Abstract
Porphyromonas gingivalis, the major causative bacterium of periodontitis, contributes significantly to elevated proteolytic activity at periodontal pockets due to the presence of both, bacteria and host, predominantly neutrophil-derived, serine proteases. Normally the activity of the latter enzymes is tightly regulated by endogenous proteins, including elafin, a potent neutrophil elastase and proteinase 3 inhibitor released from epithelial cells at site of inflammation. Here we have found that all three gingipains (HRgpA, RgpB and Kgp) were able to degrade elafin but RgpB was far more efficient than other gingipains. RgpB already inactivated efficiently the inhibitory activity of elafin at subnanomolar concentrations through a proteolysis limited to the Arg22-Cys23 peptide bond within the surface loop harbouring the inhibitor active site. Notably, elafin resisted inactivation by several Staphylococcus aureus-derived serine- and cysteine proteases confirming this protein's high stability for proteolytic degradation. Therefore, we concluded that elafin inactivation by RgpB represents a specific pathogenic adaptation of P. gingivalis to disturb the protease-protease inhibitor balance in the infected gingival tissue. This contributes to enhanced degradation of host proteins and generation of a pool of peptides serving as nutrients for this asaccharolytic pathogen.
Keywords: protease, inflammation, gingivitis, elastase, proteinase 3, innate immunity
Introduction
Periodontitis is the most prevalent inflammatory disease driven by microorganisms inhabiting the subgingival bacterial plaque. Although more than 600 bacterial species were found in this dwelling only a handful are considered pathogenic, including Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola (Kuru et al., 1999). Out of this trio, P. gingivalis is the best investigated pathogen with respect to virulence traits, which are apparently involved in initiation and maintenance of pathogenic conditions driving chronic inflammation. It is believed that formidable forces of innate immunity frustrated by inability to eradicate irritating infection are directly responsible for periodontal tissues degradation and eventual teeth loss (Socransky et al., 1998;Oliver and Brown, 1993;Cutler et al., 1995;Haffajee et al., 1998). Cysteine proteases of P. gingivalis, referred to as gingipains, are the major virulence factors manipulating mechanisms of host defenses against infection (Potempa et al., 2000).
Gingipains are produced by three genes (rgpA, rgpB, and kgp) encoding strictly Arg-Xaa (RgpA and RgpB) and Lys-Xaa (Kgp) peptide bond specific proteases, respectively (Mikolajczyk-Pawlinska et al., 1998). Due to posttranslational processing and modifications gingipains are secreted in many different molecular forms, both soluble and associated with the outer membrane of the bacterial surface (Potempa et al., 2003). Although gingipains can directly degrade components of the connective tissue, they can inflict far more damage by usurping functions of tightly regulated host proteases of coagulation, fibrinolysis, complement, receptor signaling and kinin-release pathways (Potempa and Pike, 2009). Moreover, by inactivation of endogenous proteinase inhibitors (Travis and Potempa, 2000a) gingipains may contribute to uncontrolled proteolytic activity at the periodontitis sites infected with P. gingivalis.
One of the hallmarks of periodontitis is infiltration of the periodontal tissue by large number of neutrophils. Neutrophils, an indispensable arm of innate immunity, can do a lot of collateral damage to the connective tissue if powerful proteases, including human neutrophil elastase (HNE), proteinase 3 and cathepsin G, are released from disintegrated cells. As a countermeasure blood plasma leaking into inflammatory sites (edema) provides high quantities of endogenous inhibitors. Also, some inhibitors are produced locally reinforcing anti-proteolytic activity of serum. Among those inhibitors elafin secreted by epithelial cells under pro-inflammatory stimuli is an reversible inhibitor of neutrophil elastase and proteinase 3 (Wiedow et al., 1990a). The inhibitory potential characterized by Ki in a subnanomolar range and high levels of expression (Wiedow et al., 1990b;Guyot et al., 2005a;Alkemade et al., 1994b;Pfundt et al., 1996) makes elafin the major elastase and proteinase 3 inhibiting agent in inflamed epithelial tissues (Alkemade et al., 1994a). Unlike plasma inhibitors susceptible to oxidative and proteolytic inactivation, elafin with four disulfide bridges stabilizing the reactive centre loop is an extremely stable molecule resistant to proteolytic degradation in its native, unreduced form (Alkemade et al., 1994;Guyot et al., 2005). Nevertheless, enhanced local expression of elafin in the inflamed connective tissue of the periodontium is apparently not sufficient to control neutrophil proteases and gingival crevicular fluid contains the free elastase activity. Significantly, the level of elastase activity was found to correlate not only with the P. gingivalis infection but also with the presence of Rgps in gingival crevicular fluid (Mailhot et al., 1998). Therefore, our goal was to investigate whether gingipains, in comparison to proteases from Staphylococcus aureus, can interfere with the inhibitory activity of elafin and elucidate a mechanism of inhibitor interaction with gingipains.
Results
Specific inactivation of elafin by RgpB
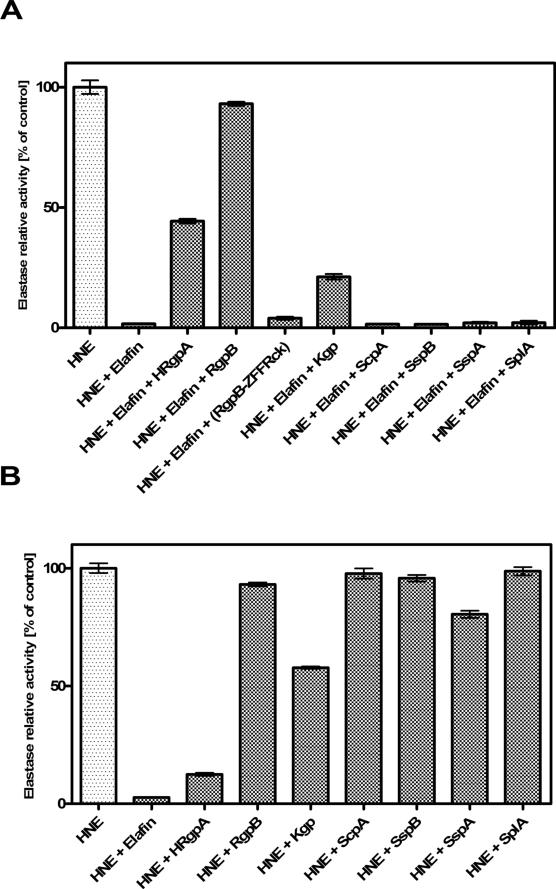
Proteases from P. gingivalis and S. aureus were investigated for their ability to affect elafin-dependent inhibition of HNE. All tested proteases of P. gingivalis were able to inactivate elafin but with varying efficiency. Under experimental conditions applied (Figure 1), only Arg-specific gingipain B (RgpB) inactivated elafin completely with no residual anti-HNE activity of elafin left after 2h preincubation. This effect was dependent on the proteolytic activity of RgpB as demonstrated by complete preservation of elafin inhibitory capacity when the inhibitor was preincubated with Z-Phe-Phe-Arg-chloromethyloketone-treated RgpB.
Figure 1. Elafin is inactivated by proteases of P. gingivalis but not S. aureus.
Elafin was preincubated with a bacterial protease at a 1:1 molar ratio for 2 h at 37°C before human neutrophil elastase (HNE) was added (the molar ratio of HNE : elafin = 1 : 2). The residual elastase activity was determined with MeoSuc-AAPV-pNa as substrate. Presented results are normalized to percentage of the appropriate control. Incubation with P. gingivalis-derived cysteine proteases (HRgpA, RgpB and Kgp) significantly decreased the inhibitory potential of elafin but only RgpB did not interfere with activity of elastase (B). ZFFRck indicates protease inhibitor Z-Phe-Phe-Arg-chloromethyloketone. Staphylococcal proteases (ScpA, SspA, SspB and SplA) did not affect inhibitory activity of elafin. In (B) the influence of tested bacterial proteases on the activity of elastase is shown.
Bars represent mean of triplicate measurement +/− SEM. Experiments shown are representative for at least 3 replicates.
HRgpA and Kgp had incremental effect on anti-elastase capacity of elafin diminishing it by 60% and 80%, respectively. Notably, in contrast to RgpB these gingipains directly inactivated HNE to some extent. After preincubation with HRgpA and Kgp, the HNE activity was decreased by 85% and 70%, respectively (Figure 1B). Contrary to gingipains none of the tested staphylococcal enzymes affected inhibitory capacity of elafin under the experimental conditions and only glutamyl endopeptidase I (SspA) interacted directly with HNE. However, in comparison to HRgpA and Kgp the effect of SspA was far less profound. After pre-incubation with SspA, HNE lost only 15% activity (Figure 1B).
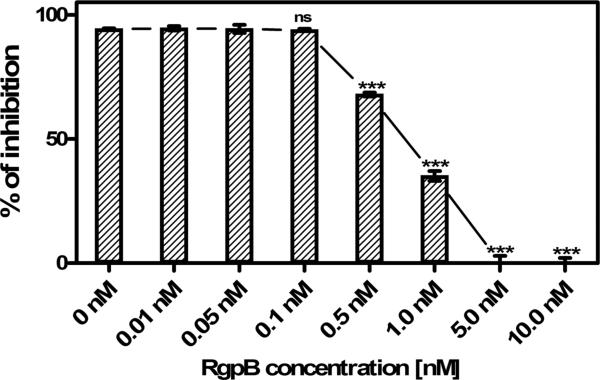
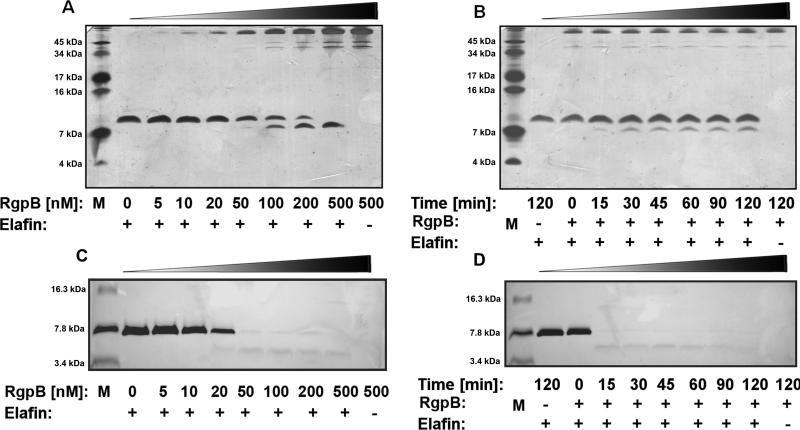
To further characterize RgpB interaction with elafin we have determined the time- and concentration dependent effect of RgpB on the elafin inhibitory activity and protein integrity (Figure 2 and 3). Already at sub-nanomolar concentration (0.5 nM) RgpB significantly diminished the elafin anti-HNE activity (Figure 2). The total inactivation of the inhibitor occurred within 2 h when elafin was incubated with 5 nM RgpB. Gingipain-exerted elafin inactivation was not correlated with the inhibitor degradation when the integrity of the elafin molecule was assessed using non-reducing SDS-PAGE analysis (Figure 3A, B). RgpB up to 50 nM did not cause any significant cleavage of elafin and only at concentrations 100 nM and higher a discrete cleavage product, stable for further degradation even at 500 nM RgpB was observed under non-reducing conditions. However, separation of reduced samples (Figure 3C, D) revealed rapid degradation within 15 min of elafin by 100 nM RgpB. A single cleavage product was noticed with the apparent molecular mass of 4.3 kDa. We concluded that elafin is inactivated by RgpB possibly due to limited proteolysis within a disulfide-bridge-bound loop of the inhibitor.
Figure 2. The gingipain RgpB inactivates elafin is a dose-dependent manner.
Elafin (10 nM final concentration) was preincubated with increasing concentrations of RgpB and the residual elastase inhibitory activity of elafin was determined as described in Figure 1 and shown as remaining percentage of inhibition in relation to elastase activity alone. Inactivation of elafin was already observed at sub-nanomolar concentrations of RgpB. Bars represent mean of triplicate measurement +/− SEM. The experiment shown is representative for at least 3 replicates. *** indicates significant difference in relation to 0 nM RgpB using Student's t-test, p< 0.05.
Figure 3. RgpB degrades elafin in a concentration- and time-dependent manner.
(A) Elafin (1 μg) was incubated with the indicated final concentration of RgpB for 2 h and the samples subjected to SDS-PAGE. (B) Elafin (10 μg) was incubated with RgpB (100 nM) and at the specific time points 10 μl aliquots were withdrawn, mixed with the sample buffer (SB) and instantly frozen. All collected samples were resolved using 15.5:1 T:C, 15% SDS-PAGE in non-reducing conditions. (B) and (C) Samples were prepared similarly, mixed with sample buffer with DTT and boiled for 5 minutes. Reduced samples were separated using peptide gel (16% acrylamide, T:C 8,5:1). The protein bands were stained with silver. M – molecular mass marker.
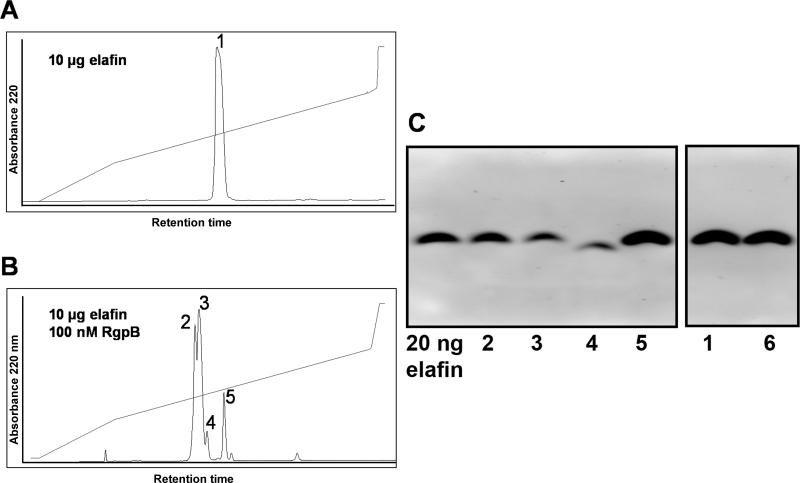
To determine the cleavage site(s), elafin was incubated with RgpB and applied to HPLC reverse-phase chromatography (Figure 4). Collected fractions were analysed by mass spectrometry and tested for elastase inhibition (Table 1). The HPLC separation after RgpB treatment revealed two major fractions (number 2 and 3 in Figure 4B) eluting shortly before the fraction of residual intact elafin (number 5 in Figure 4B). A peptide in fractions no. 2 and no. 3 exhibited a molecular mass of 6016.5 Da (Table 1) precisely matching the molecular mass of native elafin with intact disulphide bridges and hydroxylation at one single amino group position. This form of elafin exhibited no inhibitory activity even at five molar excess over elastase. Hydroxylation and lost of the inhibitory activity of elafin did not occur spontaneously as shown in control experiments (Figure 4A, Table 1). In addition to the major cleaved product, a minor one was identified in fraction 4 (Figure 4B). This fraction constitutes 7.6% of the starting elafin and contains an inhibitory inactive peptide with the molecular mass of 5364.6 Da matching that of the major form of proteolytically modified elafin additionally truncated at Lys6. This cleavage site is rather unusual for Arg-specific RgpB, however, as our proteases were purified from native bacteria, a trace contamination of our enzyme preparation with Kgp (Lys-specific gingipain) is possible.
Figure 4. HPLC analyses of RgpB-inactivated elafin.
10 μg of elafin were separated by RP-HPLC (A). The same amount of elafin was incubated with 100 nM RgpB for 2 h at 37°C and separated accordingly by RP-HPLC (B). Protein peaks were collected and the amount of protein in each peak was calculated based on peak area. Fractions were analyzed by western blotting (50 ng protein loaded per lane) using specific anti-elafin antibodies (C). Numbers describing individual lanes correspond to peak numbers in the HPLC chromatogram. An additional lane (lane 6) contains elafin incubated for two hours in reaction buffer alone. Only in lane 4, a band with molecular mass lower than native elafin is visible. On all other lanes, the visible band has the same mobility as intact elafin. Some differences are observed in antibody affinity, as bands 2, 3 and 4 are noticeably weaker than control ones (5, 6), despite the fact that all lanes were loaded with the same protein amount.
Table 1. Analyses of HPLC fractions (Figure 4) of RgpB-treated and non-treated Elafin.
Samples and fraction numbers are indicated as shown in Figure 4. Protein amount was calculated by area under peak from indicated fraction. 10 μg of elafin were initially applied on RP-HPLC. Molecular weight was determined by ESI-MS analyses and elastase inhibition by 5-molar excess of elafin fraction compared to HNE.
| Sample | Fraction number # | Protein Amount μg | Molecular Mass | Elastase Inhibition % |
|---|---|---|---|---|
| Elafin, untreated | 1 | 10 | 5997.5 | 99 |
| Elafin + RgpB | 2 | 2.5 | 6016.5 | 2 |
| Elafin + RgpB | 3 | 4.63 | 6016.5 | 3 |
| Elafin + RgpB | 4 | 0.76 | 5364.6 | 2 |
| Elafin + RgpB | 5 | 1.07 | 5997.8 | 98 |
| Elafin, buffer treated | 6 | 10 | 5998.0 | 99 |
Non-reducing SDS-PAGE (data not shown) and the western blot analysis (Figure 4C) of HPLC resolved elafin-derived peptides revealed bands with the same mobility as the intact elafin with the notable exception of the N-terminally truncated elafin in fraction no. 4. However, SDS-PAGE resolution of elafin and elafin degradation products under reducing conditions discriminated the intact and the cleaved inhibitor (Figure 3C, D). This finding confirms that elfin with a single peptide bond cleaved is inactive as the elastase inhibitor despite the stable structure maintained by disulfide bridges.
RgpB is known to cleave exclusively the arginine-Xaa peptide bond. To determine which of the two Arg-Xaa peptide bonds present in the elafin polypeptide chain is cleaved, fractions of RgpB-treated elafin (fractions no. 2, 3, and 4 in Table 1) were reduced with DTT and subsequently alkylated. Samples were subjected again to reverse-phase-HPLC and the resulting fractions were analyzed by mass spectrometry. The observed masses of RgpB-generated peptides in fraction 2 and 3 matched the molecular mass of the elafin fragments derived from cleavage at the Arg22-Cys23 peptide bond (Table 2) corresponding to the apparent molecular mass of the cleavage product identified in the reducing SDS-PAGE. On the other hand, the peptide in the fraction no. 4 corresponded to Arg22-hydrolysed elafin, N-terminally truncated at Lys6. Notable, no elafin fragments indicating the cleavage at Arg31 were detected.
Table 2. Observed molecular masses of reduced and alkylated fractions of RgpB treated Elafin.
Fraction numbers are the same as in Table 1. Fractions were reduced, alkylated and separated by HPLC. Resulting fractions were measure by ESI-MS. Calculated molecular masses of reduced and alkylated cleavage products of elafin are given. Elafin product describes the amino acid position of the correlating theoretical molecular mass.
| Fraction number # | Observed Molecular Mass | Theoretical Molecular Mass | Elafin Product |
|---|---|---|---|
| 2 | 2347.4 | 2347.81 | Elafin 1-22 |
| 2 | 4132.79 | 4133.88 | Elafin 23-57 |
| 3 | 2348.3 | 2347.81 | Elafin 1-22 |
| 3 | 4132.8 | 4133.88 | Elafin 23-57 |
| 4 | 1696.11 | 1695.06 | Elafin 7-22 |
| 4 | 4131.98 | 4133.88 | Elafin 23-57 |
Discussion
Elafin is a very potent inhibitor of the neutrophil serine proteases elastase and proteinase 3. The structure of elafin consists of a single four-disulfide core protein domain, with the reactive site loop expanding to the outside. The rigid, strongly stabilised core renders elafin unusually stable and resistant to proteolysis (Guyot et al., 2005b), which is considered as an adaption to function in a proteolytic milieu. Elafin expression is induced in inflamed epithelial tissues playing an important role in the regulation of elastase and proteinase 3 activities at sites of inflammation. Besides having tissue destructive activity, neutrophil serine proteases are able to promote inflammation by processing pro-inflammatory cytokines and activating various inflammatory receptors (Meyer-Hoffert, 2009;Wiedow and Meyer-Hoffert, 2005). Uncontrolled neutrophil protease activity results in enhanced local inflammation and tissue destruction providing peptides which are indispensable nutrients for asaccharolytic P. gingivalis. Although P. gingivalis proteases, including gingipains, can directly degrade many host proteins here we show that peptidic nutrients generation can be accomplished in a more efficient indirect way. As high elastase and proteinase 3 activities were reported in gingival crevicular fluid (GCF) (Cox and Eley, 1989b;Uitto et al., 2003), we presumed that P. gingivalis can interfere with control mechanisms of proteolysis in the periodontal tissue utilizing the host inflammatory proteases elastase and proteinase 3 to facilitate local overall proteolytic activity. We describe herein proteolytic inactivation of elafin by the gingipain RgpB and propose a potential novel virulence trait for this bacterial enzyme. Compared to other proteases from P. gingivalis and S. aureus this inactivation was highly specific and efficient. Though we could detect some proteolytic inactivation of elastase activity by HRgpA and Kgp, this partial inactivation might be of low relevance in the in vivo situation since overall elastase activity is elevated in the GCF (Cox and Eley, 1989a). In the gingival pocket elafin might be likely exposed to both elastase and RgpB simultaneously. Preliminary experiments suggest that RgpB is not able to cleave elafin efficiently in complex with elastase (data not shown). However, the local concentration of elafin is likely to be higher than those of HNE in the gingival pocket. Under these conditions RgpB might cleave free elafin. Moreover, as other gingipains are efficient in inactivation of HNE, the HNE-elafin complex might be destroyed in order to release free elafin, now accessible to RgpB-mediated cleavage. Importantly, it must be considered, that high concentrations of elastase substrates like elastin compete with each other and elastase inhibitors in the in vivo situation. Moreover, pre-elafin (trappin-2) may be covalently bound via transglutamination to extracellular proteins, including laminin, fibronectin, fibrinogen or elastin. Elafin as well as its proform remain potent inhibitors of HNE and proteinase 3 and various authors have proposed that such anchoring may provide a local protection from excessive proteolysis by neutrophil proteinases. Whether pre-elafin is inactivated by RgpB needs further experimental confirmation.
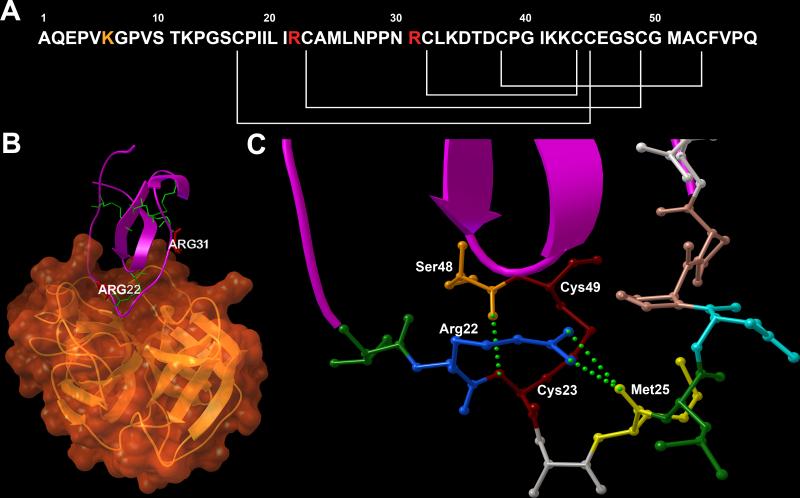
HPLC separation of the RgpB/Elafin mixture revealed three distinct cleavage products with no elastase inhibitory activity. As two cleavage products were expected for each of the two arginine residues of the elafin amino acid sequence, the presence of the third one was surprising and later deciphered as additional cleavage after Lys6. Most interestingly, both dominant HPLC peaks contained elafin inactivated by hydrolysis after Arg22, which were possibly separated on the basis of slight structural differences occurring after the cleavage. The hydrolysis after the Arg22 residue might result in the formation of two distinct folds of the loop region. Analysis of the elafin structure in the complex with porcine pancreatic elastase revealed a complex structural organisation of the expanded reactive site loop (RSL) (Tsunemi et al., 1996b). The RSL, expanding from Leu20 to Leu26, is covalently attached to the rigid core region of the molecule by the disulfide bond between Cys23 and Cys49, however this bond takes an unusual, expanded conformation (Figure 5). As revealed by solution NMR experiments (Francart et al., 1997), this extended conformation is enforced by two Pro28 and Pro29 residues, which provide a rigid scaffold for the expanding loop. This fold is further supported by a network of hydrogen bonds. Two η-amino groups of Arg22; P3 residue, according to Schechter and Berger nomenclature (Schechter and Berger, 1967) form hydrogen bonds with the P1’ Met25. Furthermore, Cys22 forms a hydrogen bond with Ser48, additionally stabilising the fold of RSL. The observed cleavage by RgpB breaks up this network by reformatting the carboxyl group of Arg22. This may lead to the destruction of hydrogen bonds between Arg22 – Met25, leaving this region unstable. Subsequently, this event might lead to a lower interaction in the P2-P3’ fragment of RSL, which may lead to the restoration of optimal conformation of the Cys23-Cys45 disulfide bridge and possibly might result in the collapse of this fragment into the core region and to the inactivation of the protease inhibitory activity. The single hydrolysis inactivation of elafin at nanomolar levels of RgpB concentration may be interpreted as specific, evolutionary designed interaction that targets a delicate and sensitive balance between protease and its inhibitor in healthy tissue.
Figure 5. RgpB inactivates elafin by cleavage after Arg22.
(A) The amino acid sequence of the elafin molecule; white lines mark Cys-Cys bridges, Arg22 and Arg31 are highlighted in red and Lys6 in orange. The arrangement of disulfide bonds, which stabilize the inhibitor structure, does not allow dissociation of fragments formed by hydrolysis of both Arg-Cys-peptide bonds. (B) 3-D representation of elafin (violet) in the complex with pancreatic elastase (orange). Both arginine residues are placed at locations closely interacting with the enzyme molecule and are critical for complex formation. Cys-Cys bonds are shown in green. (C) Structure of the reactive site loop of elafin, interacting with the enzyme active site. Aminoacid residues are coloured according to RasMol scheme, the Cys22-Cys49 bridge is marked in burgundy and hydrogen bonds are presented as green spheres. Structure obtained from www.pdb.org with accession code 1FLE (Tsunemi et al., 1996a). 3-D images were prepared with Python Molecular Viewer (Sanner, 1999).
Inactivation of host protease inhibitors by bacterial enzymes was described for SERPIN family members in numerous reports (Travis and Potempa, 2000b), where cleavage within the RSL, often at the P1-P1’ peptide bond, leads to complete inactivation of the inhibitor. Inhibitors of the whey acidic protein family, including elafin and SLPI, are stable proteins and due to their rigid core structure often unsusceptible to proteolytic cleavage. So far, proteolysis was only reported for SLPI by RgpB (Into et al., 2006) and for elafin by dust mite proteases (Brown et al., 2003), but these studies did not report the cleavage site.
Taken together, gingipains play an important role in modulating the balance between host proteases and their inhibitors at the infection site. The herein described distinctive inhibition of elafin by RgpB may add to the pathophysiology of gingivitis. Notably, elafin resisted inactivation by several Staphylococcus aureus-derived serine- and cysteine proteases confirming this protein high stability for proteolytic degradation. Therefore, we concluded that elafin inactivation by RgpB represents a specific pathogenic adaptation of P. gingivalis to disturb the protease-protease inhibitor balance in the infected gingival tissue. This leads to enhanced degradation of host proteins and generation of a pool of peptides serving as nutrients for this asaccharolytic pathogen.
Material and Methods
Materials
Reagents were obtained from Sigma–Aldrich (Taufkirchen, Germany) unless otherwise indicated. Enzymes included: Human Neutrophil Elastase (HNE) - EC-Number 3.4.21.37 (Elastin Products, Owensville, Missouri); Staphopain A (ScpA) and B (SspB) - MEROPS ID C47.001 and C47.002, respectively; Glutamyl Endopeptidase I (SspA) – MEROPS ID S01.269; staphylococcal Serine-Protease-Like A protease (SplA); Gingipains R (HRgpA, RgpB) – MEROPS ID C25.001 and Gingipain K (Kgp) – MEROPS ID C25.002. Elafin was provided by Proteo Biotech AG, Kiel, Germany.
The HNE active site was titrated with recombinant eglin C (Ciba Geigy, Basel, Switzerland). HNE proved to be more than 80% catalytically active. HNE was dissolved in 0.1 M sodium acetate (pH 5.0). Proteolytic activity was measured after each experiment by substrate assay with MeoSuc-Ala-Ala-Pro-Val-p-nitroanilide as described below. Staphylococcal enzymes were purified according to procedures described in (Potempa et al., 1988;Drapeau, 1978;Massimi et al., 2002) and P. gingivalis proteases were obtained as recently described (Potempa and Nguyen, 2007).
Elastase inhibition assays
For initial inactivation assays HNE was diluted in TET buffer – 50 mM Tris-HCl, pH 7.6 containing 5 mM EDTA and 0.05% Tween-20 (Staphopain A, Staphopain B, SplA, glutamyl endopeptidase I (V8 protease)), or in TNCT buffer – 0.2 M Tris-HCl, 0.15 M NaCl, 5 mM CaCl2, 0.05% Tween-20 (HRgpA, RgpB, Kgp). Cysteine proteinases were activated with 2 mM DTT in TET (ScpA, SspB) or with 10 mM L-Cysteine in 0.2 M HEPES 5 mM CaCl2, pH 8.0 buffer (HRgpA, RgpB, Kgp) by 15 min incubation in 37°C and further diluted in buffer specified above without reducing agents. Elafin was preincubated in the same buffer with a set of bacterial proteases in concentration 200 nM vs. 200 nM (1:1 molar ratio) for 2 h. Thereafter the prepared solution of elastase was added to each sample and after 5 min the substrate MeoSuc-Ala-Ala-Pro-Val-pNa was added. The final mixture contained 25 nm HNE, 50 nM elafin, 50 nM respective bacterial enzyme and 1 mM substrate. The colour development was monitored as p-nitroaniline release at 405 nm using microplate absorbance reader (Sunrise, TECAN Group Inc, Männedorf, Switzerland). Concentration dependency was measured similarly using final concentrations of RgpB in the range 0 to 10 nM, 10 nM elafin and 5 nM elastase in a final volume of 200 μl.
SDS-PAGE
Electrophoresis was run in a Schagger/von Jagow tricine system using 15% acrylamide, with T:C 15.5:1 ratio (Schagger et al., 1988). A total of 1 μg of elafin was applied per well. Protein size was indicated using the molecular marker SeeBlue Plus2 (Invitrogen, Hamburg, Germany). Protein bands were visualized using silver staining.
Western blot was performed by electrotransfer of proteins to PVDF membrane (Millipore, Schwalbach, Germany), followed by overnight blocking with 2% bovine albumine in Tris-buffered saline, 0.05% Tween-20 (TTBS). Blocked membrane was incubated for 1h with primary antibody (1:500, biotinylated goat anti-human Trappin-2; (BAF-1747, R&D, Wiesbaden-Nordenstadt, Germany)), then washed 3 times with TTBS and streptavidin-HRP was applied for 1h (1:20000 (RPN-1231 GE Healthcare, Munich, Germany)). ECL+ (GE Healthcare, Munich, Germany) was used as the chemiluminescence substrate and images were acquired using Diana imager (Raytest, Straubenhardt, Germany).
High Performance Liquid Chromatography
Preparations: A containing 10 μg of elafin and 100 nM activated RgpB in a total volume of 50 μl, B: 10 μg elafin and C: 100 nM RgpB in TNCT buffer were incubated separately for 2 h at 37°C. All samples were applied on the reverse-phase C2/C18 HPLC column μRPC SC 2.1/100 (GE Healthcare, Munich, Germany). The column was developed with an acetonitrile gradient using 0.1% TFA in H2O (solution A) and 80% acetonitrile in solution A (solution B) starting with 0% solution B changing to 25% after 10 min, 70% after 40 min and 100% after 41 min. The fractions were collected automatically (SMART, Pharmacia, Uppsala, Sweden) and applied to mass spectrometry (see below). Identified peptides were tested for anti-elastase activity (see above) and further reduced by incubation with 5 mM DTT at 37°C for 1 h. After reduction, samples were re-run on the same HPLC column to separate resulting linearized peptides and gathered fractions were again analyzed using mass spectrometry. Protein concentrations of HPLC fractions were calculated from peak areas and a 5-fold molar excess of each fraction was used to test inhibitor activity as described above.
Reduction and Alkylation
The fractions were lyophilized and the concentrations were adjusted to 4 μg of protein in 1 μl of ddH2O. The protein fractions (20 μg) were reduced in a mixture of 85 μL 100 mM NH4HCO3 and 10 μl 100 mM DTT for 20 min at 65°C. After reduction proteins were alkylated with 11 μl iodacetamide for 20 min at ambient temperature in the dark. After reduction and alkylation the mixture was separated on an HPLC column (C2/C18) as described above and analyzed by mass spectrometry.
Mass Spectrometry
Protein and peptide mass determinations were performed by electrospray mass ionization spectrometry (ESI-MS)-analyses using a Quadrupol-Time-of-Flight-Hybrid-Mass-spectrometer (Q-TOF™ II, Waters Micromass, Milford, Massachusetts) equipped with an orthogonal electrospray source (Z-spray) operated in positive ionisation mode. For MS analysis, 10 μl of HPLC-fractions were lyophilized and diluted with 100 μl carrier solvent (50% acetonitrile, 50% water, 0.2% formic acid) and infused into the electrospray source at a rate of 20 μl/min. Sodium iodide was used for m/z calibration at a range of 80-2000. The capillary potential was set to 2.55 or 3.00 kV and the cone voltage to 40 V; cone temperature was set to 100°C; desolvation temperature was 150°C; the ESI gas was nitrogen. The charge-to-mass ratio of ions was scanned in different ranges between 200 to 2000 and 700 to 2000.
Data processing
Acquisition and data analysis were all performed using the MassLynx 3.5 software package (Waters Micromass, Milford, Massachusetts). Mass spectra were averaged typically over 100-150 scans (scan time 1 s; interscan time 0.1 s). The multiply charged raw data of intact proteins were background subtracted and deconvoluted using MaxEnt1 to obtain singly charged ion mass spectra to determine average molecular masses of intact proteins. The raw combined spectral data from small peptides were background-subtracted and subjected to Maximum Entropy 3 (‘MaxEnt3’) deconvolution to determine monoisotopic molecular masses.
Acknowledgement
We acknowledge the technical assistance provided by Jutta Quitzau. This study was supported in part by grants from the Deutsche Forschungsgemeinschaft DFG (SFB 617, J.-M. Schröder), from the German Federal Ministry of Education and Research (BMBF, SkinStaph project, R. Gläser and U. Meyer-Hoffert) and grants from the Ministry of Science and Higher Education (1642/B/P01/2008/35, Warsaw, Poland) and the National Institutes of Health (DE 09761, United States, to J. Potempa).
References
- Alkemade JA, Molhuizen HO, Ponec M, Kempenaar JA, Zeeuwen PL, de Jongh GJ, Vlijmen-Willems IM, van Erp PE, van de Kerkhof PC, Schalkwijk J. SKALP/elafin is an inducible proteinase inhibitor in human epidermal keratinocytes. J.Cell Sci. 1994a;107(Pt 8):2335–2342. doi: 10.1242/jcs.107.8.2335. [DOI] [PubMed] [Google Scholar]
- Alkemade JA, Molhuizen HO, Ponec M, Kempenaar JA, Zeeuwen PL, de Jongh GJ, Vlijmen-Willems IM, van Erp PE, van de Kerkhof PC, Schalkwijk J. SKALP/elafin is an inducible proteinase inhibitor in human epidermal keratinocytes. J.Cell Sci. 1994b;107(Pt 8):2335–2342. doi: 10.1242/jcs.107.8.2335. [DOI] [PubMed] [Google Scholar]
- Brown A, Farmer K, MacDonald L, Kalsheker N, Pritchard D, Haslett C, Lamb J, Sallenave JM. House dust mite Der p 1 downregulates defenses of the lung by inactivating elastase inhibitors. Am.J.Respir.Cell Mol.Biol. 2003;29:381–389. doi: 10.1165/rcmb.2003-0060OC. [DOI] [PubMed] [Google Scholar]
- Cox SW, Eley BM. Detection of cathepsin B- and L-, elastase-, tryptase-, trypsin-, and dipeptidyl peptidase IV-like activities in crevicular fluid from gingivitis and periodontitis patients with peptidyl derivatives of 7-amino-4-trifluoromethyl coumarin. J.Periodontal Res. 1989a;24:353–361. doi: 10.1111/j.1600-0765.1989.tb00882.x. [DOI] [PubMed] [Google Scholar]
- Cox SW, Eley BM. Detection of cathepsin B- and L-, elastase-, tryptase-, trypsin-, and dipeptidyl peptidase IV-like activities in crevicular fluid from gingivitis and periodontitis patients with peptidyl derivatives of 7-amino-4-trifluoromethyl coumarin. J.Periodontal Res. 1989b;24:353–361. doi: 10.1111/j.1600-0765.1989.tb00882.x. [DOI] [PubMed] [Google Scholar]
- Cutler CW, Kalmar JR, Genco CA. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- Drapeau GR. Role of metalloprotease in activation of the precursor of staphylococcal protease. J.Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francart C, Dauchez M, Alix AJ, Lippens G. Solution structure of R-elafin, a specific inhibitor of elastase. J.Mol.Biol. 1997;268:666–677. doi: 10.1006/jmbi.1997.0983. [DOI] [PubMed] [Google Scholar]
- Guyot N, Zani ML, Berger P, Dallet-Choisy S, Moreau T. Proteolytic susceptibility of the serine protease inhibitor trappin-2 (pre-elafin): evidence for tryptase-mediated generation of elafin. Biol.Chem. 2005a;386:391–399. doi: 10.1515/BC.2005.047. [DOI] [PubMed] [Google Scholar]
- Guyot N, Zani ML, Berger P, Dallet-Choisy S, Moreau T. Proteolytic susceptibility of the serine protease inhibitor trappin-2 (pre-elafin): evidence for tryptase-mediated generation of elafin. Biol.Chem. 2005b;386:391–399. doi: 10.1515/BC.2005.047. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr., Socransky SS. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J.Clin.Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Into T, Inomata M, Kanno Y, Matsuyama T, Machigashira M, Izumi Y, Imamura T, Nakashima M, Noguchi T, Matsushita K. Arginine-specific gingipains from Porphyromonas gingivalis deprive protective functions of secretory leucocyte protease inhibitor in periodontal tissue. Clin.Exp.Immunol. 2006;145:545–554. doi: 10.1111/j.1365-2249.2006.03156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru B, Yilmaz S, Noyan U, Acar O, Kadir T. Microbiological features and crevicular fluid aspartate aminotransferase enzyme activity in early onset periodontitis patients. J Clin.Periodontol. 1999;26:19–25. doi: 10.1034/j.1600-051x.1999.260104.x. [DOI] [PubMed] [Google Scholar]
- Mailhot JM, Potempa J, Stein SH, Travis J, Sterrett JD, Hanes PJ, Russell CM. A relationship between proteinase activity and clinical parameters in the treatment of periodontal disease. J Clin.Periodontol. 1998;25:578–584. doi: 10.1111/j.1600-051x.1998.tb02491.x. [DOI] [PubMed] [Google Scholar]
- Massimi I, Park E, Rice K, Muller-Esterl W, Sauder D, McGavin MJ. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J Biol.Chem. 2002;277:41770–41777. doi: 10.1074/jbc.M207162200. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U. Neutrophil-derived serine proteases modulate innate immune responses. Front Biosci. 2009;14:3409–3418. doi: 10.2741/3462. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton PA, Chen WC, Travis J, Potempa J. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol.Chem. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- Oliver RC, Brown LJ. Periodontal diseases and tooth loss. Periodontol. 1993;2:117–127. doi: 10.1111/j.1600-0757.1993.tb00224.x. 2000. [DOI] [PubMed] [Google Scholar]
- Pfundt R, van Ruissen F, Vlijmen-Willems IM, Alkemade HA, Zeeuwen PL, Jap PH, Dijkman H, Fransen J, Croes H, van Erp PE, Schalkwijk J. Constitutive and inducible expression of SKALP/elafin provides anti-elastase defense in human epithelia. J.Clin.Invest. 1996;98:1389–1399. doi: 10.1172/JCI118926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. 2000. [DOI] [PubMed] [Google Scholar]
- Potempa J, Dubin A, Korzus G, Travis J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J.Biol.Chem. 1988;263:2664–2667. [PubMed] [Google Scholar]
- Potempa J, Nguyen KA. Purification and characterization of gingipains. Curr.Protoc.Protein Sci. 2007 doi: 10.1002/0471140864.ps2120s49. Chapter 21, Unit. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of Innate Immunity by Bacterial Proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr.Protein Pept.Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- Sanner MF. Python: a programming language for software integration and development. J.Mol.Graph.Model. 1999;17:57–61. [PubMed] [Google Scholar]
- Schagger H, Aquila H, Von Jagow G. Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal.Biochem. 1988;173:201–205. doi: 10.1016/0003-2697(88)90179-0. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem.Biophys.Res.Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J.Clin.Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Travis J, Potempa J. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochim.Biophys.Acta. 2000a;1477:35–50. doi: 10.1016/s0167-4838(99)00278-2. [DOI] [PubMed] [Google Scholar]
- Travis J, Potempa J. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochim.Biophys.Acta. 2000b;1477:35–50. doi: 10.1016/s0167-4838(99)00278-2. [DOI] [PubMed] [Google Scholar]
- Tsunemi M, Matsuura Y, Sakakibara S, Katsube Y. Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution. Biochemistry. 1996a;35:11570–11576. doi: 10.1021/bi960900l. [DOI] [PubMed] [Google Scholar]
- Tsunemi M, Matsuura Y, Sakakibara S, Katsube Y. Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution. Biochemistry. 1996b;35:11570–11576. doi: 10.1021/bi960900l. [DOI] [PubMed] [Google Scholar]
- Uitto VJ, Overall CM, McCulloch C. Proteolytic host cell enzymes in gingival crevice fluid. Periodontol. 2003;31:77–104. doi: 10.1034/j.1600-0757.2003.03106.x. 2000. [DOI] [PubMed] [Google Scholar]
- Wiedow O, Meyer-Hoffert U. Neutrophil serine proteases: potential key regulators of cell signalling during inflammation. J.Intern.Med. 2005;257:319–328. doi: 10.1111/j.1365-2796.2005.01476.x. [DOI] [PubMed] [Google Scholar]
- Wiedow O, Schroder JM, Gregory H, Young JA, Christophers E. Elafin: an elastase-specific inhibitor of human skin. Purification, characterization, and complete amino acid sequence. J.Biol.Chem. 1990b;265:14791–14795. [PubMed] [Google Scholar]
- Wiedow O, Schroder JM, Gregory H, Young JA, Christophers E. Elafin: an elastase-specific inhibitor of human skin. Purification, characterization, and complete amino acid sequence. J.Biol.Chem. 1990a;265:14791–14795. [PubMed] [Google Scholar]