Abstract
Chelicerates constitute a basic arthropod group with fossil representatives from as early as the Cambrian period. Embryonic development and the subdivision of the segmented body region into a prosoma and an opisthosoma are very similar in all extant chelicerates. The mode of head segmentation, however, has long been controversial. Although all other arthropod groups show a subdivision of the head region into six segments, the chelicerates are thought to have the first antennal segment missing. To examine this problem on a molecular level, we have compared the expression pattern of Hox genes in the spider Cupiennius salei with the pattern known from insects. Surprisingly, we find that the anterior expression borders of the Hox genes are in the same register and the same relative segmental position as in Drosophila. This contradicts the view that the homologue of the first antennal segment is absent in the spider. Instead, our data suggest that the cheliceral segment is homologous to the first antennal segment and the pedipalpal segment is homologous to the second antennal (or intercalary) segment in arthropods. Our finding implies that chelicerates, myriapods, crustaceans, and insects share a single mode of head segmentation, reinforcing the argument for a monophyletic origin of the arthropods.
Keywords: engrailed/chelicerates/development/homeobox/Drosophila
The relationships of arthropods and their mode of head segmentation are the subjects of an on-going debate (1–3). The major groups recognized are the chelicerates, myriapods, crustaceans, and insects. Traditionally, myriapods and insects have been grouped together into the Tracheata, but molecular phylogenies have suggested that the crustaceans should be seen as the sister group of insects instead (4, 5). Chelicerates, on the other hand, always have been considered as an unquestioned, basic monophyletic group. They share a distinct and conserved bauplan, the major features of which include a prosomal–opisthosomal subdivision and lack of antennae. Moreover, the analysis of the brain ganglia and the innervation of the head appendages (the chelicerae and the pedipalps) have suggested that chelicerates lack the homologue of the first antennal segment (1, 2).
Hox genes are involved in specifying the identity of segments along the anterior–posterior axis in diverse animal phyla (6, 7). Among the arthropods, Hox genes have so far been studied most intensively in an insect, namely Drosophila, in which they also were discovered originally (8, 9). Expression patterns of homologues of some Hox genes also have been studied in several other insects (10, 11) as well as in crustaceans (12, 13) and a myriapod (14). Most of these studies have dealt with genes involved in patterning the thoracic and abdominal region of the animals. These regions show a particular degree of diversification among arthropods, and changes in Hox gene expression patterns have been implicated in the generation of morphological differences in crustaceans (12, 13). Head segmentation, by comparison, is highly stereotypic among crustaceans, including two antennal segments as well as the mandibles and two pairs of maxillae as feeding apparatus. Insects and myriapods, on the other hand, have only one pair of antennae, but the segment corresponding to the second pair of antennae can be recognized in the embryo and has been termed the “intercalary segment” (Fig. 1). The expression of Hox genes in the head region of different insects is very similar, with only minor deviations (10, 11). No data are currently available on the expression pattern of anterior Hox genes in crustaceans and myriapods, but, because of the unquestioned homology of the head segments in these groups (1–3), it would seem unlikely that there are major differences. However, because of the assumed major difference in head organization in the chelicerates, it should be particularly revealing to compare Hox gene expression patterns in their head region. In this study, we have chosen the spider Cupiennius salei as a representative model. Although spiders belong to the most advanced forms of chelicerates, they can nonetheless serve as typical representatives of chelicerates in terms of embryonic development (15).
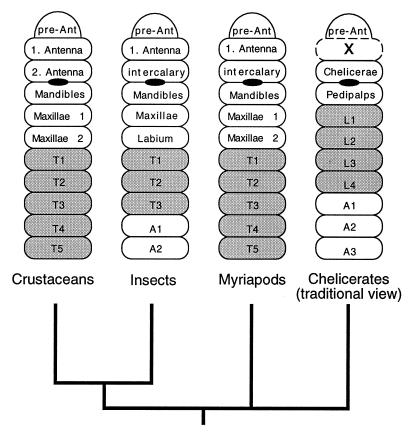
Figure 1.
Phylogenetic scheme of the major arthropod groups and modes of head segmentation. The phylogeny depicted is based on molecular data (4, 5); the depiction of the head segments represents a consensus from textbooks (1, 34). The segments are labeled according to the conventions in the respective taxa; leg-bearing segments are shaded. The position of the mouth is indicated by a black oval. The presumed missing segment in the chelicerates is marked with an “x”. The drawing is modified from Willmer (34).
MATERIALS AND METHODS
Isolation of Hox Genes.
Sequences for the Cupiennius Hox genes initially were amplified by PCR from genomic DNA (by using degenerate primers) and were cloned. One-hundred and twenty clones were sequenced and grouped according to their similarities with the respective Drosophila gene. In a second step, specific primers that were directed against specific sequences within the homeodomain then were designed for each gene or each subset of the Hox genes and were used for reverse transcription amplification experiments. Corresponding cDNAs were isolated from a cDNA library representing all embryonic stages prepared in λ ZAPII (Stratagene). Several cDNAs that were isolated from this library are incomplete at their 5′-ends, partly because they seem to have retained parts of the intron that separates the hexapeptide region from the homeobox. However, for all of the genes described here, we obtained the full homeobox sequences as well as the complete 3′-regions; for all except for UBX-1, we also obtained at least the region including the hexapeptide. A PCR fragment for the engrailed (EN) gene was obtained by a nested reverse transcription–PCR by using degenerate primers directed against sequences within or in the proximity of the homeodomain. The corresponding EN cDNA was isolated from the embryonic cDNA library by using this PCR fragment as a probe. All primer sequences and PCR conditions are available on request.
Expression Analysis.
The expression patterns were examined by in situ hybridization by using digoxigenin-labeled RNA probes. Fixation and in situ hybridization were done essentially as described for Drosophila (36) with minor modifications which are detailed in ref. 26. The vitelline membranes were removed manually by using Dumont-5 forceps. The embryos were stored in methanol at −20°C for several days before using them in an in situ hybridization. The wash steps and incubation steps were prolonged to last up to several hours. A detailed protocol is available on request. Immunohistochemical staining was done with the FP6.87 mAb that is directed against the homeodomain of Ultrabithorax (UBX) and abdominal-A (ABD-A) (24).
RESULTS
Hox Gene Sequences.
Hox gene fragments from Cupiennius were obtained by PCR using degenerate primers. The fragments were cloned and sequenced and then were used to screen an early embryonic cDNA library, which allowed us to obtain the complete homeobox sequences for each gene. Several unequivocal orthologues to other arthropod Hox genes could be assigned on the basis of sequence similarity comparisons because the spider sequences cluster unequivocally with their respective orthologues from crustaceans and insects (not shown). The genes identified in this way include orthologues of labial (LAB), Deformed (DFD), Antennapedia (ANTP), Ultrabithorax (UBX), and abdominal-A (ABD-A). Two different variants were found for UBX (see below), and we cannot exclude that there might be duplicated copies for the other genes as well because we may not have reached saturation in the initial PCR screen.
A Spider Engrailed Gene.
To obtain a molecular marker for segmental boundaries in the spider, we also cloned a homologue of the EN gene. EN is expressed at the anterior margin of the parasegments in insects and crustaceans (16). In situ hybridization with the C. salei engrailed (Cs-EN) cDNA indicates that it is expressed in the same way (Fig. 2). In the segments bearing appendages, it is always the posterior half of the appendage that is stained (Fig. 2 E), confirming the notion that the appendages are formed exactly at the parasegmental boundaries in arthropods (9). The most anterior stripe seen is that of the cheliceral segment. However, in early embryos, there is also some EN staining anterior to the cheliceral stripe (Fig. 2 E), which may correspond to remnants of a preantennal segment. A detailed analysis of the EN staining in insects also has suggested that EN is expressed in the preantennal segment, although not as a proper stripe, but merely as distinct spots (18, 19). The exact number of segments in the preantennal region is still under dispute (19, 20), and further analysis of this region also will be necessary in the spider.
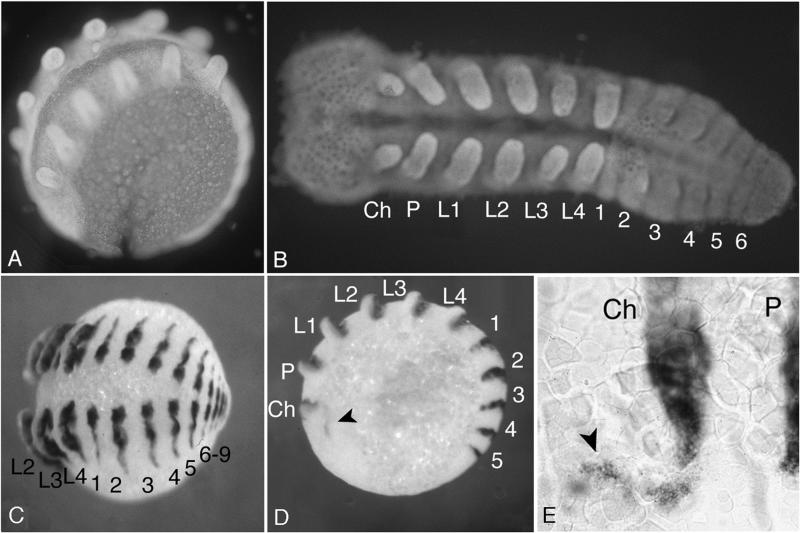
Figure 2.
Embryos of C. salei stained with 4′,6-diamidino-2-phenylindole (A and B) and with the EN probe by whole-mount in situ hybridization (C–E). Embryos were stained at midstage embryogenesis, when their general appearance is most reminiscent of the phylotypic stage of arthropods. (A) Embryo within the egg. (B) Embryo dissected free from yolk and flattened out. The anterior segments with their appendages already have formed fully while the abdominal segments are still incomplete. (C) Whole embryo stained with the EN probe. This embryo is at a later stage than the one in B and shows the final number of abdominal segments. (D) Embryo at an earlier stage showing an additional expression of EN in the head region (marked with an arrowhead), which might correspond to the preantennal segment. (E) Enlargement of the region showing the preantennal EN expression. Ch, cheliceres; P, pedipalps; L1–L4, leg 1–leg 4; 1–9, abdominal segments 1–9.
Hox Gene Expression.
The first unequivocal segment in the spider is the cheliceral segment, which currently is assumed to be the homologue of the second antennal segment in other arthropods (1–3). In this case, one should expect that the Hox gene LAB is expressed in the cheliceral segment. In Drosophila, LAB is expressed in the intercalary segment (21), which is homologous to the second antennal segment (compare Fig. 1). However, LAB clearly is not expressed in the chelicerae. Instead, we find LAB staining in the pedipalps, which lie one segment more posteriorly (Fig. 3 A and B). In fact, none of the Hox genes tested here is expressed in the chelicerae, similar to the situation found in the antennal segment of Drosophila. Hence, the most parsimonious interpretation of the LAB expression data is that the antennal segment is not missing in the chelicerates but that the chelicerae are homologous to the first antennal segment and the pedipalps to the second antennal segment of other arthropods.
Figure 3.
Cupiennius salei embryos stained with Hox gene probes. (A and B) Staining with LAB. The anterior border of staining is within the pedipalpal segment (A), and the posterior border is within the last leg segment (B). The chelicerae are completely free of staining. (C) Staining with DFD. All four leg segments are stained. (D–E) Staining with ANTP. Staining is evident directly adjacent to the last leg-bearing segment but covers the posterior half of L4 (E). (F) Staining with UBX-1. The anterior border of expression is seen within abdominal segment 1. (G) Staining with UBX-2. Expression begins in abdominal segment 2. (H) Staining with ABD-A. Expression begins within abdominal segment 3. (I and J) Close comparison of expression borders of UBX-1 (I) and UBX-2 (J) and immunostaining with the broadly cross-reacting antibody FP6.87 against UBX and ABD-A homeoboxes (24) in K; the staining of the leg (L4) is an artifact of the labeling procedure used in this case. Abbreviations as in Fig. 2.
This inference also is supported by the DFD expression pattern. In Drosophila, the anterior boundary of DFD expression marks the border between the intercalary and the mandibular segments (21). In the spider, the most anterior DFD-expression is in the first leg-bearing segment (Fig. 3 C) and not in the pedipalps, as one would have anticipated if the first antennal segment were in fact missing.
The anterior expression borders of ANTP, UBX, and ABD-A in the spider (Fig. 3 D–K) also closely match the borders known from Drosophila (Fig. 4). For ANTP, however, there are slight discrepancies in the extent of the anterior expression border between Drosophila and the locust Schistocerca (22). Although the first and most persistent expression in Drosophila is seen in parasegment (PS) 4, it appears that the expression in Schistocerca begins in PS 5 but later extends into PS 3, marking the boundary between head and thorax. A more anterior extension of the expression of ANTP also is seen in Drosophila at later stages, but this is confined to the neural epithelium (22). Thus, the ANTP boundary seems to be somewhat variable among insects, and it will be interesting to compare this also in different spiders. The border in Cupiennius, however, corresponds most closely to the early PS 4 boundary in Drosophila (Fig. 4).
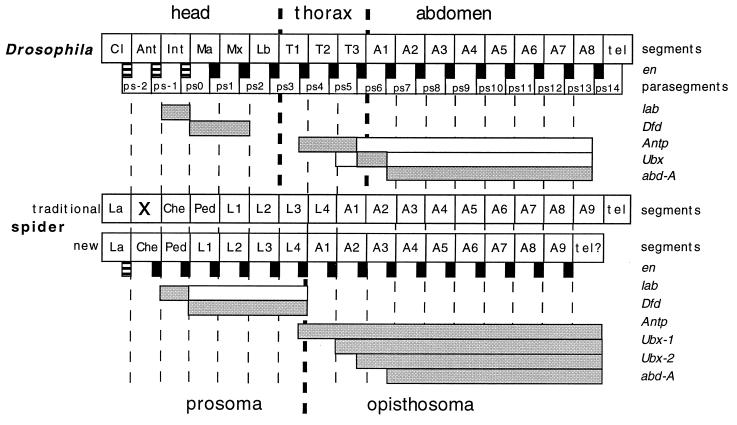
Figure 4.
Schematized interpretation of the expression boundaries of the Hox genes in Drosophila and in Cupiennius. The broken thin lines represent the segmental boundaries, and the broken thick lines represent the boundaries between the major tagmata (head, thorax, and abdomen in the fly, prosoma and opisthosoma in the spider). The EN stripes (black boxes) are located at the parasegmental boundaries in Drosophila, which also are indicated. Note that the first three EN stripes are not true stripes, but only spots of expression in the respective head segments. In the spider, we infer the same relative location of the EN stripes, where the first one again is only a spot of expression in the head (see Fig. 2E). The respective Hox gene expression domains are represented by bars. Shaded bars indicate strong expression; open bars indicate weak expression.
Another interesting peculiarity exists with respect to UBX. In Drosophila, the primary expression of UBX occurs in PS 6 (23). The anterior expression border of one of the two UBX genes in the spider is in the posterior half of the second abdominal segment (Fig. 3 G), which is equivalent to the anterior PS 6 border in Drosophila (Fig. 4). Moreover, immunostaining with a mAb antibody that broadly cross-reacts with UBX and ABD-A homeoboxes (24) reveals the same border (Fig. 3 K). However, in Drosophila, there is also a low expression of UBX in PS 5, which has a separate, specific function for this parasegment (23). Intriguingly, the second UBX gene that we isolated from the spider also is expressed slightly more anteriorly (compare Fig. 3 I and J), although a corresponding signal is not evident in the protein staining. This could either imply that the translation of this gene occurs at a very low level and is thus below the sensitivity threshold of the antibody or that translation of the second UBX gene occurs at a later stage, which in our experiments was not accessible to immunolabeling because of the formation of cuticle.
The anterior boundary of expression of ABD-A in the spider lies in the posterior half of the third abdominal segment (Fig. 3 H), which is equivalent to the anterior border of ABD-A expression in Drosophila in PS 7 (Fig. 4). All three—ANTP, UBX, and ABD-A—in Cupiennius thus are expressed clearly in a parasegmental register, as their anterior borders cover the posterior half of the appendage in the respective segments (Fig. 3 D–H). By contrast, LAB and DFD show complete staining of the respective appendages (Fig. 3 A–C), indicating that their expression domains do not follow a parasegmental register. Of interest, it is also uncertain in Drosophila whether the organization of the head region complies to a parasegmental register (19, 25). Future comparative analysis will have to show whether a segmental organization can be generally be found for the head region of arthropods.
DISCUSSION
The register of the anterior expression boundaries of the Hox genes in Cupiennius closely resembles that seen in Drosophila. This is rather surprising in view of the fact that spiders and insects represent two very different and also highly derived groups of arthropods, at least when judged on the basis of adult morphology. Embryonic development, on the other hand, is much more comparable between these groups, and the fully segmented spider embryo resembles the extended germ band stage embryo of a short germ insect. Hence, it seems possible that the Hox gene expression pattern seen in spiders and insects represents the ancestral pattern for arthropods as a whole. However, so far this inference is not supported directly by the available evidence of Hox gene expression in crustaceans, which have been studied by in situ hybridization (12) and by means of a cross-reacting mAb against the UBX/ABD-A homeodomain (12, 13, 24). The anterior UBX/ABD-A expression borders in particular are much more flexible in different basic groups of crustaceans, and it is thought that this is correlated with morphological adaptations of particular segmental regions (12, 13). Among the basic groups of crustaceans tested so far, at least the Maxillopoda appear to comply with the insect/spider pattern, in that the anterior boundary of UBX/ABD-A expression lies within the second thoracic segment (13). Although Maxillopoda so far have not been considered as a sister group to insects, it remains a possibility that insects are derived from a crustacean group that has kept the ancestral state of Hox gene expression pattern. Similarly, analysis of UBX/ABD-A expression in a centipede shows an anterior boundary in the second thoracic segment (14), which is again compatible with an assumed ancestral state of this boundary.
The similarity of the anterior expression borders contrasts with some differences in the extent of the posterior borders. The two anterior Hox genes analyzed here, LAB and DFD, have a posterior expression border at the prosomal–opisthosomal boundary, which would be the equivalent of the boundary between the first and second thoracic segment in insects (Fig. 4). LAB and DFD in Drosophila, on the other hand, are expressed in a much more confined region (21). In the spider, the posterior boundaries of ANTP, UBX, and ABD-A all extend to the end of the segmented region and are expressed strongly in the respective segments. In Drosophila, at least ANTP and UBX are expressed only weakly in the more posterior abdominal segments, which suggests an additional level of regulatory control, apparently by the BX-C genes (25). The functional significance of these differences is unclear. In vertebrates, for example, the posterior boundary of Hox gene expression is often not well defined at all (6). Still, at least in the spider, it seems possible to speculate that the posterior boundaries of the anterior genes reflect a functional diversification between the prosomal and opisthosomal body regions, in particular because we found that the expression of a spider homologue of the vertebrate Hox3 genes also is delimited by this posterior boundary (26).
The equivalent of the prosomal–opisthosomal boundary in insects would lie in PS 4—i.e., in the middle of the thorax, which does not relate to any conspicuous morphological boundary. However, a closer analysis of the molecular mechanisms that are patterning the region anterior to PS 4 in Drosophila suggests that these act by rules different from those posterior to this boundary (19, 27). Most intriguingly, in the ancestral form of embryogenesis of insects, the short-germ embryos (28), it is roughly the region anterior to PS 4 (i.e., with the beginning of the thorax region) that becomes specified during the blastoderm stage whereas the more posterior segments arise in a secondary growth process, a correlation that also was noted by Cohen and Jürgens (27). The PS 4 boundary thus might turn out to be an important ancestral border separating two different modes of segmentation in arthropods, even though it is only in the chelicerates that it is associated with a clear morphological boundary.
The setting of the Hox gene expression boundaries in Drosophila depends on many transregulatory factors, most notably the gap genes, the trithorax group genes, the polycomb genes, and crossregulation among the Hox genes themselves (26, 29, 30). The similarities of these boundaries in the spider would suggest that most of these regulatory circuits should be conserved as well, although this conclusion will have to be tested in the future.
All of the inferences above rely on the assumption that no antennal segment is missing in chelicerates. Hence, it is necessary to discuss the arguments that originally led to the conclusion of a missing antennal segment. The strongest argument in favor of a missing segment is based on classical developmental and histological observations assuming that the ganglion innervating the chelicerae is organized similar to the tritocerebral ganglion of crustaceans and insects (2). During embryogenesis, it initially is located behind the stomodaeum, then moves anteriorly during further development, and eventually fuses with the pre-oral ganglia. Several but not all chelicerates keep a post-oral commissure between their cheliceral hemiganglia when reaching adult stage (2, 31). However, if all arthropods had a homonomously segmented ancestor, one would have to conclude that remnants of the first antennal segments or the deutocerebral structures should be visible in chelicerates. This issue has long been controversial, and so far convincing evidence has not been put forward (2). An alternative and more parsimonious interpretation is that the recruitment of ancestral segments into the head region occurred in distinct steps (Fig. 5). The ancestor would have had a stomodaeum right behind the archicerebrum, similar to extant polychaetes. In a first evolutionary step, the ganglia of the pre-antennal segment would be fused, forming a pre-oral protocerebrum. Second, the ganglia of the first antennal segment would be fused partially with the protocerebrum, keeping both a pre-oral and a post-oral commissure. This situation would remain conserved in the present chelicerates. In a third step, the ganglia of the first antennal segment would be fused fully with the protocerebrum in a pre-oral position forming a deutocerebrum while the ganglia of the second antennal segment would be fused partially, retaining both pre-oral and post-oral commissures such as in the tritocerebrum of extant insects (32). In fact, such a series of transformations already was suggested by Remane (1), albeit the second step was considered a purely hypothetical intermediate stage. In view of the basal position of the chelicerates among the arthropods, it seems possible that the chelicerates have in fact retained important features of this “hypothetical” intermediate stage.
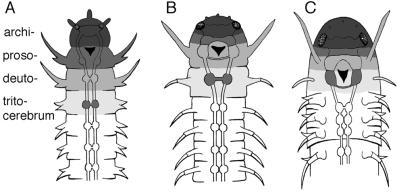
Figure 5.
Evolutionary transformation of the head region in arthropods after Remane (1). (A) An annelid-like ancestor is assumed in which the archicerebrum (darkest shading; top) is connected with the eyes, and the prosocerebrum is located behind the stomodaeum (indicated by a black triangle). (B) More advanced bauplan in which the prosocerebrum has moved anterior to the stomodaeum and has fused with the archicerebrum. The deutocerebrum also has moved to fuse partially with the prosocerebrum but has retained a post-oral commissure. Remane (1) originally proposed this as a hypothetical intermediate step toward the crustacean/insect bauplan whereas our present results suggest that this is the situation in spiders. (C) The crustacean/insect bauplan in which the deutocerebrum has fused fully with the prosocerebrum and the tritocerebrum has fused partly with the deutocerebrum, retaining a post-oral commissure. The position of the tritocerebral ganglion with its commissure and its transformations is shown darkly enhanced in each stage.
A shared mode of head segmentation among all extant arthropod groups strengthens the assumption of a monophyletic origin of arthropods. A possible polyphyly has been suggested by Manton (33), mainly based on arguments derived from functional morphology. Additional arguments for polyphyly still are being put forward (34, 35), but molecular phylogenies clearly have favored a monophyletic origin (4, 5). A broad comparative analysis of molecular embryological features, such as those described here for the Hox genes and the engrailed expression pattern, should help to resolve this question unequivocally in the future.
Acknowledgments
We thank Friedrich G. Barth (University of Vienna) for providing Cupiennius cocoons in an early phase of the project, Reinhard Schröder for help in preparing the embryonic Cupiennius cDNA library, Michael Akam for providing the FP6.87 antibody, and Markus Friedrich for suggestions on the manuscript. W.G.M.D. was supported by a fellowship of the Niels Stensen Foundation (Amsterdam) and a Marie Curie fellowship of the European Commission.
ABBREVIATION
- PS
parasegment
Note Added in Proof
Similar conclusions on the segmentidentity in head of a mite were reached by M. J. Telford and R. H. Thomas (37); see accompanying paper on pages 10671–10675 of this issue.
Footnotes
References
- 1.Remane A, Storch V, Welsch U. Systematische Zoologie. Stuttgart: Gustav Fischer Verlag; 1975. [Google Scholar]
- 2.Weygoldt P. In: Neurobiology of Arachnids. Barth F G, editor. Heidelberg: Springer; 1985. pp. 20–37. [Google Scholar]
- 3.Scholtz G. In: Arthropod Relationships. Fortey R A, Thomas R H, editors. London: Chapman & Hall; 1997. pp. 317–332. [Google Scholar]
- 4.Friedrich M, Tautz D. Nature (London) 1995;376:165–167. doi: 10.1038/376165a0. [DOI] [PubMed] [Google Scholar]
- 5.Regier J C, Shultz J W. Mol Biol Evol. 1997;14:902–913. doi: 10.1093/oxfordjournals.molbev.a025833. [DOI] [PubMed] [Google Scholar]
- 6.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 7.Akam M. Philos Trans R Soc Lond B. 1995;349:313–319. doi: 10.1098/rstb.1995.0119. [DOI] [PubMed] [Google Scholar]
- 8.Lewis E B. Nature (London) 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann T C, Seeger M A, Olsen G. In: Advances in Genetics: Genetic Regulatory Hierarchies in Development. Wright T R F, editor. Vol. 27. San Diego: Academic; 1990. pp. 309–362. [DOI] [PubMed] [Google Scholar]
- 10.Denell R E, Brown S J, Beeman R W. Semin Cell Dev Biol. 1996;7:527–538. [Google Scholar]
- 11.Rogers B T, Peterson M D, Kaufman T C. Development (Cambridge, UK) 1997;124:149–157. doi: 10.1242/dev.124.1.149. [DOI] [PubMed] [Google Scholar]
- 12.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 13.Averof M, Patel N H. Nature (London) 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 14.Grenier J K, Garber T L, Warren R, Whitington P M, Carroll S. Curr Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 15.Chabaud F, Seyfarth E-A, Reichert H. In: Brain Perception and Cognition: Proceedings of the 18th Göttingen Neurobiology Conference. Elsner N, Roth G, editors. Stuttgart: Thieme; 1990. p. 368. [Google Scholar]
- 16.Patel N. H. (1994) Development (Cambridge, U.K.) Suppl., 201- 207.
- 17.Cohen S. In: Development of Drosophila malanogaster. Bate M, Martinez-Arias A, editors. Plainview, New York: Cold Spring Harbor Lab. Press; 1993. pp. 747–841. [Google Scholar]
- 18.Schmidt-Ott U, Sander K, Technau G M. Roux′s Arch Dev Biol. 1994;203:298–303. doi: 10.1007/BF00457800. [DOI] [PubMed] [Google Scholar]
- 19.Rogers B T, Kaufman T C. Development (Cambridge, UK) 1996;122:3419–3432. doi: 10.1242/dev.122.11.3419. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt-Ott U, González-Gaitán M, Jäckle H, Technau G M. Proc Natl Acad Sci USA. 1994;91:8363–8367. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diederich R J, Pattatucci A M, Kaufman T C. Development (Cambridge, UK) 1991;113:273–281. doi: 10.1242/dev.113.1.273. [DOI] [PubMed] [Google Scholar]
- 22.Hayward D, Patel N, Rehm E J, Goodman C, Ball E E. Dev Biol. 1995;172:452–465. doi: 10.1006/dbio.1995.8030. [DOI] [PubMed] [Google Scholar]
- 23.Castelli-Gair J, Akam M. Development (Cambridge, UK) 1995;121:2973–2982. doi: 10.1242/dev.121.9.2973. [DOI] [PubMed] [Google Scholar]
- 24.Kelsh R, Weinzierl R O J, White R A H, Akam M. Dev Genet (Amsterdam) 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- 25.Struhl G, White R A H. Cell. 1985;43:507–519. doi: 10.1016/0092-8674(85)90180-1. [DOI] [PubMed] [Google Scholar]
- 26.Damen, W. & Tautz, D. (1998) Dev. Genes Evol., in press. [DOI] [PubMed]
- 27.Cohen S, Jürgens G. Trends Genet. 1991;7:267–272. doi: 10.1016/0168-9525(91)90327-M. [DOI] [PubMed] [Google Scholar]
- 28.Tautz, D., Friedrich, M. & Schröder, R. (1994) Development (Cambridge, U.K.) Suppl., 193–199.
- 29.Irish V F, Martine-Arias A, Akam M. EMBO J. 1989;8:1527–1537. doi: 10.1002/j.1460-2075.1989.tb03537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon J, Chiang A, Bender W. Development (Cambridge, UK) 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- 31.Babu K S. Zoologische Jahrbücher Anatomie. 1965;82:1–154. [Google Scholar]
- 32.Strausfeld N J. Atlas of an Insect Brain. Heidelberg: Springer; 1976. [Google Scholar]
- 33.Manton S M. The Arthropoda. Oxford: Clarendon; 1977. [Google Scholar]
- 34.Willmer P. Invertebrate Relationships: Patterns in Animal Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1990. [Google Scholar]
- 35.Fryer G. In: Arthropod Relationships. Fortey R A, Thomas R H, editors. London: Chapman & Hall; 1997. pp. 23–33. [Google Scholar]
- 36.Lehmann R, Tautz D. Methods Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- 37.Telford M J, Thomas R H. Proc Natl Acad Sci USA. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]