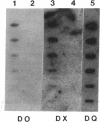
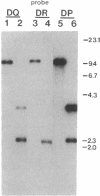
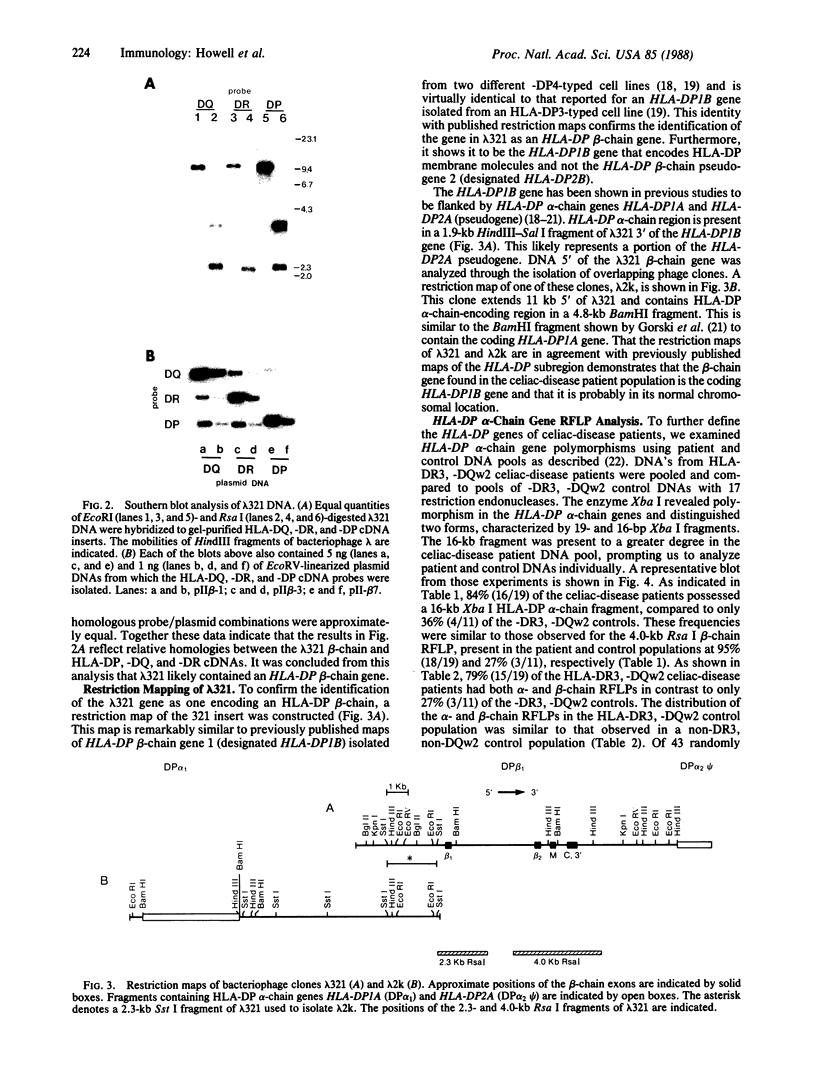
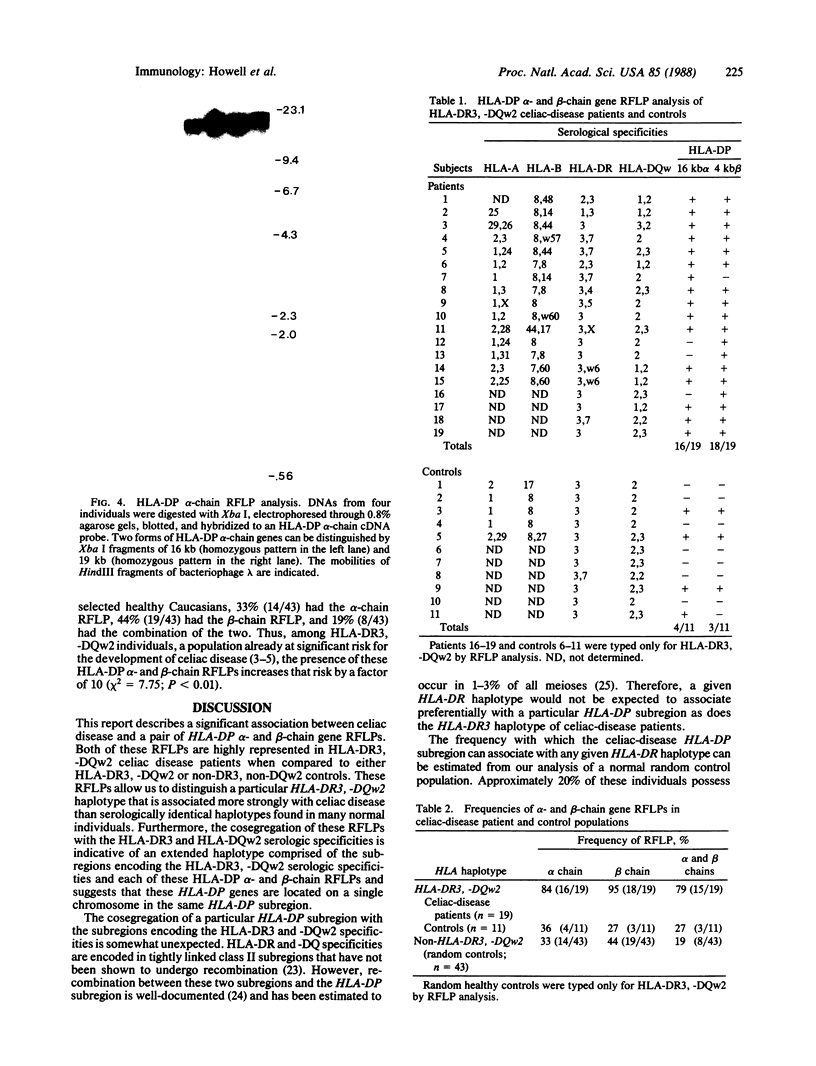
Abstract
Celiac disease has one of the strongest associations with HLA (human leukocyte antigen) class II markers of the known HLA-linked diseases. This association is primarily with the class II serologic specificities HLA-DR3 and -DQw2. We previously described a restriction fragment length polymorphism (RFLP) characterized by the presence of a 4.0-kilobase Rsa I fragment derived from an HLA class II beta-chain gene, which distinguishes the class II HLA haplotype of celiac disease patients from those of many serologically matched controls. We now report the isolation of this beta-chain gene from a bacteriophage genomic library constructed from the DNA of a celiac disease patient. Based on restriction mapping and differential hybridization with class II cDNA and oligonucleotide probes, this gene was identified as one encoding an HLA-DP beta chain. This celiac disease-associated HLA-DP beta-chain gene was flanked by HLA-DP alpha-chain genes and, therefore, was probably in its normal chromosomal location. The HLA-DP alpha-chain genes of celiac disease patients also were studied by RFLP analysis; 84% of HLA-DR3, -DQw2 patients had a 16-kb Xba I fragment that was present in only 36% of HLA-DR3, -DQw2 controls. Moreover, 79% of these patients had both alpha- and beta-chain polymorphisms in contrast to 27% of controls. Thus, celiac disease is associated with a subset of HLA-DR3, -DQw2 haplotypes characterized by HLA-DP alpha- and beta-chain gene RFLPs. Within the celiac-disease patient population, the joint segregation of these HLA-DP genes with those encoding the serologic specificities HLA-DR3 and -DQw2 indicates: (i) that the class II HLA haplotype associated with celiac disease is extended throughout the entire HLA-D region, and (ii) that celiac-disease susceptibility genes may reside as far centromeric on this haplotype as the HLA-DP subregion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alper C. A., Fleischnick E., Awdeh Z., Katz A. J., Yunis E. J. Extended major histocompatibility complex haplotypes in patients with gluten-sensitive enteropathy. J Clin Invest. 1987 Jan;79(1):251–256. doi: 10.1172/JCI112791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar A., Nepom G. T., Mickelson E., Erlich H., Hansen J. A. HLA-DP and HLA-DO genes in presumptive HLA-identical siblings: structural and functional identification of allelic variation. J Immunol. 1987 Mar 15;138(6):1947–1953. [PubMed] [Google Scholar]
- Arnheim N., Strange C., Erlich H. Use of pooled DNA samples to detect linkage disequilibrium of polymorphic restriction fragments and human disease: studies of the HLA class II loci. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6970–6974. doi: 10.1073/pnas.82.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J., Rassenti L., Smoot S., Smith K., Newby C., Hohlfeld R., Toyka K., McDevitt H., Steinman L. HLA-DQ beta-chain polymorphism linked to myasthenia gravis. Lancet. 1986 May 10;1(8489):1058–1060. doi: 10.1016/s0140-6736(86)91330-9. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Boss J. M., Strominger J. L. Cloning and sequence analysis of the human major histocompatibility complex gene DC-3 beta. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5199–5203. doi: 10.1073/pnas.81.16.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Haguenauer O., Robbins E., Massart C., Busson M., Deschamps I., Hors J., Lalouel J. M., Dausset J., Cohen D. A systematic study of HLA class II-beta DNA restriction fragments in insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A. 1985 May;82(10):3335–3339. doi: 10.1073/pnas.82.10.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. G., Kagnoff M. F. Celiac disease. Annu Rev Nutr. 1985;5:241–266. doi: 10.1146/annurev.nu.05.070185.001325. [DOI] [PubMed] [Google Scholar]
- Corazza G. R., Tabacchi P., Frisoni M., Prati C., Gasbarrini G. DR and non-DR Ia allotypes are associated with susceptibility to coeliac disease. Gut. 1985 Nov;26(11):1210–1213. doi: 10.1136/gut.26.11.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J., Rollini P., Long E., Mach B. Molecular organization of the HLA-SB region of the human major histocompatibility complex and evidence for two SB beta-chain genes. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3934–3938. doi: 10.1073/pnas.81.13.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson K., Emmoth E., Widmark E., Böhme J., Peterson P. A., Rask L. Isolation of a cDNA clone coding for an SB beta-chain. Nature. 1984 May 3;309(5963):76–78. doi: 10.1038/309076a0. [DOI] [PubMed] [Google Scholar]
- Gustafsson K., Wiman K., Emmoth E., Larhammar D., Böhme J., Hyldig-Nielsen J. J., Ronne H., Peterson P. A., Rask L. Mutations and selection in the generation of class II histocompatibility antigen polymorphism. EMBO J. 1984 Jul;3(7):1655–1661. doi: 10.1002/j.1460-2075.1984.tb02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy D. A., Bell J. I., Long E. O., Lindsten T., McDevitt H. O. Mapping of the class II region of the human major histocompatibility complex by pulsed-field gel electrophoresis. Nature. 1986 Oct 2;323(6087):453–455. doi: 10.1038/323453a0. [DOI] [PubMed] [Google Scholar]
- Howell M. D., Austin R. K., Kelleher D., Nepom G. T., Kagnoff M. F. An HLA-D region restriction fragment length polymorphism associated with celiac disease. J Exp Med. 1986 Jul 1;164(1):333–338. doi: 10.1084/jem.164.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M. D., Resner J., Austin R. K., Kagnoff M. F. Rapid identification of hybridization probes for chromosomal walking. Gene. 1987;55(1):41–45. doi: 10.1016/0378-1119(87)90246-0. [DOI] [PubMed] [Google Scholar]
- Larhammar D., Schenning L., Gustafsson K., Wiman K., Claesson L., Rask L., Peterson P. A. Complete amino acid sequence of an HLA-DR antigen-like beta chain as predicted from the nucleotide sequence: similarities with immunoglobulins and HLA-A, -B, and -C antigens. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3687–3691. doi: 10.1073/pnas.79.12.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt H. O. The molecular basis of autoimmunity. Clin Res. 1986 Apr;34(2):163–175. [PubMed] [Google Scholar]
- Nepom B. S., Palmer J., Kim S. J., Hansen J. A., Holbeck S. L., Nepom G. T. Specific genomic markers for the HLA-DQ subregion discriminate between DR4+ insulin-dependent diabetes mellitus and DR4+ seropositive juvenile rheumatoid arthritis. J Exp Med. 1986 Jul 1;164(1):345–350. doi: 10.1084/jem.164.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom G. T., Seyfried C. E., Holbeck S. L., Wilske K. R., Nepom B. S. Identification of HLA-Dw14 genes in DR4+ rheumatoid arthritis. Lancet. 1986 Nov 1;2(8514):1002–1005. doi: 10.1016/s0140-6736(86)92614-0. [DOI] [PubMed] [Google Scholar]
- Okada K., Prentice H. L., Boss J. M., Levy D. J., Kappes D., Spies T., Raghupathy R., Mengler R. A., Auffray C., Strominger J. L. SB subregion of the human major histocompatibility complex: gene organization, allelic polymorphism and expression in transformed cells. EMBO J. 1985 Mar;4(3):739–748. doi: 10.1002/j.1460-2075.1985.tb03691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owerbach D., Lernmark A., Platz P., Ryder L. P., Rask L., Peterson P. A., Ludvigsson J. HLA-D region beta-chain DNA endonuclease fragments differ between HLA-DR identical healthy and insulin-dependent diabetic individuals. Nature. 1983 Jun 30;303(5920):815–817. doi: 10.1038/303815a0. [DOI] [PubMed] [Google Scholar]
- Servenius B., Gustafsson K., Widmark E., Emmoth E., Andersson G., Larhammar D., Rask L., Peterson P. A. Molecular map of the human HLA-SB (HLA-DP) region and sequence of an SB alpha (DP alpha) pseudogene. EMBO J. 1984 Dec 20;3(13):3209–3214. doi: 10.1002/j.1460-2075.1984.tb02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler D., Grumet F. C., Erlich H. A. Polymorphic restriction endonuclease sites linked to the HLA-DR alpha gene: localization and use as genetic markers of insulin-dependent diabetes. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8100–8104. doi: 10.1073/pnas.82.23.8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termijtelen A., Meera Khan P., Shaw S., van Rood J. J. Mapping SB in relation to HLA and GLO1 using cells from first-cousin marriage offspring. Immunogenetics. 1983;18(5):503–512. doi: 10.1007/BF00364391. [DOI] [PubMed] [Google Scholar]
- Tonnelle C., DeMars R., Long E. O. DO beta: a new beta chain gene in HLA-D with a distinct regulation of expression. EMBO J. 1985 Nov;4(11):2839–2847. doi: 10.1002/j.1460-2075.1985.tb04012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi R., Vismara D., Tanigaki N., Ferrara G. B., Cicimarra F., Buffolano W., Follo D., Auricchio S. Evidence that celiac disease is primarily associated with a DC locus allelic specificity. Clin Immunol Immunopathol. 1983 Sep;28(3):395–404. doi: 10.1016/0090-1229(83)90106-x. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Kelly A., Lee J., Carson S., Austin P., Travers P. Linkage map of two HLA-SB beta and two HLA-SB alpha-related genes: an intron in one of the SB beta genes contains a processed pseudogene. Cell. 1984 Aug;38(1):241–249. doi: 10.1016/0092-8674(84)90546-4. [DOI] [PubMed] [Google Scholar]