Abstract
Cell migration is a critical step in cancer cell invasion. Recent studies have implicated the importance of the ERK signaling pathway in cancer cell migration. However, the mechanism associated with ERK-regulated cell migration is poorly understood. Using a panel of breast cancer cell lines, we detected an excellent correlation between ERK activity and cell migration. Interestingly, we noticed that a 48-hr treatment with U0126 (specific MEK1/2 inhibitor) was needed to significantly inhibit breast cancer cell migration while this inhibitor blocked ERK activity within 1 hr. This observation suggests that ERK-dependent gene expression rather than direct ERK signaling is essential for cell migration. In further study, we found that ERK activity promoted the expression of AP1 components Fra-1 and c-Jun, both of which were necessary for cell migration. Combination of U0126 treatment and Fra-1/c-Jun knockdown did not yield further reduction in cell migration than either alone, indicating that ERKs and Fra-1/c-Jun act in the same mechanism to facilitate cell migration. In an attempt to investigate the role of Fra-1/c-Jun in cell migration, we found that ERK-Fra-1/c-Jun axis regulated slug expression in an AP1-dependent manner. Moreover, the occurrence of U0126-induced migratory inhibition coincided with slug reduction, and silencing slug expression abrogated breast cancer cell migration. These results suggest an association between ERK-regulated cell migration and slug expression. Indeed, cell migration was not significantly inhibited by U0126 treatment or Fra-1/c-Jun silencing in cells expressing slug transgene. Our study suggests that the ERK pathway regulates breast cancer cell migration by maintaining slug expression.
Keywords: ERK, slug and cell migration
Introduction
Invasion plays a critical role in tumor metastasis and is a multiple step process (1). During invasion, cells first interact with the surrounding extracellular matrix (ECM), subsequently degrade or remodel the surrounding ECM and eventually migrate through the dissolved ECM to reach adjacent tissues (1). Among all these steps, cell migration is one of the most critical rate-limiting steps and is a complex process that requires the coordinate assemble and dissemble of focal adhesion complexes (2). Major effort has been exerted on defining the mechanisms associated with focal adhesion complex turnover and identifying molecules essential to this cellular event. However, the knowledge on the signaling pathways relevant to cell migration process is still lacking.
There are at least three families of mitogen-activated protein kinases (MAPKs), namely extracellular signal-regulated kinases (ERKs), Jun N-terminus kinases (JNKs) and p38 MAPKs. In addition to their well-characterized role in cell proliferation/differentiation and cell survival/apoptosis, the MAPK signaling pathways also actively participate in cell migration. The evidences connecting JNK to cell migration include: 1) inhibiting JNK activity or ablating JNK impairs cell migration of diverse cell types including fibroblasts, endothelial and Schwann cells (3-5) and 2) JNKs phosphorylate adhesion paxillin and non-JNK-phosphorylable paxillin inhibits cell migration (6). The evidences linking p38 MAPK to cell migration include: 1) p38 inhibitor blocks integrin-mediated breast cancer cell migration (7) and 2) caldesmon is a potent p38 substrate and p38-mediated caldesmon phosphorylation is required for smooth muscle cell migration (8). The use of chemical and genetic inhibitors has also implicated the role of the ERK signaling pathway in the cell migration (9-11). However, it is poorly understood how the ERK signaling pathway is involved in cell migration.
The AP1 transcription factors are the dimers of the Fos (c-Fos, FosB, Fra-1 and Fra-2) and Jun (c-Jun, JunB and JunD) families (12, 13), and activation of AP1 is essential for cancer initiation by Ras (14) and ETV-NTRK3 fusion oncogene (15). Among AP1 factors, Fra-1 is frequently elevated in metastatic breast, glioma and thyroid cancers (16-18). Fra-1 may play a role in cell migration since ectopically expressing Fra-1 enhances cell migration and blocking Fra-1 reduces cell migration (19, 20). As Fra-1 is expressed in an ERK-dependent manner (21-23), it is very likely that the ERK pathway regulates cell migration through Fra-1. This possibility is supported by a report in that Fra-1 was shown to act downstream of ERK to facilitate cell migration (24).
Slug is a member of the snail family and is upregulated in metastatic breast cancer and mesothelioma (25, 26). Recent evidences strong suggest that slug acts as an potent inducer of cell movement (27). For example, skin explants from slug-null mice have retarded migration rates of keratinocytes (28). Forced expression of slug in corneal explants resulted in higher rates of corneal epithelial cell migration (29). As slug is capable of regulating integrin expression (30), it may impact cell migration by modulating cell-substratum interaction.
In this study, we observed an excellent correlation between ERK activity and cell migration among breast cancer cell lines. However, our results indicate that ERK-dependent gene expression rather than direct ERK signaling is involved in cell migration. To determine the potential involvement of AP1 dimer in ERK-regulated cell migration, we showed that Fra-1 and c-Jun expression is dependent on ERK activity. Although prolonged treatment of U0126 (an MEK1/2 inhibitor), Fra-1 and c-Jun shRNAs were all capable of inhibiting MDA-MB-231 and MDA-MB-436 cell migration, combined treatment of all three did not show additive effect on the reduction in cell migration. This observation supports the notion that ERK and AP1 work in the same signaling axis to facilitate cell migration. To define the role of Fra-1/c-Jun in cell migration, we performed a microarray analysis with total RNA isolated from Fra-1/c-Jun knockdown MDA-MB-231 cells and identified slug as a Fra-1/c-Jun-regulated gene. Silencing slug expression diminished MDA-MB-231 and MDA-MB-436 cell migration and forced slug expression effectively reversed the inhibition in cell migration caused by prolonged U0126 treatment or Fra-1/c-Jun shRNA. Our results suggest that the ERK-Fra-1/c-Jun axis regulates breast cancer cell migration by maintaining slug expression.
Materials and Methods
Immunoblotting
Immunoblotting was performed as previously described (31). To determine the effect of U0126 on ERK phosphorylation, Fra-1, c-Jun, Fra-2 or slug, cells were treated with 5μM U0126 for various times followed by immunoblotting. To determine shRNA effect, cells were infected with lentiviral vectors containing shRNAs for 4 days and then lysed for immunoblotting. The antibodies used in immunoblotting were all obtained from (Santa Cruz Biotechnology, Santa Cruz, CA).
Cell migration assay
Cell migration was assayed by Transwells and wound healing assays as described previously (31). For Transwell assay, cells were detached with 10mM EDTA-containing PBS, then resuspended in serum-free medium and allowed migration for 4 hrs. To determine the effect of U0126 on cell migration, cells were treated with 5μM U0126 for various times in medium containing 10% FCS. To determine the effect of Fra-1, c-Jun or slug knockdown, cells were infected with lentiviral vector containing luciferase (control), Fra-1, c-Jun or slug shRNA for 4 days prior to the analysis of cell migration. To determine the effect of murine slug expression on U0126-caused inhibition in cell migration, cells were infected with lentiviral vector encoding murine slug cDNA for 2 days and then treated with 5μM U0126 for another 2 days followed by assaying cell migration. To determine the effect of murine slug expression on Fra-1/c-Jun shRNA-caused inhibition in cell migration, cells were first infected with lentiviral vectors containing Fra-1 and c-Jun shRNA for 2 days and then again infected with lentiviral vector encoding murine slug cDNA for 2 days before assaying cell migration.
Microarray analysis
Total RNA was isolated from control (luciferase shRNA) or Fra-1/c-Jun-knockdown MDA-MB-231 cells using Trizol (Invitrogen). The microarray was performed using Illumina whole genome BeadArray technology. The microarray data were initially processed using the lumi package in R (32). The data were then transformed using the variance stabilizing transformation and robust spline normalization (33). The processed data were analyzed using the LIMMA package in R (34).
Analysis of slug promoter activity
The construction of wild-type and AP1-mutant slug promoter reporter plasmids and luciferase assay were described in the Supplementary Data. The luciferase was analyzed using Duel Luciferase System (Promega, Madison, WI) according to manufacturer's protocol. To determine the effect of U0126 on slug or RSV promoter activities, cells were transfected with the reporter gene constructs for 1 day and 5μM U0126 then added to cells for another 2 days prior to luciferase activity assay. To determine the effect of silencing Fra-1/c-Jun on promoter activity, the reporter gene construct was transfected into Fra-1/c-Jun shRNA-expressing cells for 2 days followed by assaying luciferase activity.
In vitro invasion assay, tumor outgrowth and lung metastasis
In vitro cell invasion was assayed using matrigel invasion chamber (Cellbio Labs, San Diego, CA) according to manufacturer's protocol. Control or slug-knockdown MDA-MB-231 cells were added into matrigel-coated chamber and allowed to invade for 24 hrs.
In vivo tumor development was done by subcutaneously injecting control or slug-knockdown MDA-MB-231 cells (3 × 106 cells/mouse) at 4th mammary fat pad area of female athymic nude mice (6-7 weeks of age, Harlan Laboratories, Indianapolis, IN). Xenografts were measured using a caliper and tumor volume (V) calculated by equation: V = (L × W2) × 0.5, where L is the length and W is the width of a xenograft. To determine metastasis to the lung, mice were sacrificed at 8 weeks after injection and lungs removed for formalin fixation. The sections were cut from the fixed lung tissue and subjected to hematoxylin and eosin (H&E) staining. The metastatic lesions were visualized under microscope.
Statistical Analysis
Statistical analyses of cell migration and matrigel invasion were done by student t test using Microsoft Excel software. Comparison between animal receiving control and slug knockdown cells were made by Student-Newman-Keuls test using the SPSS software program (SPSS Inc., Chicago, IL). P < 0.05 was considered statistically significant.
Results
ERK-dependent gene expression rather than direct ERK signaling is required for cell migration
To investigate the potential role of the ERK signaling pathway in cell migration, we examined both ERK activity and cell migration in six breast cancer cell lines. Immunoblotting with anti-phosphor-ERK(Thr202/Tyr204) mAb showed that ERK phosphorylation (ERK activity) was high in BT-549, MDA-MB-231 and MDA-MB-436 but low in MCF-7, T47D and ZR-75-1 lines (Fig.1A). Transwell migration assay showed that lines with high ERK activity were migratory while lines with low ERK activity were poorly migratory (Fig.1A). These results demonstrate an excellent correlation between ERK activity and cell migration.
Fig.1. ERK-dependent gene expression is required for breast cancer cell migration.
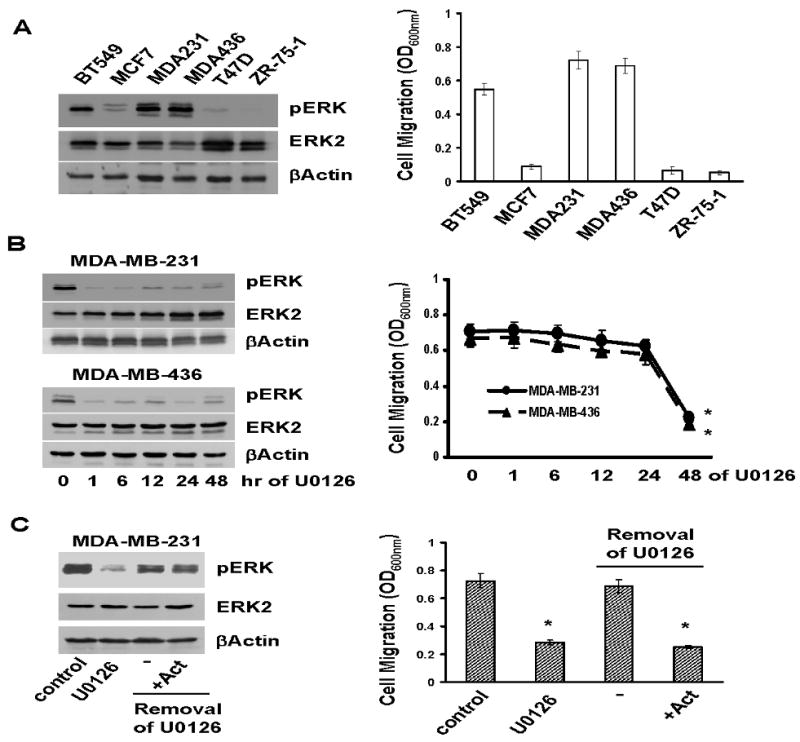
A. A portion of overnight-cultured BT549, MCF7, MDA-MB-231, MDA-MB-436, T47D and ZR-75-1 cells were lysed and lysates subjected to immunoblotting to detect phosphor-ERK, ERK2 and βactin with the respective antibodies. The remaining portion of cells was analyzed for cell migration using Transwells as described in “Materials and Methods”. Data are means ± S.E. B. MDA-MB-231 and MDA-MB-436 cells were treated with 5μM U0126 for various times. A portion of cells was analyzed for the levels of phosphor-ERK, ERK2 and βactin and the remaining portion analyzed for cell migration. Data are means ± S.E (*, p < 0.005 vs 0-hr). C. MDA-MB-231 cells were treated with 5μM U0126 for 2 days, then washed with serum-free medium three times to remove U0126, and incubated in the absence or the presence of 2 μg/ml actinomycin for 4 h. A portion of cells were analyzed for phosphor-ERK, ERK2 and βactin levels and the remaining portion assayed for cell migration. Data are means ± S.E. (*, p < 0.005 vs control).
We next investigated the importance of ERK activity in cell migration by treating MDA-MB-231 and MDA-MB-436 cells with 5μM U0126 (a specific MEK1/2 inhibitor) for various times. One-hour U0126 treatment was enough to block over 90% of ERK phosphorylation (Fig.1B); however, cell migration was only substantially inhibited after 2-day U0126 treatment (Fig.1B). The time disparity between the rapid inhibition of ERK phosphorylation (<1 hr) and delayed onset of cell migration inhibition (∼2 days) strongly suggests that the ERK does not participate in cell migration through direct signaling.
The ERK signaling pathway regulates the expression of various genes (35), we thus tested whether ERK-dependent gene expression was required for breast cancer cell migration. MDA-MB-231 cells were first treated with U0126 for 2 days and U0126 was then removed by several washes. Cells were cultured in 10% FCS-containing medium with or without 2μg/ml actinomycin (RNA synthesis inhibitor) for 4 hrs followed by the analyses of ERK phosphorylation and cell migration. ERK activity was completely restored 4 hrs after removal of U0126 and actinomycin had no effect on ERK re-activation (Fig.1C). Removal of U0126 restored cell migration (Fig.1C); however, the presence of actinomycin prevented cell migration (Fig.1C). As actinomycin prevents de novo gene expression, these results suggest that ERK-regulated gene expression is required for cell migration.
Fra-1/c-Jun and ERKs regulate breast cancer cell migration in the same mechanism
Fra-1 is regulated in an ERK-dependent manner (21-23) and has been indicated to be important for cell migration (19, 20). To investigate whether Fra-1 and other AP1 components were functionally linked to ERK in the regulation of cell migration, we first examined the levels of Fra-1, Fra-2 and c-Jun in breast cancer cell lines. Fra-1 and c-Jun levels were high in BT-549, MDA-MB-231 and MDA-MB-436 lines (Fig.2A), and these lines also displayed high ERK activity and were migratory (Fig.1A). In contrast, Fra-1 and c-Jun levels were low in MCF-7, T47D and ZR-75-1 lines (Fig.2A), and these lines also exhibited low ERK activity and were poorly migratory (Fig.1A). Fra-2 was seen in all but T47D line and its levels were not correlated to ERK activity (Fig.2A). This observation demonstrates an excellent correlation between the levels of Fra-1/c-Jun and ERK activity/cell migration in breast cancer cells.
Fig.2. ERK-Fra-1/c-Jun axis is involved in cell migration.

A. BT549, MCF7, MDA-MB-231, MDA-MB-436, T47D and ZR-75-1 cells were lysed and lysates subjected to immunoblotting to detect Fra-1, c-Jun, Fra-2 and βactin with the respective antibodies. B. MDA-MB-231 and MDA-MB-436 cells were treated with 5μM U0126 for various times and then lysed for immunoblotting to detect Fra-1, c-Jun, Fra-2 and βactin. C. MDA-MB-231 and MDA-MB-436 cells were either infected with control lentiviral vector (luciferase shRNA) or vector containing Fra-1, c-Jun shRNA or both. Some Fra-1/c-Jun shRNA expressing cells were further treated with 5μM U0126 for 2 days. Cells were detached with 10 mM EDTA and then assayed for cell migration as described in “Materials and Methods”. Data are means ± S.E. (*, p < 0.005 vs control; #, p < 0.01 vs control).
We next investigated whether ERK activity was required for Fra-1/c-Jun expression by treating MDA-MB-231 and MDA-MB-436 cells with 5μM U0126 for various times. Immunoblotting with the respective antibodies showed that the levels of Fra-1 and c-Jun were greatly decreased at 8 hrs and nearly abolished at 24 hrs (Fig.2B), suggesting that ERK activity is required for high levels of Fra-1 and c-Jun in migratory breast cancer cells. However, U0126 did not significantly alter Fra-2 expression in both lines (Fig.2B). This corroborates the observation that there is no clear correlation between Fra-2 and ERK activity in breast cancer cells (Fig.2A).
To determine the importance of Fra-1/c-Jun in breast cancer cell migration, we designed two shRNAs each for Fra-1 and c-Jun and expressed them in MDA-MB-231 and MDA-MB-436 cells. Though all shRNAs effectively suppressed their target gene expression (Fig.S1), they did not affect the status of ERK phosphorylation (Fig.S1), which agrees that ERK is in upstream to regulate Fra-1/c-Jun expression. Transwell migration assay showed that silencing either Fra-1 or c-Jun reduced over 60% of cell migration (Fig.2C), but Fra-1 and c-Jun shRNAs together did not lead to greater inhibition in cell migration than either alone (Fig.2C). Similar results were also obtained with wound healing assay (Fig.S2). These results indicate that Fra-1 and c-Jun are involved in cell migration with the same mechanism. To rule out the potential off-target effect of shRNAs, we expressed murine Fra-1 and c-Jun in Fra-1/c-Jun-knockdown MDA-MB-231 cells and found their expression was able to rescue cell migration to the control level (Fig.S3). In a parallel experiment, we examined the combinational effect of inhibiting ERK activity and silencing Fra-1/c-Jun on cell migration. MDA-MB-231 and MDA-MB-436 cells expressing both Fra-1 shRNA1 and c-Jun shRNA2 were treated with U0126 for 2 days followed by the analysis of cell migration. Additional U0126 treatment exhibited only slightly greater inhibitory effect on cell migration than Fra-1/c-Jun knockdown alone (Fig.2C and Fig.S2). Since the expression of Fra-1 and c-Jun is ERK-dependent (Fig.2B), these results suggest that the ERK signaling pathway may indirectly participate in cell migration by promoting Fra-1/c-Jun expression.
ERK-Fra-1/c-Jun axis regulates slug expression
U0126 abolished over 90% Fra-1/c-Jun expression in less than 1 day (Fig.2B) but did not significantly inhibit cell migration until 2-day treatment (Fig.1B). As Fra-1 and c-Jun are the components of AP1 dimer, we reasoned that a particular AP1-regulated gene might be essential for cell migration. To search for such gene, we performed a microarray analysis with total RNA collected from control and Fra-1/c-Jun-knockdown MDA-MB-231 cells. Using more than 2 fold difference as the cut-off standard, we detected that the levels of 15 genes were elevated and 43 genes decreased in Fra-1/c-Jun-knockdown cells in comparison with the control (Table S1). Among these genes, the expression of slug was decreased more than three fold (Table S1).
Slug has been previously described as an inducer of cell movement (27), we thus hypothesized that Fra-1/c-Jun facilitated cell migration by regulating slug expression. To test this hypothesis, we treated MDA-MB-231 and MDA-MB-436 cells with U0126 for various times followed by immunoblotting to detect slug. Slug expression was significantly inhibited following 2-day U0126 treatment in both lines (Fig.3A) and this time point coincided with U0126-caused inhibition in cell migration (Fig.1B). In parallel, we analyzed the levels of slug in Fra-1- or c-Jun-knockdown cells and found that silencing either Fra-1 or c-Jun was able to downregulate slug expression (Fig.3B).
Fig.3. ERK-Fra-1/c-Jun axis regulates slug expression.

A. MDA-MB-231 and MDA-MB-436 cells were treated with 5μM U0126 for various times and then lysed for immunoblotting to detect slug and βactin with the respective antibodies. B. MDA-MB-231 cells were infected with lentiviral vector containing luciferase shRNA (control), Fra-1 or c-Jun shRNA for 4 days and then lysed for immunoblotting to detect slug and βactin.
To further investigate the mechanism associated with ERK/Fra-1/c-Jun-regulated slug expression, we constructed a slug promoter reporter plasmid by linking the slug promoter to a luciferase gene and transfected this plasmid into MCF-7, MDA-MB-231, MDA-MB-436 and ZR-75-1 cells. Assaying luciferase activity showed that slug promoter activity was much greater in MDA-MB-231 and MDA-MB-436 cells (displaying high ERK activity and Fra-1/c-Jun expression) than MCF-7 and ZR-75-1 cells (displaying low ERK activity and Fra-1/c-Jun expression) (Fig.4A). In the subsequent experiments, we transfected MDA-MB-231 and MDA-MB-436 cells with the slug or RSV promoter reporter plasmid for 1 day followed by 2-day U0126 treatment. U0126 inhibited over 70% of slug promoter activity in comparison with the untreated cells in both lines but reduced only less than 20% RSV promoter activity (Fig.S4A). Similarly, forced expression of dominant negative MEK1 led to 60-70% reduction in slug promoter activity but only marginal decrease in RSV promoter activity (Fig.4B and Fig.S4A). These results suggest that the activity of ERK is specifically required for slug gene transcription.
Fig.4. Slug is expressed in an AP1-dependent manner.
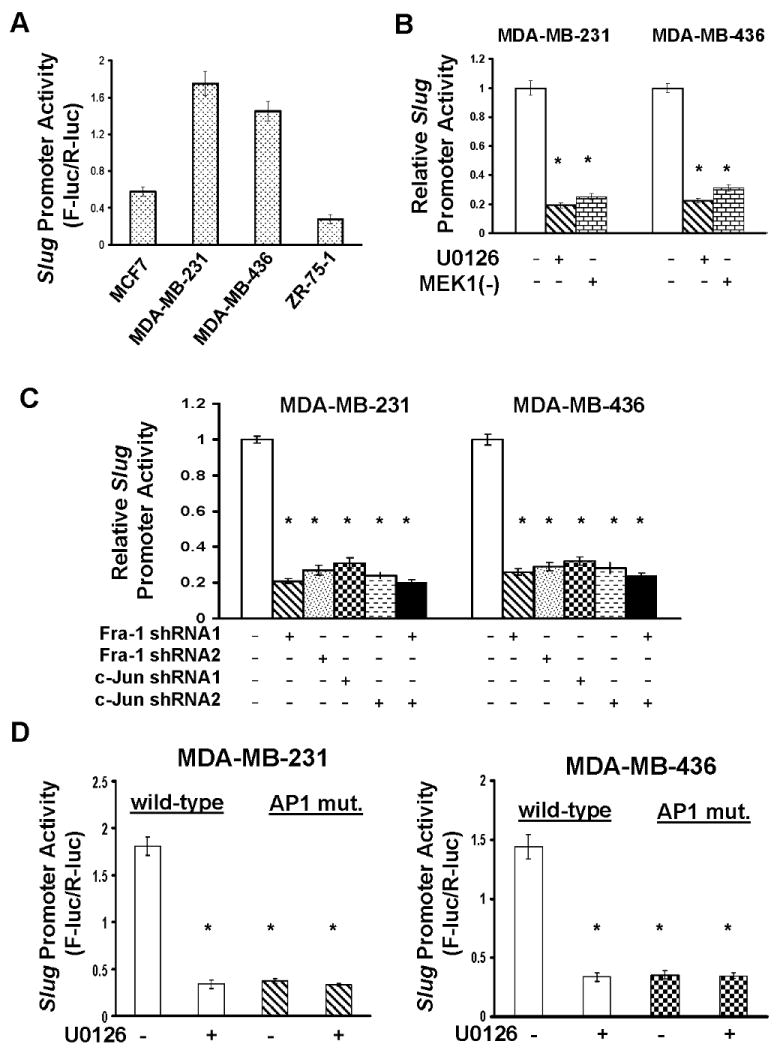
A. MCF7, MDA-MB-231, MDA-MB-436 and ZR-75-1 cells were transfected with the slug promoter luciferase reporter plasmid for 2 days, then lysed and cell lysates measured for luciferase activity as described in “Materials and Methods”. Data are means ± S.E. B. MDA-MB-231 and MDA-MB-436 cells were transfected with the slug promoter plasmid for 1 day and 5μM U0126 then added to cells for another 2 days. In a parallel expreriment, the slug promoter plasmid was co-transfected into cells with dominant negative MEK1 expression vector [MEK1(-)] for 3 days. Cells were lysed and cell lysates analyzed for luciferase activity. Data are means ± S.E. (*, p < 0.005 vs control). C. The slug promoter plasmid was transfected into MDA-MB-231 and MDA-MB-436 cells that expressed Fra-1 or c-Jun shRNA or both for 2 days. Cells were lysed and cell lysates analyzed for luciferase activity. Data are means ± S.E. (*, p < 0.005 vs control). D. MDA-MB-231 and MDA-MB-436 cells were transfected with the slug promoter plasmid or plasmid containing slug promoter with mutation in AP1 consensus site for 1 day and 5μM U0126 added to cells for another 2 days. Cells were lysed and cell lysates measured for luciferase activity. Data are means ± S.E. (*, p < 0.005 vs cells with wild-type promoter/no U0126).
To determine the importance of Fra-1/c-Jun in slug gene transcription, the slug or RSV promoter construct was transfected into Fra-1- or c-Jun-knockdown MDA-MB-231 and MDA-MB-436 cells. Silencing Fra-1 or c-Jun alone abolished 60-70% of slug promoter activity in comparison with the control and silencing them simultaneously did not further diminish slug promoter activity (Fig.4C). In contrast, Fra-1 or c-Jun shRNA had little effect on RSV promoter activity (Fig.S4B). These results indicate that Fra-1 and c-Jun work in the same mechanism to regulate slug transcription. In parallel, we overexpressed Fra-1 and c-Jun in MCF7 and ZR-75-1 cells and subsequently measured slug promoter activity. Forced Fra-1/c-Jun expression led to only 15-20% increase in slug promoter activity in (Fig.S4C). Interestingly, expressing Fra-1/c-Jun together with constitutively active MEK1 greatly enhanced slug promoter activity (Fig.S4C). As the ERK signaling pathway is known to affect Fra-1/c-Jun phosphorylation status, these results suggest that in addition to the presence of Fra-1/c-Jun, ERK-mediated Fra-1/c-Jun phosphorylation may also be important for slug expression.
In further studies, we performed mutagenesis at the potential AP1 consensus sequence (GTGACTTCA→GTAGATTCA) in the slug promoter and determined how this mutation affected slug promoter activity in MDA-MB-231 and MDA-MB-436 cells. The mutation in AP1 site resulted in over 70% reduction in slug promoter activity in both lines (Fig.4D) and additional treatment of U0126 did not further reduce the activity of the slug mutant promoter (Fig.4D). These results suggest that the ERK-Fra-1/c-Jun axis regulates slug expression through the AP1 site in the slug promoter.
Slug expression is required for cell migration
To determine the role of slug in cell migration, we designed two slug shRNAs and found both slug shRNAs were capable of effectively suppressing slug expression (Fig.5A). Transwell migration assay showed that silencing slug led to MDA-MB-231 and MDA-MB-436 cells migrating at about 40% of the rate observed with control cells (Fig.5B). Similar results were also obtained with wound healing assay (Fig.S5). As forced murine slug expression completely reversed the inhibition in cell migration caused by slug shRNA (Fig.S5), these results show that the presence of slug is essential for cell migration.
Fig.5. Slug is essential for ERK-Fra-1/c-Jun axis-regulated cell migration.
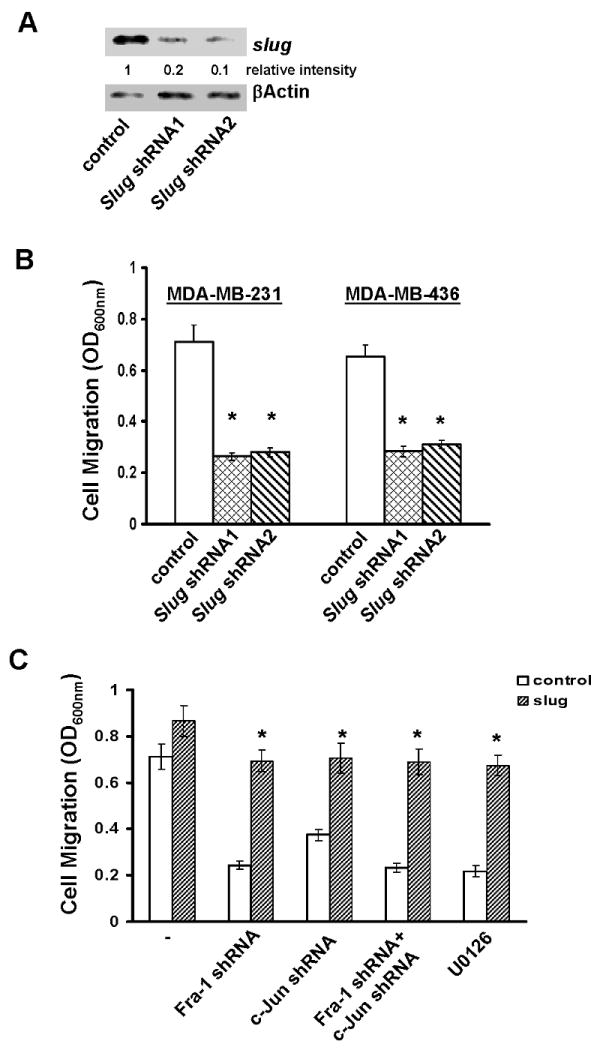
A. MDA-MB-231 cells were infected with lentiviral vector containing luciferase shRNA (control) or slug shRNA for 4 days and then lysed for immunoblotting to detect slug and βactin with the respective antibodies. B. MDA-MB-231 and MDA-MB-436 cells were infected with control (luciferase shRNA) or slug shRNA-containing lentiviral vector for 4 days, then detached with 10 mM EDTA and assayed for cell migration as described in “Materials and Methods”. Data are means ± S.E. (*, p < 0.005 vs control). C. MDA-MB-231 cells were infected with empty lentiviral vector (control) or vector encoding murine slug cDNA for 4 days. Control cells or cells with stable murine slug cDNA expression were further infected with lentiviral vector containing luciferase shRNA, Fra-1 shRNA or c-Jun shRNA or both for 4 days. In a parallel experiment, control and murine slug cDNA-expressing cells were treated with 5μM U0126 for 2 days. Cells were detached and analyzed for cell migration. Data are means ± S.E. (*, p < 0.005 vs control).
We next expressed murine slug transgene in Fra-1/c-Jun-knockdown MDA-MB-231 cells and then analyzed migration of these cells. While forced slug expression moderately enhanced the migration of parental cells, it elevated migration of Fra-1/c-Jun-knockdown cells to those seen in the control cells (Fig.5C). Similarly, forced slug expression also largely rescued U0126-caused inhibition in cell migration (Fig.5C). These results suggest that the ERK-Fra-1/c-Jun axis regulates cell migration by maintaining slug expression.
Slug expression is required for in vitro cell invasion and in vivo metastasis
To investigate the role of slug in cell invasion, we examined how silencing slug affected in vitro invasiveness of MDA-MB-231cells by performing matrigel invasion assay. MDA-MB-231 cells exhibited robust invasion but the expression of slug shRNAs blocked over 80% of their abilities to invade matrigel (Fig.6A). Similar results were also obtained with MDA-MB-436 cells (data not shown). Subsequently, we examined the importance of slug in in vivo tumor outgrowth and metastasis by subcutaneously injecting control or slug-knockdown MDA-MB-231 cells in the mammary fat pad of nude mice. We detected palpable tumors within 7 days in both groups and daily monitoring up to 8 weeks revealed no statistically significant difference in tumor outgrowth between them (Fig.S6). When lung sections from euthanized mice were analyzed by H&E staining, we observed significant metastatic lesions in the lungs of animals receiving control cells (5.3±2.4 lesions/section) (Fig.6B). In contrast, metastastic lesion was not detected from any animal receiving slug-knockdown cells (Fig.6B). Taken together, these results suggest that slug is needed for both invasion and metastasis but tumor outgrowth.
Fig.6. Slug is essential for in vitro invasion and lung metastasis.
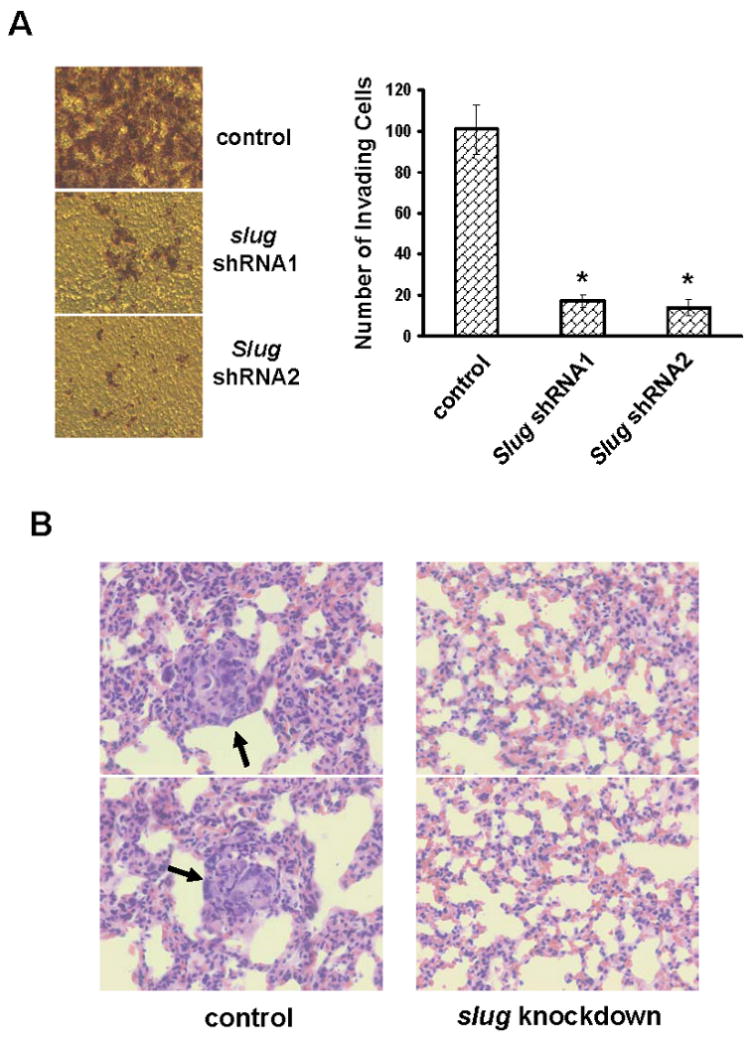
A. Control or slug-knockdown MDA-MB-231 cells were added into invasion chambers and allowed to invade for 24 hrs. The cells on the undersurface of chambers were stained and visualized under microscope. Data are means ± S.E. (*, p < 0.001 vs control). B. The lungs from mice receiving either control or slug-knockdown MDA-MB-231 cells for 8 weeks were fixed and sectioned. The sections were subjected to H&E staining and visualized under microscope. The arrows point at metastatic lesions and the images are in 40× magnification.
Discussion
The enzymatic ERK activity is increased in invasive breast carcinomas as compared to normal tissues or benign lesions (36, 37). As invasive cancer cells are migratory, it is reasonable to postulate that the enzymatic ERK activity could be involved in cancer cell migration. This notion is supported by our observation that the levels of ERK activity correlated extremely well with the migratory status of breast cancer cells (Fig.1A). Recent studies have implicated the involvement of the ERK pathway in cell migration (9). However, the mechanism associated with ERK-regulated cell migration is not clear. We showed that a 48-hr U0126 treatment was required to significantly inhibit MDA-MB-231 and MDA-MB-436 cell migration, whereas ERK activity was diminished within 1 hr of U0126 treatment (Fig.1B). This observation suggests that the direct ERK signaling is unlikely to be responsible for breast cancer cell migration. Using actinomycin to block de novo RNA synthesis, we found that ERK-dependent gene expression is necessary for MDA-MB-231 cell migration (Fig.1C). In search for the genes essential for breast cancer cell migration, we turned our attention to the components of AP1 complex because the expression of AP1 component Fra-1 is regulated by the ERK signaling pathway (21) and Fra-1 acts as a downstream effector of ERK to facilitate colon carcinoma cell migration (24). In our studies, we detected high levels of Fra-1/c-Jun only in migratory breast cancer cell lines (Fig.2A) and the expression of Fra-1/c-Jun was diminished by 16-hr U0126 treatment (Fig.2A). To determine the importance of Fra-1/c-Jun in cell migration, we found that silencing either Fra-1 or c-Jun significantly decreased breast cancer cell migration (Fig.2C). Importantly, combination of Fra-1/c-Jun knockdown and U0126 treatment did not yield any further inhibition in cell migration than either Fra-1/c-Jun knockdown or U0126 treatment alone (Fig.2C), strongly suggesting that ERK and Fra-1/c-Jun act in the same mechanism to facilitate cell migration. Our findings confirmed the importance of Fra-1 in cell migration as reported by previous studies (19, 20, 24) and also showed the necessity of c-Jun for cell migration (Fig.2C). As Fra-1 is known to interact with c-Jun to form AP1 heterodimer to promote gene expression, the requirement of both Fra-1 and c-Jun suggest that Fra-1/c-Jun AP1 dimer-driven gene expression is essential for breast cancer cell migration. In fact, we noticed that U0126-induced Fra-1/c-Jun downregulation occurred at least 1 day earlier than U0126-induced inhibition in cell migration (16 hrs vs 48 hrs). This observation also supports the notion that Fra-1/c-Jun regulates cell migration by promoting a particular gene expression through an AP1-dependent manner.
Recent studies have implicated that slug acts as an inducer of cell movement (27). In our studies, we showed that the expression of slug is regulated by the ERK-Fra-1/c-Jun signaling axis through the AP1 consensus sequence in the slug promoter (Fig. 3 and 4). Moreover, we found that U0126-induced inhibition in cell migration coincided with U0126-induced reduction of slug expression (Fig.1 and Fig.3) and that silencing slug expression abrogated breast cancer cell migration (Fig.5A and B), cell invasion and spontaneous metastasis (Fig.6A and B). More importantly, the ability of U0126 or Fra-1/c-Jun shRNAs to inhibit cell migration is lost in cells expressing slug transgenes (Fig.5C). Our results strongly support the role of slug in cell migration. Recent studies have reported that slug and its related protein snail can regulate integrin expression (30, 38). Since integrins are essential for cell-matrix substratum interaction and cell migration, we reason that the ERK-Fra-1/c-Jun/slug axis impacts breast cancer cell migration through the regulation of the integrins. This possibility is buttressed by a previous study in which AP-1 was shown to regulate integrin expression through the ERK pathway (39).
In conclusion, our data show that the ERK signaling pathway facilitates breast cancer cell migration by regulating slug expression in an AP1-dependent mechanism. This knowledge suggests that anti-breast cancer approaches may be developed by targeting the ERK-AP1-slug axis.
Supplementary Material
Acknowledgments
Grant support: NIH grant CA093926 and HL083335
References
- 1.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Rev Cancer. 2003;3:362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 2.Wehrle-Haller B, Imhof BA. Actin, microtubules and focal adhesion dynamics during cell migration. Int J Biochem Cell Biol. 2003;35:39–50. doi: 10.1016/s1357-2725(02)00071-7. [DOI] [PubMed] [Google Scholar]
- 3.Javelaud D, Laboureau J, Gabison E, Verrecchia F, Mauviel A. Disruption of basal JNK activity differentially affects key fibroblast functions important for wound healing. J Biol Chem. 2003;278:24624–8. doi: 10.1074/jbc.M301942200. [DOI] [PubMed] [Google Scholar]
- 4.Meadows KN, Bryant P, Vincent PA, Pumiglia KM. Activated Ras induces a proangiogenic phenotype in primary endothelial cells. Oncogene. 2004;23:192–200. doi: 10.1038/sj.onc.1206921. [DOI] [PubMed] [Google Scholar]
- 5.Yamauchi J, Chan JR, Shooter EM. Neurotrophin 3 activation of TrkC induces Schwann cell migration through the c-Jun N-terminal kinase pathway. Proc Natl Acad Sci. 2003;100:14421–6. doi: 10.1073/pnas.2336152100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–23. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- 7.Klekotka PA, Santoro SA, Zutter MM. alpha 2 integrin subunit cytoplasmic domain-dependent cellular migration requires p38. MAPK J Biol Chem. 2001;276:9503–11. doi: 10.1074/jbc.M006286200. [DOI] [PubMed] [Google Scholar]
- 8.Goncharova EA, Vorotnikov AV, Gracheva EO, et al. Activation of p38 MAP-kinase and caldesmon phosphorylation are essential for urokinase-induced human smooth muscle cell migration. Biol Chem. 2002;383:115–26. doi: 10.1515/BC.2002.012. [DOI] [PubMed] [Google Scholar]
- 9.Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–28. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 10.Rajalingam K, Wunder C, Brinkmann V, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nature Cell Biology. 2005;7:837–43. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 11.Bove PF, Hristova M, Wesley UV, Olson N, Lounsbury KM, van der Vliet A. Inflammatory levels of nitric oxide inhibit airway epithelial cell migration by inhibition of the kinase ERK1/2 and activation of hypoxia-inducible factor-1 alpha. J Biol Chem. 2008;283:17919–28. doi: 10.1074/jbc.M709914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim biophys Acta. 1991;1072:129–57. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 13.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–61. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Mechta F, Lallemand D, Pfarr CM, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–47. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Tognon CE, Godinho FJ, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–58. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamberger AM, Methner C, Lisboa BW, et al. Expression pattern of the AP-1 family in breast cancer: association of fosB expression with a well-differentiated, receptor-positive tumor phenotype. Int J Cancer. 1999;84:533–8. doi: 10.1002/(sici)1097-0215(19991022)84:5<533::aid-ijc16>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 17.Debinski W, Gibo DM. Fos-related antigen 1 modulates malignant features of glioma cells. Mol Cancer Res. 2005;3:237–49. doi: 10.1158/1541-7786.MCR-05-0004. [DOI] [PubMed] [Google Scholar]
- 18.Chiappetta G, Tallini G, De Biasio MC, et al. FRA-1 expression in hyperplastic and neoplastic thyroid diseases. Clin Cancer Res. 2000;6:4300–6. [PubMed] [Google Scholar]
- 19.Kustikova O, Kramerov D, Grigorian M, et al. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol Cell Biol. 1998;18:7095–105. doi: 10.1128/mcb.18.12.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tkach V, Tulchinsky E, Lukanidin E, Vinson C, Bock E, Berezin V. Role of the Fos family members, c-Fos, Fra-1 and Fra-2, in the regulation of cell motility. Oncogene. 2003;22:5045–54. doi: 10.1038/sj.onc.1206570. [DOI] [PubMed] [Google Scholar]
- 21.Casalino L, De Cesare D, Verde P. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol Cell Biol. 2003;23:4401–15. doi: 10.1128/MCB.23.12.4401-4415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vial E, Marshall CJ. Elevated ERK-MAP kinase activity protects the FOS family member FRA-1 against proteasomal degradation in colon carcinoma cells. J Cell Sci. 2003;116:4957–63. doi: 10.1242/jcs.00812. [DOI] [PubMed] [Google Scholar]
- 23.Young MR, Nair R, Bucheimer N, et al. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol Cell Biol. 2002;22:587–98. doi: 10.1128/MCB.22.2.587-598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vial E, Sahai E, Marshall CJ. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 25.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–96. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Catalano A, Rodilossi S, Rippo MR, Caprari P, Procopio A. Induction of stem cell factor/c-Kit/slug signal transduction in multidrug-resistant malignant mesothelioma cells. J Biol Chem. 2004;279:46706–14. doi: 10.1074/jbc.M406696200. [DOI] [PubMed] [Google Scholar]
- 27.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 28.Savagner P, Kusewitt DF, Carver EA, et al. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J Cell Physiol. 2005;202:858–66. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- 29.Chandler HL, Colitz CM, Lu P, Saville WJ, Kusewitt DF. The role of the slug transcription factor in cell migration during corneal re-epithelialization in the dog. Exp Eye Res. 2007;84:400–11. doi: 10.1016/j.exer.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Turner FE, Broad S, Khanim FL, et al. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281:21321–31. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, New L, Pan Z, Han J, Nemerow GR. Urokinase plasminogen activator/urokinase-specific surface receptor expression and matrix invasion by breast cancer cells requires constitutive p38alpha mitogen-activated protein kinase activity. J Biol Chem. 2000;275:12266–72. doi: 10.1074/jbc.275.16.12266. [DOI] [PubMed] [Google Scholar]
- 32.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 33.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 35.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–44. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivaraman VS, Wang H, Nuovo GJ, Malbon CC. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J Clin Invest. 1997;99:1478–83. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller H, Flury N, Eppenberger-Castori S, Kueng W, David F, Eppenberger U. Potential prognostic value of mitogen-activated protein kinase activity for disease-free survival of primary breast cancer patients. Int J Cancer. 2000;89:384–8. doi: 10.1002/1097-0215(20000720)89:4<384::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 38.Haraguchi M, Okubo T, Miyashita Y, et al. Snail regulates cell-matrix adhesion by regulation of the expression of integrins and basement membrane proteins. J Biol Chem. 2008;283:23514–23. doi: 10.1074/jbc.M801125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eriksson M, Arminen L, Karjalainen-Lindsberg ML, Leppa S. AP-1 regulates alpha2beta1 integrin expression by ERK-dependent signals during megakaryocytic differentiation of K562 cells. Exp Cell Res. 2005;304:175–86. doi: 10.1016/j.yexcr.2004.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


