ER, Wilson JM, Wolfe JH. Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol Ther. 2008;16:1710–1718. doi: 10.1038/mt.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allocca M, Mussolino C, Garcia-Hoyos M, Sanges D, Iodice C, Petrillo M, et al. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J Virol. 2007;81:11372–11380. doi: 10.1128/JVI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabinowitz JE, Bowles DE, Faust SM, Ledford JG, Cunningham SE, Samulski RJ. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J Virol. 2004;78:4421–4432. doi: 10.1128/JVI.78.9.4421-4432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, et al. Structure of Adeno-Associated virus serotype 8, a gene therapy vector. J Virol. 2007 doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol. 2006;80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smuda JW, Carter BJ. Adeno-associated viruses having nonsense mutations in the capsid genes: growth in mammalian cells containing an inducible amber suppressor. Virology. 1991;184:310–318. doi: 10.1016/0042-6822(91)90847-5. [DOI] [PubMed] [Google Scholar]
- 41.Chen CL, Jensen RL, Schnepp BC, Connell MJ, Shell R, Sferra TJ, et al. Molecular characterization of adeno-associated viruses infecting children. J Virol. 2005;79:14781–14792. doi: 10.1128/JVI.79.23.14781-14792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Bueno A, Mateu MG, Almendral JM. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J Virol. 2003;77:2701–2708. doi: 10.1128/JVI.77.4.2701-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci U S A. 2005;102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Bueno A, Villarreal LP. Almendral JM. Parvovirus variation for disease: a difference with RNA viruses? Curr Top Microbiol Immunol. 2006;299:349–370. doi: 10.1007/3-540-26397-7_13. [DOI] [PubMed] [Google Scholar]
- 45.Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T, et al. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taymans JM, Vandenberghe LH, Haute CV, Thiry I, Deroose CM, Mortelmans L, et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- 47.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 48.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.