Abstract
Arginine deiminase (ADI)-based arginine depletion is a novel strategy under clinical trials for the treatment of malignant melanoma with promising results. The sensitivity of melanoma to ADI treatment is based on its auxotrophy for arginine due to a lack of argininosuccinate synthetase (AS) expression, the rate-limiting enzyme for the de novo biosynthesis of arginine. We show here that AS expression can be transcriptionally induced by ADI in melanoma cell lines A2058 and SK-MEL-2 but not in A375 cells; and this inducibility was correlated with resistance to ADI treatment. The proximal region of the AS promoter contains an E-box that is recognized by c-Myc and HIF-1α and a GC-box by Sp4. By chromatin immunoprecipitation asssays, we demonstrated that under noninduced conditions, the E-box was bound by HIF-1α in all the three melanoma cell lines. Under arginine depletion conditions, HIF-1α was replaced by c-Myc in A2058 and SK-MEL-2 cells but not in A375 cells. Sp4 was constitutively bound to the GC-box regardless of arginine availability in all three cell lines. Overexpressing c-Myc by transfection upregulated AS expression in A2058 and SK-MEL-2 cells; whereas co-transfection with HIF-1α suppressed c-Myc-induced AS expression. These results suggest that regulation of AS expression involves interplay among positive transcriptional regulators c-Myc and Sp4, and negative regulator HIF-1α that confers resistance to ADI treatment in A2058 and SK-MEL-2 cells. Inability of AS induction in A375 cells under arginine depletion conditions was correlated by the failure of c-Myc to interact with the AS promoter.
Keywords: Melanoma, Argininosuccinate Synthetase, Arginine Deiminase, c-Myc, HIF-1α
Introduction
The incidence of malignant melanoma continues to increase in the past decade. Conventional immunotherapy-, chemotherapy-based or combination of these therapies yield an overall response rate of <25%. Thus, novel strategies for treating melanoma have been sought (1, 2). It has been reported that majority of melanomas and hepatocellular carcinomas (HCC) are auxotrophic for arginine because of their lack of argininosuccinate synthetase (AS) expression (3–5). Arginine is synthesized from citrulline through two-step reactions catalyzed by argininosuccinate synthetase (AS) and argininosuccinate lyase (AL), in which AS is the rate-limiting enzyme. In light of these unique properties, arginine deiminase (ADI), a bacterial enzyme that converts arginine to citrulline and ammonium, resulting in arginine deprivation, was developed for treatment of melanoma and HCC. To suppress its immunogenicity and enhance its stability in circulation, the clinically used ADI (ADI-PEG20) was conjugated with polyethylene glycol (PEG). In the laboratory setting, exposure of melanoma cells to ADI results in cell death, whereas normal cells that express AS are able to survive (6–9). Phase II clinical trials using ADI-PEG20 has been conducted in melanoma and HCC. Arginine was successfully degraded from serum by ADI-PEG20, and a response has been seen (6–9). In some patients, however, insensitivity to ADI treatment was associated with the induced expression of AS (10), suggesting that induction of AS expression under ADI treatment may contribute to failure of ADI treatment. However, the mechanisms by which AS was expressed in ADI-treated patients have not been elucidated.
The current study was initiated to investigate the mechanism by which AS is induced in three melanoma cell lines under arginine deprivation conditions, either by treating these cells with ADI-PEG20 or by culturing them in arginine-free medium. We found that the inducibility of AS expression was correlated with ADI resistance and that expression of AS was reversibly regulated by arginine availability. Further investigations revealed that induction of AS expression by arginine depletion involves the positive transcription regulator c-Myc and negative regulator HIF-1α which recognize the E-box in collaboration with Sp4 which recognizes the GC-box at the AS promoter in A2058 and SK-MEL-2 cells. Failure of AS induction in A375 melanoma cells was associated with the inability of c-Myc to interact with the E-box. Our study reveals the important roles of c-Myc/HIF-1α/Sp4 in the regulation of AS expression that confer ADI-PEG20 resistance in melanoma cells.
Results
Induction of AS expression by arginine depletion is associated with ADI resistance in melanoma cell lines
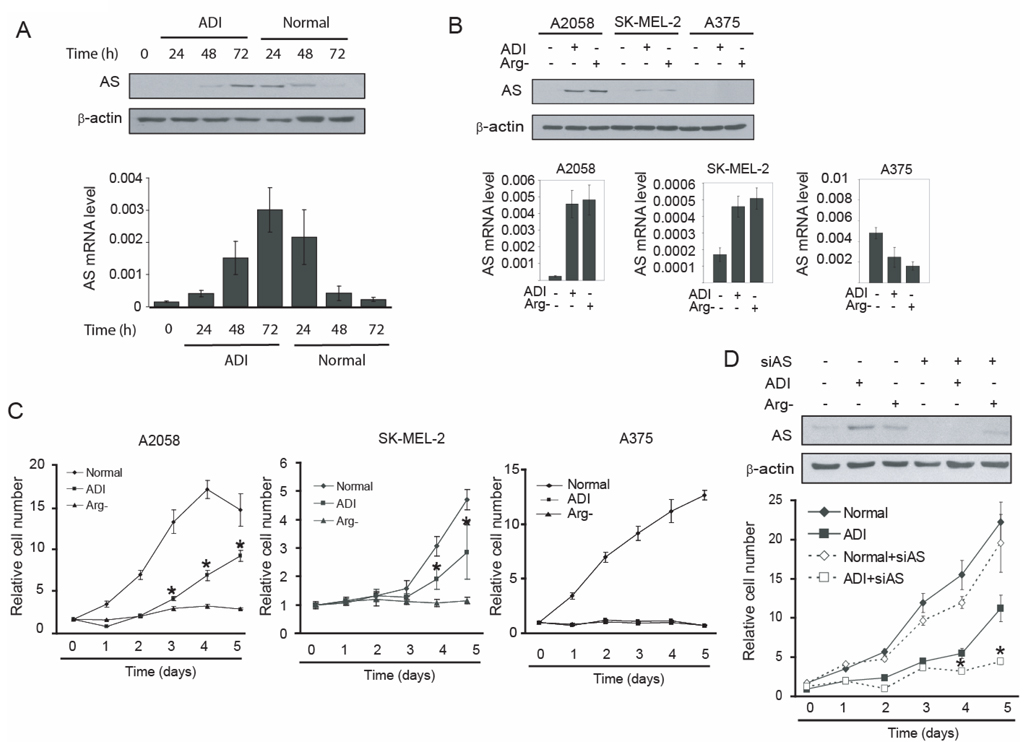
We first analyzed the effects of ADI treatment on AS expression in A2058 melanoma cells. A2058 cells were maintained in ADI-containing or arginine-free medium for various lengths of time up to 72 h, the culture medium was then replaced with complete medium. AS expression level was analyzed by Western blotting and real-time PCR. AS expression appeared 48–72 h after ADI treatment and gradually decreased after removal of ADI (Fig. 1A). Similar results were observed when A2058 cells were maintained in arginine-free medium (data not shown). These results indicated that levels of AS expression are controlled by arginine availability.
Fig. 1.
Induction of AS expression by arginine depletion contributes to ADI resistance. A. Kinetic study of AS induction by arginine depletion. A2058 cells were maintained in medium containing 0.05 µg/ml ADI for the time intervals as indicated. Thereafter, cells were cultured in normal medium for an additional 24, 48 and 72 h. The cells were harvested and AS expression levels were analyzed by Western blotting or real-time PCR. The AS mRNA levels are represented as the expression level relative to β-actin. B. A2058, SK-MEL-2, and A375 cells were maintained in normal medium, medium containing 0.05 µg/ml ADI, or arginine-free medium for 72 h and AS expression levels were analyzed by Western blotting and real-time PCR. The AS mRNA levels are represented as expression level relative to β-actin. C. Proliferation of A2058, SK-MEL-2 and A375 cells after ADI treatment or cultured in arginine-free medium. Cells (100 each) were seeded on 96-well plates and cultured in normal medium, medium containing 0.05 µg/ml ADI or arginine-free medium. Cells were fixed by 10% trichloroacetate every 24 h and the growth rate was analyzed by SRB assay. D. A2058 cells were transfected with control or AS siRNA (100 nM each) and maintained in normal medium, medium containing 0.05 µg/ml ADI, or arginine-free medium and cell growth rate was analyzed. Seventy two hours after transfection of AS siRNA, AS expression levels were examined by Western blotting 72 hr after transfection. All the error bars represent standard deviation by student’s t-test from three independent experiments. (* p<0.01)
We then determined the expression of AS in two other melanoma cell lines, SK-MEL-2 and A375. Like A2058 cells, these cells have intact AS genes in their genomes by sequencing (data not shown). The cells were maintained in ADI-containing or arginine-free medium for 72 h. AS expression was induced in SK-MEL-2 cells but the level of induction was lower than that in A2058 cells. AS expression was not detected in A375 cells even under induction conditions (Fig. 1B).
These three melanoma cell lines were subjected to sensitivity test under arginine-deprivation culturing conditions, whereas the regular medium (DMEM) we used contained 0.48 mM ariginine. A2058, SK-MEL-2 and A375 cells cultured under arginine-free medium showed very little proliferative activity up to 5 days. ADI treatment completely inhibited the growth of A375 cells for the same period of time, but A2058 and Sk-MEL-2 cells showed significant growth in the presence of ADI (Fig. 1C).These data indicate that AS inducibility correlates with ADI resistance. Indeed, we were able to establish ADI-resistant cell lines from A2058 and from SK-MEL-2 cell lines but not from A375 cell lines (data not shown). We also noticed that A2058 and SK-MEL-2 cells could partially maintain growth under ADI treatment but not in the arginine-free medium, yet both culturing conditions can induce AS expression (Fig. 1B). These results suggest that induction of AS alone is not sufficient to support cell growth under complete arginine-deprivation conditions.
To address the role of AS induction in ADI resistance, we used siRNA to knockdown AS mRNA. As shown in Fig. 1D, transfection with AS siRNA effectively suppressed AS induction in ADI-containing medium or arginine-free medium; and there was a concomitant reduction in cell growth, supporting the role of AS expression in ADI resistance.
Identification of arginine deprivation-responsive cis-elements in the AS promoter
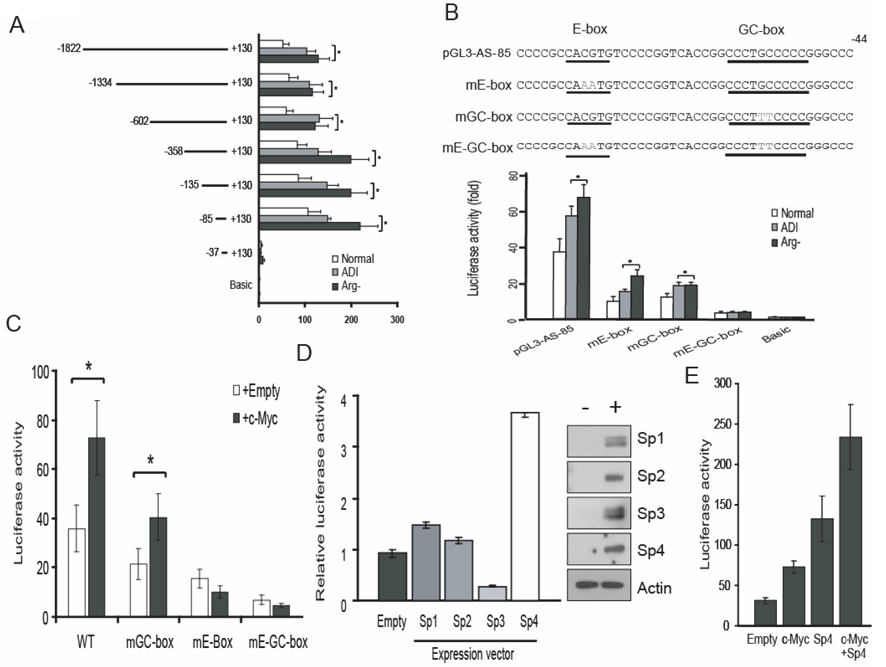
To identify the cis-elements within the AS promoter that respond to arginine availability, we first constructed a reporter recombinant pGL3-AS-1822 which contains nucleotides (nt) −1822 to +300 of the AS promoter linked to the bacterial leuciferase reporter gene. We also constructed several AS promoter deletion reporters by progressively removing the upstream AS sequences in pGL3-AS-1822. These deletion reporters were transfected into A2058 cells and their promoter activity was analyzed. As shown in Fig. 2A, the reporters with deleted sequences down to −85 nt still retained responsiveness to arginine depletion; but further deletion to −37 nt markedly reduced both basal and arginine depletion-responsive activities, suggesting that −85 to −37 nt contain sequence important for both basal and inducible AS expression. By using computational analysis described previously (11), we found an E-box and a GC-box sequences between −85 and −37 nt of the AS promoter (Fig. 2B).
Fig. 2.
Analysis of arginine-deprivation responsive cis-elements on the AS promoter. A. Deletion analysis of the AS promoter. A2058 cells were transfected with pGL3-AS recombinants encoding various lengths of AS promoter sequences and pRLII. Seventy-two hours after transfection, the promoter activities of each construct were assayed. B. Mutation analysis of the AS promoter. The point mutations were introduced on the E-box and/or GC-box of pGL3-AS-85 as indicated. A2058 cells were transfected with mutant AS promoter constructs and promoter activity was assayed. The AS promoter activity was normalized with that of the empty pGL3 vector. The error bars represent standard deviation by student t-test from three independent experiments. (* p<0.01). C. E-box-dependent activation of AS promoter activity by c-Myc. A2058 cells were transfected with wild-type, GC-box mutant, E-box mutant or double mutant of pGL3-AS-85 with empty or c-Myc expression vector and maintained in normal medium, medium containing 0.05 µg/ml ADI or arginine-free medium for 72 h. The AS promoter activity was analyzed and was normalized with that of the pGL3 vector. The error bars represent standard deviation from three independent experiments. D. A2058 cells were cotransfected with pGL3-AS-85 and expression vectors for Sp1, Sp2, Sp3 and Sp4. The promoter activity and the expression of transfected vectors were analyzed. The error bars represent standard deviation from three independent experiments. (* p<0.01). Expression levels of the transfected (+) Sp1, Sp2, Sp3 and Sp4 were determined by Western blottings using their respective antibodies. Representative expression levels of β-actin were used as loading control. E. Additive effect of c-Myc and Sp4 in AS promoter activity. A2058 cells were transfected with pGL3-AS-85 and expression vectors for c-Myc and Sp4 alone, or c-Myc plus Sp4 and AS promoter activity was analyzed. The AS promoter activity was normalized with that of the empty pGL3 vector. The error bars represent standard deviation from three independent experiments.
To determine whether these sequences are involved in AS induction, we introduced mutations into both the E-box and GC-box in pGL3-AS-85 (Fig. 2B, underscores) and analyzed their effects on AS promoter activity. Mutations at either the E-box or the GC-box alone reduced AS promoter activity and compound mutations further reduced the promoter activity in response to arginine depletion (Fig. 2B, bottom). These results suggest that both E-box and GC-box sequences play important roles in the induction of AS by arginine depletion.
Two basic helix-loop-helix transcription factors, c-Myc and HIF-1α, were considered as strong candidates for interacting with the E-box sequence. We first analyzed whether c-Myc indeed plays a role in regulating AS expression. Wild-type or mutant pGL3-AS-85 reporter recombinants were co-transfected with empty or c-Myc-expression vector. As shown in Fig. 2C, cotransfection with c-Myc expression vector up-regulated reporter activity of wild-type and GC-box mutated pGL3-AS-85, but not of the mutant E-box or double mutant constructs. These results indicate that c-Myc positively regulates AS promoter activity through its interaction with the E-box.
The Sp1 transcription factor family is well known to regulate their target genes through interactions with the GC-box. To investigate the impact of Sp1-transcription factors on the AS promoter, we analyzed the effect of co-transfection of expression vectors encoding Sp1, Sp2, Sp3 and Sp4 on AS promoter activity. Co-transfection with Sp4 recombinant showed a 4-fold induction of reporter expression levels, whereas cotransfection with Sp3 recombinant reduced >50% of reporter expression (Fig. 2D). Cotransfection with Sp1- and Sp2-recombinants increased reporter activity by <1.5-fold (Fig. 2D, left panel). Expression levels of transfected Sp1, Sp2, Sp3 and Sp4 were comparable in these transfections (Fig. 2D, right panel). These results suggested that Sp3 and Sp4 can modulate AS promoter activity. The role of Sp4 in transcriptional upregulation of AS was also demonstrated by the additive increases in reporter expression when c-Myc and Sp4 expression recombinants were co-transfected (Fig. 2E).
Induction of c-Myc expression and its cross-talk with Sp4 in the arginine depletion-induced AS expression in A2058 cells
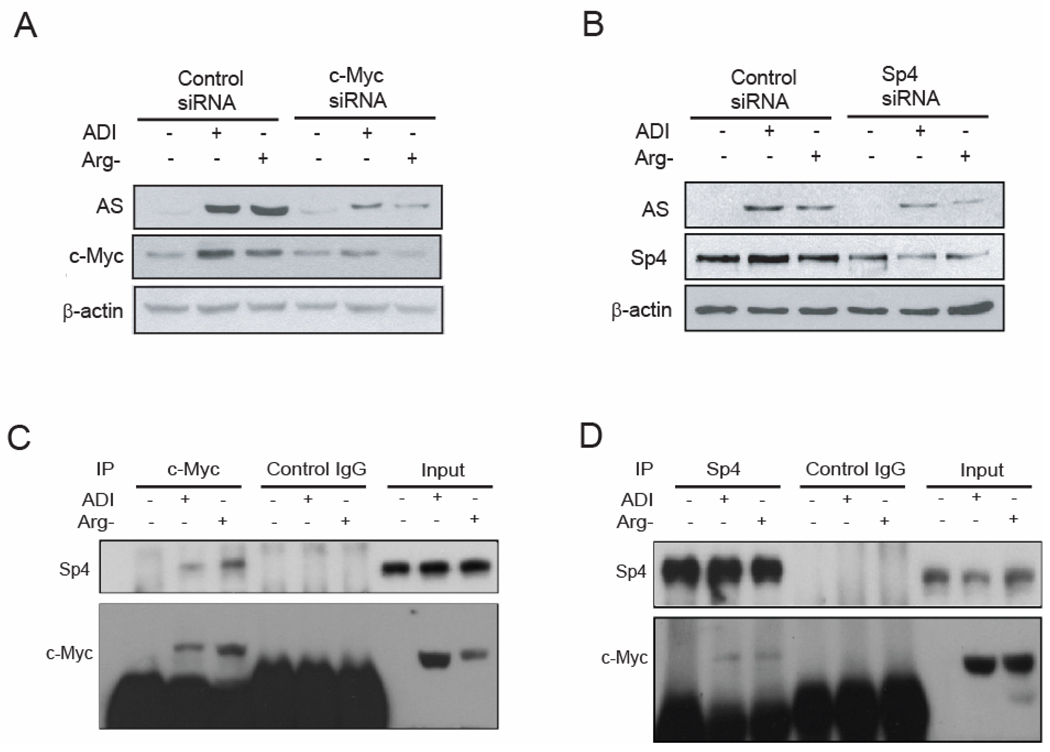
To address the roles of c-Myc, Sp4 and HIF-1α (see below) in regulating AS expression under the stress of reduced arginine in melanoma cells, we first focused on A2058 cells. We found that levels of c-Myc but not of Sp4 were increased in cells grown in medium-containing ADI or in arginine-free medium. Upregulation of c-Myc was also observed at the mRNA level (data not shown). We then investigated the effects of c-Myc or Sp4 expression on AS induction. A2058 cells were transfected with c-Myc siRNA, Sp4 siRNA or scramble siRNA. Cells were maintained in normal, ADI, or arginine-free medium thereafter for 48 hours. As expected, in the presence of control siRNA, c-Myc but not of Sp4 (Fig. 3B) protein levels were upregulated by arginine depletion. Knockdown of c-Myc by siRNA to < 50% in the ADI-treated or arginine-free medium-cultured cells resulted in > 80% suppression of AS induction, suggesting that c-Myc is a positive regulator for AS upregulation (Fig. 3A). Knockdown Sp4 to about 30% of the control level by siRNA also inhibited AS induction (Fig. 3B), suggesting that Sp4 is also involved in AS induction. The extent of AS suppression by Sp4 siRNA seemed to be much reduced in comparison with the effect of c-Myc knockdown (Fig. 3A). However, knockdown of Sp3 levels to about 50% of control by siRNA did not significantly altered AS expression levels (data not shown). We repeatedly observed that the c-Myc siRNA probe we used was less effective in suppressing its target RNA in A2058 cells grown under regular conditions than in those grown under induced conditions. This was probably due to the low abundance of c-Myc mRNA in cells grown under non-induced conditions, thereby providing reduced target size for the attack by siRNA. Nonetheless, this difference should not affect the result that knockdown of c-Myc in the ADI- or arginine-free medium-treated cells was associated with down-regulation of AS expression.
Fig. 3.
c-Myc is induced by arginine-depletion and positively regulates AS promoter activity through the E-box in A2058 cells. A. Cells were transfected with 100 nM of scramble (contol) or c-Myc siRNA. Cells were maintained in normal, ADI-containing, or arginine-free medium for 48 h. Expression levels of AS, c-Myc and β-actin were analyzed by Western blotting. B. Cells were transfected with control or Sp4 siRNA and maintained in normal medium, medium containing 0.05 µg/ml ADI or arginine-free medium for 72 h. Expression levels of AS, Sp4 and β-actin were analyzed by western blotting. C. The interaction between c-Myc and Sp4 was analyzed by co-immunoprecipitation using anti-c-Myc antibody followed by Western blotting using anti-Sp4 and anti-c-Myc antibodies. D. The interaction between Sp4 and c-Myc was analyzed by co-immunoprecipitation using anti-Sp4 antibody followed by Western blotting using anti-c-Myc and anti-Sp4 antibodies.
We then explored the possibility of physical interactions between c-Myc and Sp4. c-Myc protein was immunoprecipitated from A2058 cell lysate by anti-c-Myc antibody and the precipitate was subjected to Western blotting using anti-c-Myc and anti-Sp4 antibodies. Fig. 3C shows that Sp4 was co-immunoprecipitated with c-Myc from the lysate of cells treated with ADI or cultured in arginine-free medium, but not from cells maintained in normal medium. The interactions between c-Myc and Sp4 were also demonstrated by using anti-Sp4 antibody in reciprocal immunoprecipitation which showed that c-Myc was co-immunoprecipitated with Sp4 from the lysate of cells treated with ADI or cultured in arginine-free medium but not from cells maintained in the regular medium (Fig. 3D). These results demonstrated the physical interactions between c-Myc and Sp4 in A2058 cells cultured under arginine-depleted conditions.
HIF-1α is a negative regulator for arginine depletion-induced AS expression in A2058 cells
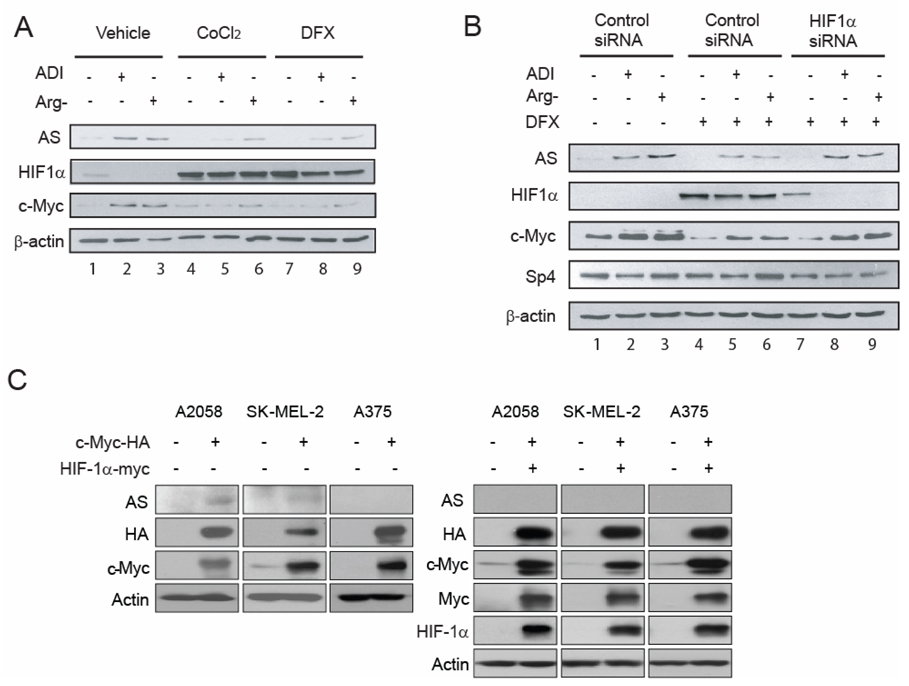
Arnt/HIF-1β is another potential interacting transcription for the E-box sequence. HIF-1β forms heterodimer with HIF-1α and modulates target gene expression under hypoxic condition. Given that the physiologic role of HIF1α is often opposite to that of c-Myc (12–14), we explored the possible involvement of HIF-1α in AS gene expression. Fig. 4A shows that treating A2058 cells with ADI or growing them in arginine-free medium resulted in down regulation of HIF-1α (compared among lanes 1 to 3; results were clearly observed with prolonged exposure of the autoradiograph, data not shown). Induction of HIF-1α using hypoxia mimics CoCl2 or DFX resulted in accumulated HIF-1α levels, and elevated HIF-1α was associated with downregulation of AS expression. Strikingly, downregulation of HIF-1α in the hypoxia mimics-treated A2058 cells by siRNA reversed the inhibitory effects of HIF-1α on AS expression in the arginine-depleted conditions (Fig. 4B). These results demonstrated that HIF-1α plays a negative role in the regulation of AS expression under arginine-depleting conditions. Moreover, we found that in the CoCl2- and DFX-treated A2058 cells, accumulation of HIF-1α in the ADI-treated or arginine deficiency-cultured A2058 cells was associated with downregulation of c-Myc (Fig. 4A, compare lanes 5, 6 and 8, 9 with 2, 3). The downregulation of HIF-1α by siRNA abolished the inhibitory effect of DFX on AS induction through the recovery of c-Myc induction (Fig. 4B, comparing lanes 8, 9 with 5, 6). These results suggest that HIF-1α also exerts a suppressive role in the regulation of AS induction through downregulation of the c-Myc pathway in A2058 cells.
Fig, 4.
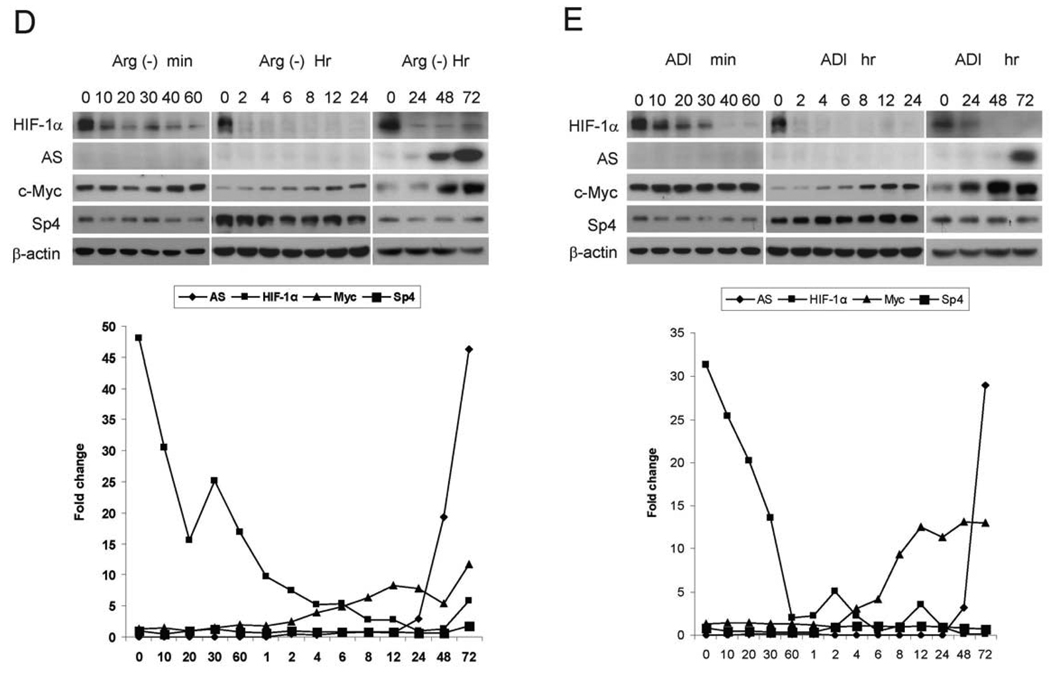
Roles HIF-1α in the regulation of AS expression. A, Arginine deprivation suppresses HIF-1α expression. A2058 cells were maintained in normal medium, medium containing 0.05 µg/ml ADI, or arginine-free medium with or without 100 µM DFX or 100 µM CoCl2. Seventy-two hours after treatment, cells were harvested and subjected for Western blotting. B, A2058 cells were transfected with 100 nM scramble (control) or HIF-1α siRNA and maintained in normal medium, medium containing 0.05 µg/ml ADI or arginine-free medium with 100 µM DFX. Untransfected A2058 cells maintained in normal medium, medium containing 0.05µg/ml ADI or arginine-free medium were used for control. Levels of AS, HIF-1α, c-Myc, Sp4 and β-actin were measured by Western blottings. C, A2058, SK-MEL-2 and A375 cells were transfected with c-Myc or co-transfected with HIF-1α expression vector and pCGN (control) vector, and cell lystates were analyzed by Western blot. As an expression control, the level of total expression of each protein was examined by immunoblotting with an antibodies against whole protein as indicated or β-actin (loading control). D and E, Kinetics of HIF-1α, AS, c-Myc and Sp4 expression in A2058 cells grown in arginine-free medium (D) or 0.05 µg/ml ADI-containing medium (E) for the time courses as indicated.
We next investigated the effects of overexpression of c-Myc and HIF-1α on the expression of AS. We transfected HA-tagged c-Myc recombinant DNA into A2058, SK-MEL-2, and A375 cells. Overexpression of c-Myc induced AS expression in A2058 and SK-MEL-2 cells, but not in A375 cells (Fig. 4C, left panel). Co-transfection of Myc-tagged HIF-1α with HA-c-Myc recombinants suppressed c-Myc-induced AS expression (Fig. 4C, right). These results support the positive role of c-Myc and negative role of HIF-1α in regulation of AS expression.
To determine the expression kinetics of transcription regulators and the induction of AS, we analyzed the time course of HIF-1α, c-Myc, and Sp4 expression in reference to AS expression in A2058 cells after treatment with ADI or under arginine-free culture conditions. We chose A2058 cells because they express comparably higher steady-state levels of HIF-1α than do SK-MEL-2 and A375 cells (see below, Fig. 5A). With prolonged exposure of autoradiographs, HIF-1α signals in the Western blots could be clearly visualized. Figs. 4D and 4E show that reduction of HIF-1α was seen within 10 min after the treatment; whereas induction of c-Myc started 4 hr after the treatment, and rapid surge of AS levels was seen 48–72 hrs thereafter. Levels of Sp4 did not change throughout the entire time-course analysis. These results indicate that induction of AS expression by arginine-deprivation in A2058 cells is a delayed effect, initiated by the down-regulation of HIF-1α and followed by the upregulation of c-Myc.
Fig. 5.
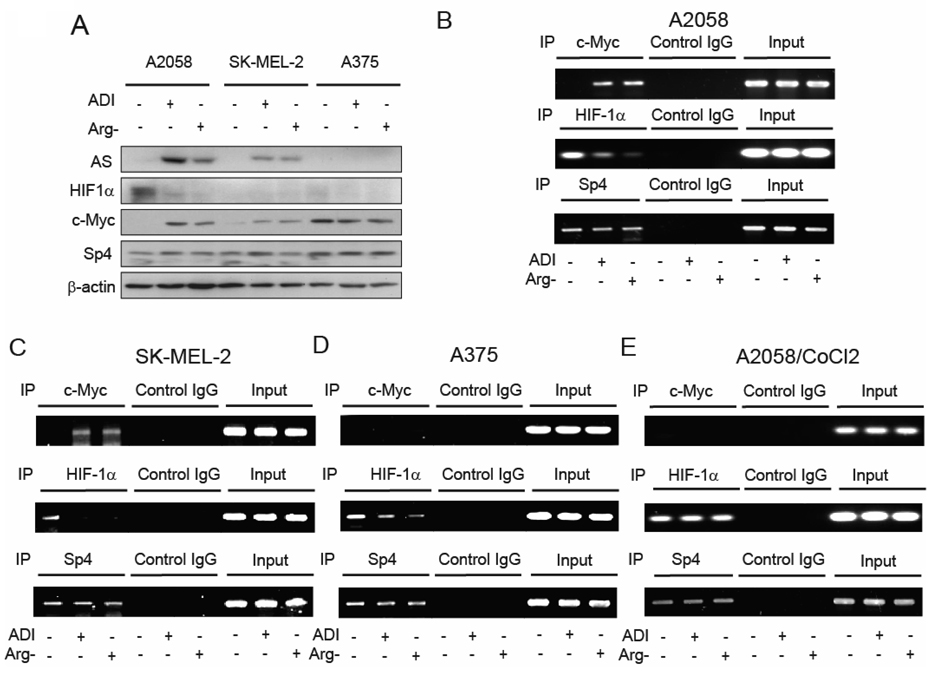
Mchanistic investigation of c-Myc, HIF-1α, and Sp4 in the regulation of AS expression in melanoma cells under arginine-available and -deficient conditions. A. Western blotting analyses of expression levels of AS, HIF-1α, c-Myc and Sp4 in A2058, SK-MEL-2 and A375 cells grown in the regular, ADI-containing (0.05 µg/ml), and arginine-free medium for 72 hours. B – D: Chromatin immunoprecipitation assay of the interactive transcriptional regulators with AS promoters under arginine depletion conditions. B and C, c-Myc displaces HIF-1α binding from AS promoter in response to arginine depletion in A2058 and SK-MEL-2 cells, respectively. D, Dissociation of HIF-1a from but no concomitant association of c-Myc to the AS promoter in response to arginine depletion in A375 cells. E, Stabilization of HIF-1α by the hypoxia mimic CoCl2 resulted in persistent association between HIF-1α and the AS promoter and preventing c-Myc to interact with the promoter in A2058 cells cultured under arginine-deficient conditions The antibodies used for ChIP and control IgG are indicated at the top. Input, genomic DNA prior to immunoprecipitation. AS promoter sequences from ChIP were quantified by PCR assay.
Mechanistic analyses of c-Myc, HIF-1α, and Sp4 in the regulation of AS expression in melanoma cell lines in response to arginine deprivation
We then investigated the mechanistic aspects of c-Myc, HIF-1α and Sp4 in the induced AS expression in A2058, SK-MEL-2 and A375 cells. Expression of HIF-1α, c-Myc, and Sp4 in these three cell lines were determined by Western blottings. Our results showed that: (i) comparing with A2058 cells, the steady-state levels of HIF-1α were much reduced in SK-MEL-2 and A375 cells, regardless of arginine availability (Fig. 5A); (ii) like A2058 cells, levels of c-Myc were low in SK-MEL-2 cells under noninduced conditions but were induced in the ADI- and arginine-free medium-treated cells. Furthermore, when induced levels of c-Myc between A2058 cells and SK-MEL-2 cells were compared, it appears that levels of c-Myc were correlated with levels of AS induction. These results are consistent with the positive role of c-Myc in the regulation of AS expression in these two cell lines; (iii) levels of c-Myc were high in A375 cells grown in the regular medium but were lower under arginine-depleting conditions; and (iv) levels of Sp4 were constitutively expressed in all the three melanoma cell lines, regardless of arginine availability.
To gain mechanistic insights into how these transcription factors regulate AS expression in response to arginine depletion, we carried out chromatin immunoprecipitation (ChIP) analysis. A2058 cells grown in the regular medium, or arginine-free medium, or treated with ADI-PEG20 were fixed with formaldehyde, sonicated, and the sheared chromatin was immunoprecipitated by antibodies against c-Myc, HIF-1α or Sp4. The precipitated DNA fragments were renatured, purified, and subjected to PCR analyses. Significant enrichment of AS promoter sequence was observed in the chromatin fractions of ADI-treated and arginine-free-cultured cells pull-down by anti-c-Myc antibody. In contrast, reduction of AS promoter sequence was observed in the samples when anti-HIF-1α antibody was used. No significant difference of AS sequence when Sp4 antibody was used (Fig. 5B). These results suggest that when A2058 cells were cultured under the noninduced conditions, the AS promoter was mainly bound by HIF-1α but not by c-Myc. When these cells were switched to ADI-containing or arginine-free medium, down-regulation of HIF-1α resulted in the replacement of c-Myc occupancy at the AS promoter. Similar results were observed in SK-MEL-2 cells (Fig. 5C).
We hypothesized that AS expression in A375 cells could not be induced by ADI treatment was because c-Myc was unable to participate in transcriptional activation at the AS promoter level. To test this hypothesis, we performed ChIP analyses. As shown in Fig. 5D, although dissociation of HIF-1α from the AS promoter was seen in A375 cells cultured under arginine-depleted conditions, no engagement of c-Myc to the AS promoter was found under these conditions. These results support the hypothesis that failure of induced AS expression in A375 cells under arginine-depleting conditions is because of the inability of c-Myc to interact with AS promoter cultured under arginine-deficient conditions.
We further demonstrated that accumulation of HIF-1α in the CoCl2-treated A2058 cells persists the binding of HIF-1α at the AS promoter and prevents the displacement by c-Myc (Fig. 5E). Alternatively, these results may be explained by that the absence of c-Myc in the engagement AS promoter is due to reduced levels of c-Myc in the CoCl2-treated cells resulting from stabilization of HIF-1α (Fig. 4A). Nonetheless, the present results, at the very least, support that persistent occupancy of AS promoter by HIF-1α is associated with the suppressive effect of AS induction by arginine deprivation, thus supporting the negative role of HIF-1α in the regulation of AS expression.
Discussion
The finding that human malignant melanoma is auxotrophic for arginine provides a molecular basis for the development of targeted therapy of this disease based on arginine deprivation using recombinant ADI (6). In this study, we found that in A2058, SK-MEL-2 and A375 melanoma cell lines, AS expression levels were very low. The expression of AS could be induced in the former two cells lines but not in the last one by ADI. The inducibility of AS expression is correlated with the resistance to ADI. These results have a clinical reminiscence that some melanoma patients do not response to ADI treatment in clinical trials, underscoring the importance of elucidating the mechanism of induced expression of AS by ADI treatment.
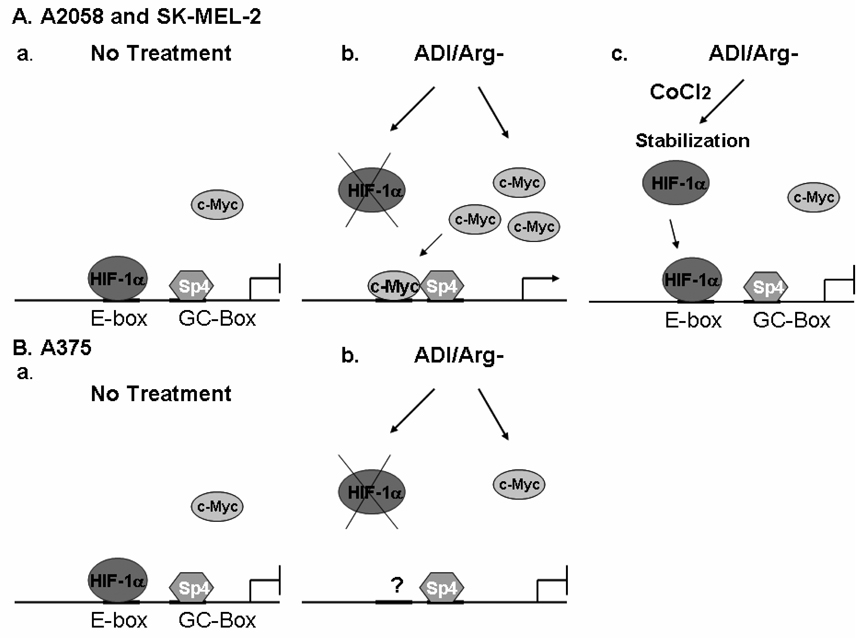
The association between AS expression levels and arginine availability was first noted more than 40 years ago (15). Multiple mechanisms have been reported for the regulation of AS expression, including transcriptional regulation (16–20) and epigenetic regulation (21, 22). However, many of these studies used nonmelanoma cells which express high basal levels of AS, so the results are not applicable to cancer chemotherapy using ADI because these cells used were not sensitive to ADI treatment. In the present study, we identified an E-box and a GC-box as arginine deprivation-responsive elements that control AS expression and elucidated that c-Myc/HIF-1α are the E-box transcription regulators and Sp4 is the major GC-box factor. Importantly, we describe that c-Myc and HIF-1α function as a positive and a negative regulator, respectively, and that a switch from HIF-1α to c-Myc binding to the E-box is accompanied with the induction of AS expression by arginine deprivation in A2058 and SK-MEL-2 cells (Fig. 6A). We further demonstrated that failure of AS induction in A375 cells by ADI was associated with the inability of c-Myc to interact with the AS promoter (Fig. 6B). To our knowledge, this is the first molecular description of how AS gene expression is induced in some but not all melanoma cells that bears clinical relevance to cancer chemotherapy using ADI-PEG20 (10).
Fig. 6.
Models depicting induction of AS expression in A2058 and SK-MEL-2 cells by arginine depletion through HIF-1α displacement by c-Myc (A). a. Under the regular culture conditions, HIF-1α acts as a repressor by binding to the E-box of AS gene. b. Under arginine-depleting conditions, HIF-1α is degraded and c-Myc is upregulated and directly binds to the E-box that drives transcription through interaction with the transcription factor Sp4 which is bound to the GC-box of AS promoter. c. In the presence of hypoxia mimics, CoCl2, HIF-1α is stabilized and remains bound at the E-box thereby preventing the induced AS expression by arginine depletion. B. Failure of c-Myc binding to AS promoter in A375 cells is associated with inability of AS expression under arginine-depleting conditions a. Under the regular culture conditions, HIF-1α acts as a repressor by binding to the promoter of AS gene. b. Under arginine-depleting conditions, HIF-1α is degraded but c-Myc does not bind to the E-box.
c-Myc and HIF-1α are among the most important oncoproteins in cancer biology. c-Myc plays a central role in a transcriptional regulation network that regulates cell growth, differentiation, apoptosis and metabolism signaling (23, 24). Likewise, HIF-1α plays a critical role in tumor angiogenesis, glycolysis, and metastasis, redox signaling and response to radiation and chemotherapy (25–28). All of these pathways are critically involved in the growth, proliferation, invasiveness and responsiveness to therapy of cancer cells. For these reasons, regulation mechanisms of c-Myc and HIF-1α have been extensively studied. However, the mechanisms by which arginine starvation modulates c-Myc and HIF-1α which in turn regulate AS expression, are currently unknown. It is plausible that PI3K/Akt signaling may be involved. Indeed, several studies have documented the cross-regulatory mechanisms among c-Myc and HIF-1α and PI3K/Akt signaling. (i) It has been described previously that constitutively activated Akt upregulates HIF-1α expression in melanoma in vivo (29), suggesting a feedback mechanism of Akt activity through HIF-1α level that can be abrogated by hyperactivation of Akt. (ii) It has been demonstrated that PI3K/Akt signaling regulates c-Myc promoter activity, mRNA stability, and protein stability through its downstream genes, GSK3β and β-catenin (30, 31). (iii) Akt phosphorylates and sequesters the transcription factors FOXO3a, Mad-1, and Miz-1, which inhibit the transactivation of c-Myc target genes (32–34). (iv) Moreover, it has been demonstrated that HIF-1α can inhibit c-Myc activity by direct interaction with c-Myc, transactivation of c-Myc corepressor Mxi1 or promoting proteosome-dependent degradation of c-Myc (24, 35). (v) Energy starvation by means of amino acid deprivation has profound effects on the function of HIF-1α and c-Myc (36) relevant to mitochondrial physiology. Expression of HIF-1α has been shown to affect two mitochondrial enzymes, pyruvate dehydrogenase (PDK1) and cytochrome C oxidase (37–39). In contrast, c-Myc promotes mitochondrial respiration through mitochondrial biogenesis (40). Moreover, it has been reported that disruption of HIF-1α activity using RNAi resulted in increased mitochondrial biogenesis and O2 consumption. Conversely, targeted disruption of c-Myc diminished mitochondrial DNA and O2 consumption (24). These results suggest that HIF-1α and c-Myc are oppositely regulated through mitochondrial metabolism. These observations, together with our finding that HIF-1α can also regulate c-Myc levels, illustrate a complex interregulatory network involving HIF-1α and c-Myc through PI3K/Akt signaling in the regulation of AS expression in response to arginine availability. This complex mechanism may explain, at least in part, the observed delayed induction of AS expression by arginine deprivation.
The complex regulatory mechanism of AS expression by arginine deprivation is compounded by the observations that c-Myc interacting transcription factor Sp4 is also involved. We demonstrated that Sp4 and Sp3 are the GC-box interacting factors. Previous studies on regulation of AS gene expression have showed that Sp1 interaction with the GC-box is important for transactivation of the AS promoter (16, 17, 41). Our present results demonstrate that Sp1 shows only a minor stimulatory function on the AS promoter as compared with Sp4 (Fig. 2D). Further investigation using Sp4 siRNA strategy supports the functional role of Sp4 in the regulation of AS expression. Moreover, co-IP results demonstrated a physical interaction between Sp4 and c-Myc. It is important to note that roles of other members of Sp1 family were not examined in these previous studies (16, 17), nor were siRNA and co-transfection assays performed.
This study also raised several important issues that have yet-to-be resolved: (i) Induction of AS expression by arginine deprivation in some melanoma cells is apparently initiated by the rapid disappearance of HIF-1α, because accumulation of HIF-1α by hypoxia mimics blocked the subsequent events leading to suppression of AS induction. It is likely that arginine deprivation induces HIF-1α degradation because HIF-1α Mrna levels were not correspondingly reduced (unpublished results). HIF-1α is a highly unstable protein (t½ ~ 10 min under normaxia conditions). The underlying mechanisms that accelerate HIF-1α degradation under arginine-depleted conditions remain to be determined. Alternatively, arginine deprivation may also suppress the translation of HIF-1α (ii) Our results demonstrated that apparently dissociation of HIF-1α from the E-box is not sufficient and that c-Myc has to be engaged in transcriptional activation of AS expression as demonstrated in A375 cells. The precise mechanism(s) by which c-Myc fails to interact with the AS promoter in A375 cells under arginine depletion conditions are currently unknown. It is plausible that the E-box in A375 chromatin may not be in open configuration due to an epigenetic mechanism such as DNA methylation (22) or masked by yet-to-be determined transcriptional repressors. Alternatively, c-Myc in A375 cells may be sequestered, rendering it unable to transcriptionally activate AS expression under arginine deprivation conditions. (iii) The regulatory mechanisms underlying the delayed AS induction by arginine deprivation remain to be critically investigated. (iv) Finally, the roles of HIF-1α, c-Myc, and Sp4 in the regulation of AS expression that associated with ADI sensitivity in malignant melanoma chemotherapy remain to be determined. Studies aimed at addressing these issues are currently underway.
In summary, we have revealed the roles of three transcriptional regulators, c-Myc, HIF-1α, and Sp4, in the regulation of AS expression in response to arginine deprivation in three melanoma cell lines. Among which, the accessibility of c-Myc to interact with the AS promoter is critical for the induction of AS expression. Lack of AS expression has been considered a biomarker for sensitivity to ADI treatment in melanomas. Our present study shows that inducibility of AS expression is associated with sensitivity to ADI treatment. These results suggest that mere screening of AS expression levels in melanoma patients prior to ADI treatment is not sufficient. Our present study has gone one step further and elucidated the mechanisms of AS induction. The identification of positive and negative regulators should provide an important rationale for the development of effective treatment modalities for malignant melanoma through modulation of AS expression.
Materials and methods
Reagent, Cell culture and Antibodies
ADI-PEG20 (specific activity: 5~10 IU/mg) was obtained from Polaris Pharmacologies Inc. Deferoxamine (DFX), cobalt chloride, and sulforhodamine B (SRB) were purchased from Sigma (St. Louis, MO) and LY294002 was purchased from Cayman Chemical (Ann Arbor, MI).
A2058, SK-MEL-2 and A375 melanoma cells were purchased from American Type Culture Collection Center (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). Arginine-free medium was purchased from Invitrogen (Carlsbad, CA) and was supplemented with 10% FBS that had been dialyzed against phosphate-buffered saline (PBS). For arginine depletion, cells were washed with PBS and maintained in DMEM containing 0.05 µg/ml ADI-PEG20 or in arginine-free medium.
For cell growth assay, 100–500 cells were seeded in 96-well plates for various lengths of time. Cells were fixed with 10% tri-chloro acetate, washed with distilled water, and stained with 0.4% SRB. The excess SRB dye was washed with 0.1% acetic acid and the remaining SRB was dissolved in 10 mM Tris buffer. The relative number of cell was measured by absorbance at 405 nm.
Mouse anti β-actin, anti-HA (Sigma), mouse anti-AS (BD Bioscience), mouse anti-myc (9E10, Roche), rabbit anti-Sp1 (Santa Cruz Biotechnology), rabbit anti-Sp2 (Santa Cruz Biotechnology), rabbit anti-Sp3 (Santa Cruz Biotechnology), rabbit anti-Sp4 (Santa Cruz Biotechnology, CA), rabbit anti-c-Myc (N262, Santa Cruz Biotechnology, CA), mouse anti c-Myc (C33, Santa Cruz Biotechnology, CA), mouse anti HIF1α (BD Bioscience, Cat no 610958, Lot 23664) were purchased commercially.
Plasmid DNA construction and siRNA transfection
The expression vector for pCGN-HA-c-Myc was generously provided by Dr. William P. Tansey (42). The pCMV4-Sp2 vector was generously provided by Dr. Jonathan M. Horowitz (43). The Sp3 expression vector was described previously (44). The expression vector for pCMV-Sp4 was generously provided by Dr. Manohar Ratnam (45). The constitutive active (CA) or dominant negative (DN) forms of HA-Akt expression constructs were described previously (46).
The proximal promoter region of AS gene (from −1822 to +300) was cloned by PCR using GC-rich PCR system (Roche). The used primers are F: 5’-ACGCGTCCTCACTGTCCCTCATGTCACTGG-3’, R: 5’-CTCGAGCGGGGCGGGCGTCTTCTAC-3’. The deletion contracts were created by PCR using same reverse primer and following forward primers; −1334: 5’-ACGCGTCCTGTGAGAGGGCTGAGGGGGCC-3’, −846: 5’-ACGCGTCTTTGGAGCCGGCAGACCAAGCTG-3’, −602: 5’-ACGCGTGGGATGAAAGCCGGGCCTTCCCGC-3’, −358: 5’-ACGCGTGCCACCCTCCGCCCCTGAGTTACA-3’, −85: 5’-ACGCGTGCGCGCGTCCCCGCCACGTGT-3’, −37: 5’-ACGCGTCCTGTGCTTATAACCTGGGAT-3’. The amplified DNA fragments were cloned using TOPO PCR system (Invitrogen (Carlsbad, (CA)) and sequence integrity was analyzed. The promoter region were digested by MluI/XhoI and ligated with pGL3 vector. The point mutations were introduced into E-box and GC-box sequence by using mutagenesis kit (Stratagene, CA).
The siRNAs for HIF1α (5’-GAUUAACUCAGUUUGAACU-3’) and AS (5’-GCAAUGACCUGAUGGAGUA-3’) were synthesized by Sigma, c-Myc siRNA was purchased from Cell Signaling and control siRNA and Sp4 siRNA were from Santa Cruz Biotech (Santa Cruz, CA). The siRNA and plasmid DNA were transfected using lipofectamine 2000 (Invitrogen, CA) according to the manufacturer’s instruction.
Immunoprecipitation and Immunoblotting
All immunoprecipitation experiments were performed as described previously (47). Briefly, protein samples were incubated with an antibody and 25 µl 50% protein A sepharose slurry, and Protein A beads were collected. Immunoprecipitates were resolved by SDS–PAGE and analyzed by immunoblotting. For immunoblot analysis, the protein samples were subjected to SDS–PAGE and transferred onto PVDF membranes. The membranes were blocked, incubated with primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies, and visualized by an enhanced chemiluminescence kit.
RT-PCR
Total RNA was isolated with trizole reagent (Invitrogen) following manufacturer’s instruction. The cDNA was synthesized from 1 µg of total RNA by using a Super Script II system (Invitrogen). The synthesized cDNA was subjected to standard PCR or realtime PCR using following oligonucleotide primers. c-Myc: forward: 5’-CCTACCCTCTCAACGACAGC-3’, reverse: 5’-ACTCTGACCTTTTGCCAGGA-3’, ASS: forward: 5’-AGGCACCATCCTTTACCATG-3’. reverse: 5’- CTGCACTTTCCCTTCCACTC-3’, β-actin: forward: 5’-GAGGCCCAGAGCAAGAGAG-3’, reverse: 5’-AGAGGCGTACAGGGATAGCA-3’.
Chromatin immunoprecipitation (ChIP) Assay
ChIP assay was carried out with a ChIP assay kit (Upstate Biotechnology) following manufacturer’s instruction. A GC-rich PCR system (Roche) was used for PCR analysis using following AS promoter-specific primers (forward: 5’-TGAGTTACATGGGTCGCAGCCACTG-3’, reverse: 5’-GCCCATCCCAGGTTATAAGCACAGG-3’) or primers for GAPDH promoter (forward: 5’-TGTCTGCCCTAATTATCAGG-3’, reverse: 5’-GGAAAGTTGGGGAACTTCTC-3’). The quantitative analyses on enrichment of precipitated DNA were also performed as described previously (48).
Luciferase assay
Luciferase assay was carried out using dual promoter gene reporter kit (Promega, WI) following manufacture’s instruction. In brief, cells cultured in 24 well dishes were transfected with 0.2 µg of pGL3 vector and 2 ng of pRLII vector using lipofectamine 2000 (Invitrogen) following manufacture’s instruction. After incubation in normal medium containing 0.05 µg/ml ADI or in arginine-free medium, cells were lysed with passive lysis buffer. Cell lysates were cleared by centrifugation and supernatants were used for dual luciferase assay. The obtained luciferase activity was normalized with that of empty pGL3 vector.
Acknowledgments
This work was supported by NCI grants 1RO1 CA79085 and CA89541, and M. D. Anderson Institutional CTT/TI-3D Grant (M. T. K.) and R01 CA109578 (L. F.) and a Merit research grant from VA (N. S.). We would like to thank Polaris Inc. for providing ADI-PEG20.
Abbreviations list
- AS
Argininosuccinate synthetase
- ADI
Arginine deiminase
- HIF-1α
Hypoxia inducible factor 1 alpha
- PI3K
Phosphoinositide 3-kinase
- DFX
deferoxamine
- SRB
sulforhodamine B
- ChIP
chromatin immunoprecipitation.
Reference
- 1.Eberle J, Kurbanov BM, Hossini AM, Trefzer U, Fecker LF. Overcoming apoptosis deficiency of melanoma-hope for new therapeutic approaches. Drug Resist Updat. 2007;10:218–234. doi: 10.1016/j.drup.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Tawbi HA, Kirkwood JM. Management of metastatic melanoma. Semin Oncol. 2007;34:532–545. doi: 10.1053/j.seminoncol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Yoon CY, Shim YJ, Kim EH, et al. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. Int J Cancer. 2007;120:897–905. doi: 10.1002/ijc.22322. [DOI] [PubMed] [Google Scholar]
- 4.Wheatley DN. Arginine deprivation and metabolomics: important aspects of intermediary metabolism in relation to the differential sensitivity of normal and tumour cells. Semin Cancer Biol. 2005;15:247–253. doi: 10.1016/j.semcancer.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Dillon BJ, Prieto VG, Curley SA, et al. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer. 2004;100:826–833. doi: 10.1002/cncr.20057. [DOI] [PubMed] [Google Scholar]
- 6.Cheng PN, Lam TL, Lam WM, et al. Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. 2007;67:309–317. doi: 10.1158/0008-5472.CAN-06-1945. [DOI] [PubMed] [Google Scholar]
- 7.Izzo F, Marra P, Beneduce G, et al. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: results from phase I/II studies. J Clin Oncol. 2004;22:1815–1822. doi: 10.1200/JCO.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 8.Ascierto PA, Scala S, Castello G, et al. Pegylated arginine deiminase treatment of patients with metastatic melanoma: results from phase I and II studies. J Clin Oncol. 2005;23:7660–7668. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- 9.Ni Y, Schwaneberg U, Sun ZH. Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 2008;261:1–11. doi: 10.1016/j.canlet.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Feun L, Savaraj N. Pegylated arginine deiminase: a novel anticancer enzyme agent. Expert Opin Investig Drugs. 2006;15:815–822. doi: 10.1517/13543784.15.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 12.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koshiji M, To KK, Hammer S, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 15.Schimke RT. Enzymes of Arginine Metabolism in Mammalian Cell Culture. I. Repression of Argininosuccinate Synthetase and Argininosuccinase. J Biol Chem. 1964;239:136–145. [PubMed] [Google Scholar]
- 16.Brasse-Lagnel C, Fairand A, Lavoinne A, Husson A. Glutamine stimulates argininosuccinate synthetase gene expression through cytosolic O-glycosylation of Sp1 in Caco-2 cells. J Biol Chem. 2003;278:52504–52510. doi: 10.1074/jbc.M306752200. [DOI] [PubMed] [Google Scholar]
- 17.Brasse-Lagnel C, Lavoinne A, Loeber D, et al. Glutamine and interleukin-1beta interact at the level of Sp1 and nuclear factor-kappaB to regulate argininosuccinate synthetase gene expression. Febs J. 2007;274:5250–5262. doi: 10.1111/j.1742-4658.2007.06047.x. [DOI] [PubMed] [Google Scholar]
- 18.Brasse-Lagnel C, Lavoinne A, Fairand A, Vavasseur K, Husson A. IL-1beta stimulates argininosuccinate synthetase gene expression through NF-kappaB in Caco-2 cells. Biochimie. 2005;87:403–409. doi: 10.1016/j.biochi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Guei TR, Liu MC, Yang CP, Su TS. Identification of a liver-specific cAMP response element in the human argininosuccinate synthetase gene. Biochemical and biophysical research communications. 2008;377:257–261. doi: 10.1016/j.bbrc.2008.09.118. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin BL, Pendleton LC, Levy MM, Solomonson LP, Eichler DC. Tumor necrosis factor-alpha reduces argininosuccinate synthase expression and nitric oxide production in aortic endothelial cells. American journal of physiology. 2007;293:H1115–H1121. doi: 10.1152/ajpheart.01100.2006. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson LJ, Smith PR, Hiller L, et al. Epigenetic silencing of argininosuccinate synthetase confers resistance to platinum-induced cell death but collateral sensitivity to arginine auxotrophy in ovarian cancer. International journal of cancer. 2009;125:1454–1463. doi: 10.1002/ijc.24546. [DOI] [PubMed] [Google Scholar]
- 22.Szlosarek PW, Klabatsa A, Pallaska A, et al. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin Cancer Res. 2006;12:7126–7131. doi: 10.1158/1078-0432.CCR-06-1101. [DOI] [PubMed] [Google Scholar]
- 23.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annual review of cell and developmental biology. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Gao P, Fukuda R, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL. Hypoxia and human disease-and the Journal of Molecular Medicine. Journal of molecular medicine (Berlin, Germany) 2007;85:1293–1294. doi: 10.1007/s00109-007-0285-z. [DOI] [PubMed] [Google Scholar]
- 26.Thomas GV, Tran C, Mellinghoff IK, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nature medicine. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. Targeting HIF-1 for cancer therapy. Nature reviews. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 28.Melillo G. Inhibiting hypoxia-inducible factor 1 for cancer therapy. Mol Cancer Res. 2006;4:601–605. doi: 10.1158/1541-7786.MCR-06-0235. [DOI] [PubMed] [Google Scholar]
- 29.Bedogni B, Welford SM, Cassarino DS, Nickoloff BJ, Giaccia AJ, Powell MB. The hypoxic microenvironment of the skin contributes to Akt-mediated melanocyte transformation. Cancer Cell. 2005;8:443–454. doi: 10.1016/j.ccr.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A, Marques M, Carrera AC. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S phase entry. Mol Cell Biol. 2006;26:9116–9125. doi: 10.1128/MCB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galmozzi E, Casalini P, Iorio MV, Casati B, Olgiati C, Menard S. HER2 signaling enhances 5'UTR-mediated translation of c-Myc mRNA. J Cell Physiol. 2004;200:82–88. doi: 10.1002/jcp.20012. [DOI] [PubMed] [Google Scholar]
- 32.Wanzel M, Kleine-Kohlbrecher D, Herold S, et al. Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nature cell biology. 2005;7:30–41. doi: 10.1038/ncb1202. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci U S A. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandramohan V, Jeay S, Pianetti S, Sonenshein GE. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J Immunol. 2004;172:5522–5527. doi: 10.4049/jimmunol.172.9.5522. [DOI] [PubMed] [Google Scholar]
- 35.Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutphin PD, Giaccia AJ, Chan DA. Energy regulation: HIF MXIes it up with the C-MYC powerhouse. Dev Cell. 2007;12:845–846. doi: 10.1016/j.devcel.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 38.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell metabolism. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Li F, Wang Y, Zeller KI, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur J Biochem. 2003;270:1887–1899. doi: 10.1046/j.1432-1033.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 42.Alarcon-Vargas D, Tansey WP, Ronai Z. Regulation of c-myc stability by selective stress conditions and by MEKK1 requires aa 127-189 of c-myc. Oncogene. 2002;21:4384–4391. doi: 10.1038/sj.onc.1205543. [DOI] [PubMed] [Google Scholar]
- 43.Moorefield KS, Fry SJ, Horowitz JM. Sp2 DNA binding activity and trans-activation are negatively regulated in mammalian cells. J Biol Chem. 2004;279:13911–13924. doi: 10.1074/jbc.M313589200. [DOI] [PubMed] [Google Scholar]
- 44.Huang N, Dardis A, Miller WL. Regulation of cytochrome b5 gene transcription by Sp3, GATA-6, and steroidogenic factor 1 in human adrenal NCI-H295A cells. Mol Endocrinol. 2005;19:2020–2034. doi: 10.1210/me.2004-0411. [DOI] [PubMed] [Google Scholar]
- 45.Shatnawi A, Tran T, Ratnam M. R5020 and RU486 act as progesterone receptor agonists to enhance Sp1/Sp4-dependent gene transcription by an indirect mechanism. Mol Endocrinol. 2007;21:635–650. doi: 10.1210/me.2006-0274. [DOI] [PubMed] [Google Scholar]
- 46.Kuo MT, Liu Z, Wei Y, et al. Induction of human MDR1 gene expression by 2-acetylaminofluorene is mediated by effectors of the phosphoinositide 3-kinase pathway that activate NF-kappaB signaling. Oncogene. 2002;21:1945–1954. doi: 10.1038/sj.onc.1205117. [DOI] [PubMed] [Google Scholar]
- 47.Tsai WB, Chung YM, Takahashi Y, Xu Z, Hu MC. Functional interaction between FOXO3a and ATM regulates DNA damage response. Nature cell biology. 2008;10:460–467. doi: 10.1038/ncb1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–4179. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]