Abstract
Background and purpose:
In several previous studies, the C-X-C chemokine receptor (CXCR)2 antagonist 1-(2-bromo-phenyl)-3-(7-cyano-3H-benzotriazol-4-yl)-urea (SB265610) has been described as binding competitively with the endogenous agonist. This is in contrast to many other chemokine receptor antagonists, where the mechanism of antagonism has been described as allosteric.
Experimental approach:
To determine whether it displays a unique mechanism among the chemokine receptor antagonists, the mode of action of SB265610 was investigated at the CXCR2 receptor using radioligand and [35S]-GTPγS binding approaches in addition to chemotaxis of human neutrophils.
Key results:
In equilibrium saturation binding studies, SB265610 depressed the maximal binding of [125I]-interleukin-8 ([125I]-IL-8) without affecting the Kd. In contrast, IL-8 was unable to prevent binding of [3H]-SB265610. Kinetic binding experiments demonstrated that this was not an artefact of irreversible or slowly reversible binding. In functional experiments, SB265610 caused a rightward shift of the concentration-response curves to IL-8 and growth-related oncogene α, but also a reduction in maximal response elicited by each agonist. Fitting these data to an operational allosteric ternary complex model suggested that, once bound, SB265610 completely blocks receptor activation. SB265610 also inhibited basal [35S]-GTPγS binding in this preparation.
Conclusions and implications:
Taken together, these data suggest that SB265610 behaves as an allosteric inverse agonist at the CXCR2 receptor, binding at a region distinct from the agonist binding site to prevent receptor activation, possibly by interfering with G protein coupling.
Keywords: allosteric, CXCR2 receptor, SB265610, radioligand binding, neutrophil, mechanism of action, inverse agonist, non-competitive
Introduction
Interleukin-8 (IL-8, CXCL8) and growth-related oncogene α (GROα, CXCL1), in addition to a host of other CXC chemokines, epithelial neutrophil-activating peptide-78 (CXCL5), neutrophil-activating protein-2 (NAP-2, CXCL7), granulocyte chemotactic protein-2 (CXCL6), GROβ (CXCL2) and GROγ (CXCL3), play an important role in the trafficking and activation of inflammatory cells. As a consequence, they are involved in a wide range of chronic diseases such as chronic obstructive pulmonary disease (COPD) and asthma (Keatings et al., 1996), cystic fibrosis (Koller et al., 1997), atherosclerosis, psoriasis and inflammatory bowel disease (Bizzarri et al., 2006). Two G protein-coupled receptors have been identified that are activated by these chemokines; C-X-C chemokine receptor (CXCR)1, which binds IL-8 and CGP-2 with high affinity, and CXCR2, which binds all the CXC chemokines mentioned previously with a range of affinities (Cerretti et al., 1993; Geiser et al., 1993). These receptors are responsible for neutrophil chemotaxis and activation, with increased numbers of neutrophils present in the sputum of COPD patients correlating with increased levels of IL-8 (Keatings et al., 1996). 1-(2-Bromo-phenyl)-3-(7-cyano-3H-benzotriazol-4-yl)-urea (SB265610; Figure 1), a small molecule antagonist of CXCR2, has been shown to inhibit neutrophil recruitment into the inflamed lung (Sarau et al., 2001), suggesting that CXCR2 antagonists may be beneficial in the treatment of inflammatory respiratory diseases.
Figure 1.
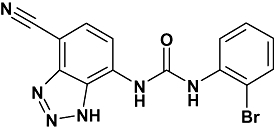
Structure of SB265610, a small molecular weight antagonist of CXCR2. CXCR, C-X-C chemokine receptor.
Antagonism of receptor activation can occur by several different mechanisms. Antagonists that compete for the same binding site as the agonist are termed competitive or orthosteric. Allosteric antagonists bind to a region distinct from the agonist binding site, potentially influencing agonist affinity. Non-competitive antagonists bind to an allosteric site but rather than influencing agonist affinity, act by directly blocking receptor activation. A special case of antagonism that is often confused with non-competitive behaviour is irreversible binding, where the ligand chemically modifies the protein, effectively reducing the population of receptors that can be activated by agonist (see Kenakin, 2006).
Several studies have concluded that SB265610 (Auten et al., 2001) and closely related analogues (Widdowson et al., 2004) act as competitive antagonists at the human CXCR2 receptor. This is perhaps surprising, as CXCR2, like all chemokine receptors, is activated by relatively large protein agonists that interact with multiple residues on the extracellular surface of the receptor. It has been shown using point mutagenesis studies of the N-terminus and first extracellular loop, that IL-8, NAP-2 and GROα utilize different residues to bind and activate the CXCR2 receptor (Katancik et al., 2000). This suggests that discovering small molecule antagonists that are competitive with each of the different agonists is likely to be difficult, if not impossible.
In support of this, there is growing evidence to suggest that many of the small molecule chemokine antagonists discovered to date behave as allosteric inhibitors. For example, CC chemokine receptor (CCR)1 and CCR3 (Sabroe et al., 2000), CCR5 (Watson et al., 2005) and also other CXCR2 antagonists (Bertini et al., 2004; Gonsiorek et al., 2007).
In order to investigate whether there is a common mechanism of action of chemokine receptor antagonists, or whether it is possible to discover competitive compounds, the mode of action of SB265610 was investigated at the CXCR2 receptor. Binding and functional approaches were utilized in both Chinese hamster ovary (CHO) cells expressing human CXCR2 receptor and human neutrophils. Furthermore, these data were directly fitted to mathematical models of ligand-receptor interaction to derive quantitative parameters that better characterize the pharmacological properties of SB265610.
Methods
CXCR2 CHO membrane preparation
Chinese hamster ovary cells stably expressing the human CXCR2 receptor were grown adherently to 90% confluency in 500 cm2 cell culture plates, in HAMS F12 media with GLUTAmax, supplemented with 10% (v·v−1) foetal bovine serum (FBS) and 400 µg·mL−1 geneticin. The cells were removed from the plates by scraping into ice-cold phosphate-buffered saline and pelleted by centrifugation. The supernatant was discarded and replaced with ice-cold buffer A (15 mmol·L−1 Tris-HCl, 2 mmol·L−1 MgCl2, 0.3 mmol·L−1 EDTA, 1 mmol·L−1 EGTA pH 7.5, 1 × Complete tablet per 50 mL buffer). The pellet was homogenized on ice with 5, 10 s bursts from an Ultra Turrax. The homogenate was centrifuged at 40 000 g for 30 min, the supernatant discarded, fresh buffer A added and the pellet re-homogenized as before, followed by a second centrifugation at 40 000 g for a further 30 min. Finally, with the supernatant discarded, the pellet was re-suspended in ice-cold buffer B (7.5 mmol·L−1 Tris-HCl, 12.5 mmol·L−1 MgCl2, 0.3 mmol·L−1 EDTA, 1 mmol·L−1 EGTA, 250 mmol·L−1 sucrose pH 7.5, 1 × Complete tablet per 50 mL buffer) and the protein concentration was determined using the method described by Bradford (1976).
Equilibrium radioligand binding studies
Binding assays were performed with two CXCR2 receptor agonists [125I]-IL-8 and [125I]-GROα and the CXCR2 receptor antagonist [3H]-SB265610. All experiments were run at room temperature (∼21°C) for 2 h in buffer containing 20 mmol·L−1 HEPES, 10 mmol·L−1 MgCl2, 100 mmol·L−1 NaCl, 1 mmol·L−1 EDTA (pH 7.4) and 0.01% (w·v−1) bovine serum albumin (BSA). [125I]-agonist assays were performed in a 0.5 mL assay volume and [3H]-SB265610 in 1.75 mL (unless otherwise stated). Binding was initiated by the addition of CHO-CXCR2 membranes (10 µg) and terminated by vacuum filtration (96-well manual harvester – PerkinElmer) onto PEI-treated GF/C plates for [125I]-agonists and non-PEI-treated GF/B plates for [3H]-SB265610. The filter plates were washed three times with ice-cold wash buffer (20 mmol·L−1 HEPES, pH 7.4) and oven-dried prior to the addition of scintillation fluid (Microscint-20). The amount of radioactivity on each filter was detected on a Topcount Scintillation counter (1 min per well read; PerkinElmer). Specific binding was defined as the difference between the binding that occurred in the presence and absence of 100 nmol·L−1 IL-8 for 125I-agonists and 1 µmol·L−1 unlabelled ligand for [3H]-SB265610. Saturation binding experiments for [125I]-IL-8 were performed in the presence and absence of a range of concentrations (1–30 nmol·L−1) of SB265610 and GTP (0.01–1 mmol·L−1). In addition, concentration inhibition curves for SB265610 were examined in the presence of a range of concentrations of [125I]-IL-8 (500–7.8 pmol·L−1).
Kinetic radioligand binding studies
Dissociation kinetics were studied for [125I]-IL-8 and [3H]-SB265610 using the ‘isotopic dilution’ method (Christopoulos, 2000). [125I]-IL-8 (40 pmol·L−1) was incubated with CHO-CXCR2 membranes for 120 min after which time, re-association of [125I]-IL-8 was prevented by the addition of 10 nmol·L−1 IL-8 (100 ×Kd) in the presence and absence of SB265610 (1 µmol·L−1) and GTP (100 µmol·L−1). SB265610 and GTP were also added in the absence of IL-8. Dissociation times of 0.5 to 45 min were used and non-specific binding was determined using 10 nmol·L−1 IL-8. [3H]-SB265610 (2 nmol·L−1) was incubated with CHO-CXCR2 membranes for 120 min in a 1 mL assay volume and dissociation was initiated by the addition of 1 µmol·L−1 SB265610 in the presence and absence of GROα and IL-8 (100 nmol·L−1). GROα and IL-8 were also added in the absence of SB265610. Dissociation times of 0.5 to 30 min were used and non-specific binding was determined using 1 µmol·L−1 SB265610. [3H]-SB265610 association kinetics were also studied by determining the observed association rates (kob) of three concentrations of [3H]-SB265610 (0.3, 1 and 3 nmol·L−1). The association was initiated by the addition of CHO-CXCR2 membranes to [3H]-SB265610 and the amount of [3H]-SB265610 bound was measured at time intervals between 0.5 and 20 min. In order to calculate the kon, the kob values were re-plotted against their respective concentrations of [3H]-SB265610 to give a straight line, the slope of which is the kon for [3H]-SB265610.
[35S]-GTPγS binding assay
[35S]-GTPγS binding to CHO-CXCR2 membranes was measured using scintillation proximity assay (SPA). All experiments were run at room temperature (∼21°C) in the same buffer used for radioligand binding [20 mmol·L−1 HEPES, 10 mmol·L−1 MgCl2, 100 mmol·L−1 NaCl, 1 mmol·L−1 EDTA (pH 7.4) and 0.01% (w·v−1) BSA] but with 10 µg·mL−1 saponin added. CHO-CXCR2 membranes (2.5 µg) were incubated with 1 µmol·L−1 GDP, 0.5 mg per well wheatgerm agglutinin (WGA) SPA beads and either IL-8 or GROα in the presence and absence of a range of concentrations of SB265610 (10 nmol·L−1 to 1 µmol·L−1) for 60 min. This initial pre-incubation was performed to allow the agonist and antagonist to equilibrate prior to the addition of [35S]-GTPγS (0.3 nmol·L−1), which was followed by a further 60 min incubation. The assay plates were centrifuged prior to detection of [35S]-GTPγS binding using a Topcount scintillation counter (30 s per well read). Additional studies were performed to investigate the effect of SB265610 on basal levels of [35S]-GTPγS binding. Concentration-response curves were constructed for SB265610 from 3 µmol·L−1 and incubated with 0.1 µmol·L−1 GDP (to increase basal activity) and CHO-CXCR2 membranes (5 µg) in the presence and absence of GROα (2 nmol·L−1).
Human neutrophil chemotaxis assay
Neutrophils were purified from peripheral blood of healthy volunteers by dextran sedimentation, Ficoll-Paque density gradient centrifugation and hypotonic lysis of residual erythrocytes (Clark, 1996). The resulting cells were washed, counted and re-suspended in assay buffer [RPMI 1640 with 2.5% FBS (v·v−1), pH 7.4] to a concentration of 2 × 106 cells·mL−1. A range of concentrations of GROα (diluted in assay buffer) were added to the lower chamber, below the insert of a 3.0 µm 24-multiwell insert plate (Beckton Dickenson). Cells were pre-incubated for 30 min at room temperature with either assay buffer alone or range of concentrations of SB265610, they were then loaded onto the inserts. The plate was incubated for 90 min at 37°C, before the inserts were removed. Cells which had migrated through the insert and into the agonist solution were measured using Flow Cytometry. Specifically, cells were gated on forward scatter/side scatter parameters by counting the total number of events occurring in each sample, over a 20 s time frame. The maximum migration was measured by gating a sample containing a known concentration of cells (5 × 105) and the minimum response measured as the amount of cell migration in the presence of assay buffer alone. The minimum amount of migration was subtracted from each sample and then the data were expressed as a percentage of the maximum migration.
Materials
Chinese hamster ovary cells stably expressing the human CXCR2 receptor were purchased from Euroscreen (Brussels, Belgium). [125I]-interleukin-8 ([125I]-IL-8) and [125I]-growth-related oncogene-α ([125I]-GROα) both human recombinant proteins with a specific activity of 2000 Ci·mmol−1 were purchased from GE Healthcare (Chalfont St Giles, UK), along with [35S]-GTPγS (specific activity∼1000 Ci·mmol−1) and WGA coated SPA beads. [3H]-SB265610 was prepared in-house by Pharma DMPK – Isoptope Laboratories. Human recombinant IL-8 and SB265610 were prepared in-house. The 96-well polypropylene plates and 500 cm2 cell culture plates were purchased from Fisher Scientific (Loughborough, UK). The 96-well GF/B and GF/C plates were purchased from Millipore (Watford, UK). White 96-well Optiplates were purchased from PerkinElmer Life and Analytical Sciences (Boston, MA, USA). Complete tablets were purchased from Roche Diagnostics (Lewes, UK). Tris-HCl, MgCl2, EDTA, EGTA, HEPES, NaCl, BSA, saponin, GDP, GTP and GTPγS were obtained from SigmaChemical Co. Ltd. (Poole, UK). All cell culture reagents and human recombinant GROα were purchased from GIBCO (Invitrogen, Paisley, UK).
Calculations and data analysis
Analysis was performed using Prism 4.0 (GraphPad Software Inc., San Diego, CA, USA). Specific binding data from [125I]-IL-8 and [3H]-SB265610 competition binding assays and [35S]-GTPγS binding assays were analysed by non-linear regression, sigmoidal dose-response (variable slope) according to the following equation:
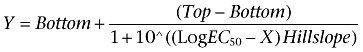 |
(1) |
Where Y is the amount of radioligand bound (cpm) or % of specific binding, Top denotes maximal asymptotic binding and Bottom denotes the minimal asymptotic binding. Saturation binding isotherms were analysed by non-linear regression according to a hyperbolic, one-site binding model, and individual estimates for total receptor number (Bmax) and radioligand dissociation constant (Kd) were calculated. The following equation was used, where [A] is the concentration of radioligand:
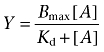 |
(2) |
Sets of [125I]-IL-8 saturation binding isotherms in the absence or presence of increasing concentrations of SB265610 were globally fitted to the following equation that enables estimation of antagonist KB based on its ability to inhibit the maximal binding of radioligand (Kenakin, 2004):
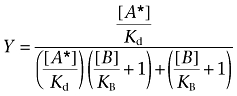 |
(3) |
Where [A*] is the concentration of radioligand, the concentration of non-labelled ligand is [B], and Kd and KB are their respective equilibrium dissociation constants. This is an empirical equation, used only to determine the KB from data where a reduction in binding sites is observed.
[125I]-IL-8 and [3H]-SB265610 dissociation data were fitted to a single phase exponential decay function to obtain koff, t1/2 values where quoted, were obtained was using the following equation:
| (4) |
[3H]-SB265610 association data were fitted to a single phase exponential association function to calculate an observed rate constant kob. The association rate constant, kon, was calculated using the following equation:
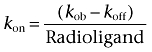 |
(5) |
where the koff value used was predetermined from dissociation rate experiments.
GROα and IL-8 concentration-response curves in the presence and absence of SB265610 from [35S]-GTPγS binding and human neutrophil chemotaxis experiments were analysed using a model (Kenakin, 2006; Price et al., 2005) combining the allosteric ternary complex model (ATCM,Figure 2) of Ehlert (1988) with the operational model of Black and Leff (1983). The form used in these studies was described by May et al. (2007):
Figure 2.
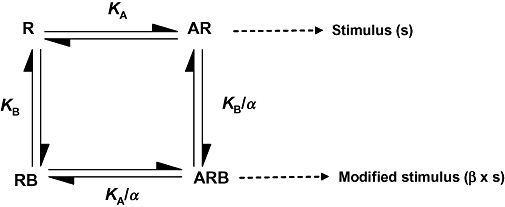
Allosteric ternary complex model (ATCM). This model describes the interaction between orthosteric ligand A, and an allosteric modulator B, in terms of their respective equilibrium dissociation constants (KA, KB), and a cooperativity factor, α, that denotes the magnitude and direction of the allosteric effect on ligand binding affinity. This has been extended to include a proportionality factor, β, which accounts for allosteric effects of the modulator on orthosteric ligand intrinsic efficacy (May et al., 2007).
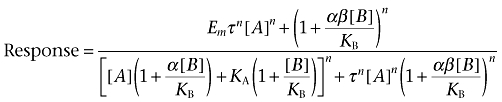 |
(6) |
Where Emax is the maximal response of the system, α denotes the cooperativity factor that governs the magnitude and direction of the allosteric interaction between the two ligands when they both occupy the receptor, β is the proportionality constant that quantifies the change in stimulus imparted to the receptor by the agonist in the presence of the modulator, KA is the equilibrium dissociation constant for the agonist-receptor complex, τ is the ratio of the receptor density divided by the transducer function for the system (defined as KE) and n is the slope of the curve.
Results
Binding studies
The binding of [125I]-IL-8 and [125I]-GROα to the human CXCR2 receptor was saturable, yielding equilibrium dissociation constants of 0.132 ± 0.02 (n= 5) and 0.106 ± 0.01 nmol·L−1 (n= 3), respectively, and maximal receptor binding of 254.3 ± 14.7 (n= 5) and 348.3 ± 68.5 fmol·mg−1 protein (n= 3). There was no significant difference between the number of receptors bound by IL-8 and GROα (P > 0.05; unpaired t-test) and the Kd values showed that IL-8 and GROα have similar affinity for the CXCR2 receptor. The CXCR2 antagonist SB265610 was able to fully displace [125I]-IL-8 and [125I]-GROα (both at 20 pmol·L−1) yielding pIC50 values of 8.41 ± 0.08 and 8.47 ± 0.03 (n= 3) respectively. However, as shown in Figure 3A the IC50 values for SB265610 did not change with elevations in the concentration of [125I]-IL-8. Saturation binding experiments for [125I]-IL-8 in the presence of increasing concentrations of SB265610 produced saturation binding isotherms with significantly reduced Bmax values and no effect on the affinity of [125I]-IL-8, as shown in Table 1. In order to determine a KB for SB265610, these data were fit to an equation that enables estimation of antagonist KB based on its ability to inhibit the maximal binding of radioligand (Equation 3) shown in Figure 3B; global fitting yielded a Kd value for [125I]-IL-8 of 0.118 nmol·L−1 (0.104–0.131 nmol·L−1, 95% CL) and a KB value for SB265610 of 5.4 nmol·L−1 (4.378–6.521 nmol·L−1, 95% CL).
Figure 3.
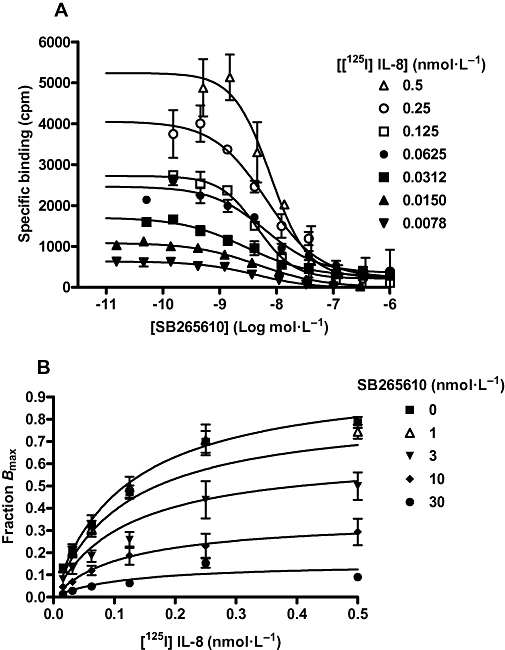
Effect of SB265610 on [125I]-IL-8 binding in CHO membranes expressing human CXCR2 receptor. (A) Competition binding between a range of concentrations of [125I]-IL-8 and SB265610. Data shown are representative of three separate experiments performed in duplicate. (B) The effect of SB265610 on [125I]-IL-8 saturation binding. Data are the mean ± s.e.mean of at least three separate experiments. Data were analysed using Equation 3 as described in Methods; Kd and KB values were determined. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; IL-8, interleukin-8.
Table 1.
Bmax and Kd values from [125I]-IL-8 saturation binding isotherms performed in the presence and absence of SB265610 in CHO membranes expressing human CXCR2 receptor
| SB265610 (nmol·L−1) | Bmax (fmol·mg−1) | Kd (nmol·L−1) | n |
|---|---|---|---|
| 0 | 254.3 ± 14.7 | 0.132 ± 0.02 | 5 |
| 1 | 289.9 ± 14.8 | 0.141 ± 0.01 | 3 |
| 3 | 170.2 ± 22.2** | 0.146 ± 0.01 | 4 |
| 10 | 96.1 ± 6.1** | 0.118 ± 0.03 | 3 |
| 30 | 31.9 ± 6.8** | 0.111 ± 0.02 | 3 |
Data are mean ± s.e.mean for the indicated number of experiments.
P < 0.01 (one-way anova followed by Dunnett's multiple comparison test) indicates Bmax value significantly different from Bmax in the absence of SB265610.
CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; IL-8, interleukin-8.
A reduction of maximal agonist binding sites may result from irreversible binding or even very slow dissociation from the receptor. In order to examine this possibility, the binding characteristics of SB265610 were determined. Binding of [3H]-SB265610 to the CXCR2 receptor was saturable (Figure 4A), yielding an equilibrium dissociation constant of 1.17 ± 0.23 nmol·L−1 (n= 3) and maximal receptor binding of 10.82 ± 1.41 pmol·mg−1 protein. The dissociation rate of [3H]-SB265610 was determined by the addition of a high concentration of SB265610 to pre-equilibrated CXCR2-[3H]-SB265610 at time = 0, with bound [3H]-SB265610 monitored at various time intervals after. The dissociation was rapid and monophasic (Figure 4B), yielding a koff value of 0.348 ± 0.029 min−1 (n= 4), calculated using Equation 4. Figure 4C shows a family of [3H]-SB265610 association curves, each curve yielding an observed on-rate (kob) from which the kon was calculated using the koff determined from dissociation experiments and Equation 5. The mean kon was determined to be 1.57 ± 0.22 × 108 mol·L−1 min−1 (this represents the mean from three separate experiments, each comprising three different [3H]-SB265610 concentrations). If [3H]-SB265610 binding follows the law of mass action, a plot of kob versus radioligand concentration should yield a straight line with a slope equal to the association rate and a y-intercept (at x= 0) equal to the dissociation rate (Motulsky and Christopoulos, 2003). When kob values were plotted against radioligand concentration (Figure 4D), the data were consistent with a straight line (r2= 0.93), indicating simple, bimolecular binding of [3H]-SB265610 to the CXCR2 receptor. Values obtained from this plot were 1.63 × 108 mol·L−1 min−1 and 0.318 min−1 for kon and koff, respectively, which are in good agreement with those calculated above. Furthermore, the kinetically derived Kd (koff/kon) calculated from the mean values for individual experiments (1.51 nmol·L−1) or from the linear plot (1.95 nmol·L−1) are in close agreement with the value obtained from [3H]-SB265610 saturation experiments (1.17 ± 0.41 nmol·L−1). Finally, the ability of the chemokine agonists to compete with [3H]-SB265610 was investigated. Even at relatively high concentrations, neither IL-8 (3 µmol·L−1) nor GROα (1 µmol·L−1) was able to displace [3H]-SB265610 (data not shown).
Figure 4.
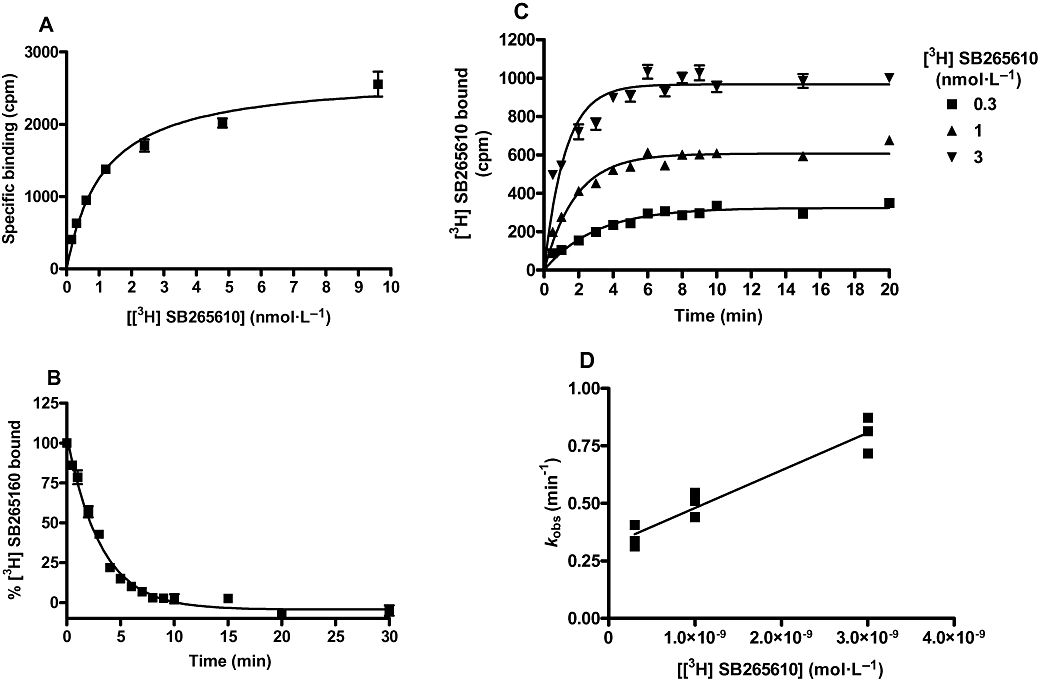
Characterization of [3H]-SB265610 in CHO membranes expressing human CXCR2 receptor. (A) [3H]-SB265610 saturation binding isotherm to determine Kd and Bmax. (B) Dissociation curve for [3H]-SB265610 to determine the koff. (C) A family of [3H]-SB265610 association curves for the determination of kob Graphs (A), (B) and (C) are representative of ≥3 separate experiments. (D) A plot of kob against concentration of [3H]-SB265610, the data shown are the individual kob from three separate experiments. The slope of the line represents the kon for [3H]-SB265610 and the y-intercept the koff. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor.
The observed reduction in agonist binding does not, therefore, result from irreversible binding, but instead may be a consequence of removing high-affinity receptor sites in the membrane preparation. As mentioned previously, agonist binding studies have shown that [125I]-IL-8 maximally labels 254.3 ± 14.7 fmol·mg−1 protein, only 2.3% of the total number of receptors bound by [3H]-SB265610 (10.82 ± 1.41 pmol·mg−1 protein). This is likely to be because G proteins, which stabilize the high-affinity conformation, are limiting in this system.
To investigate this hypothesis, GTP was included in the assay to uncouple pre-bound receptor-G protein complexes. Binding of [3H]-SB265610 was unaffected by the inclusion of 100 µmol·L−1 GTP (data not shown). The effect of GTP on the IL-8 saturation isotherm is shown in Figure 5. Concentrations of GTP up to 100 µmol·L−1 produced concentration-dependent reductions in Bmax with no effect on the affinity of [125I]-IL-8; the presence of 1 mmol·L−1 GTP produced no further decrease in Bmax. GTP 1 mmol·L−1 reduced the Bmax by 50% indicating that at least 50% of the receptors bound by [125I]-IL-8 were coupled to G protein. It was not possible to increase the concentration of GTP to 10 mmol·L−1 as this increased non-specific binding to levels above total binding.
Figure 5.
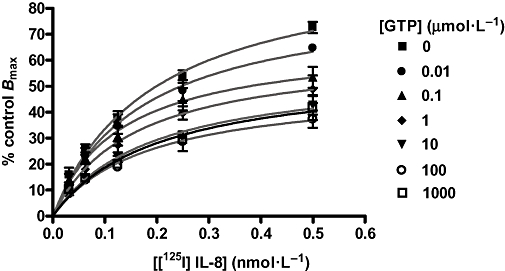
Effect of GTP on [125I]-IL-8 saturation binding in CHO membranes expressing human CXCR2 receptor. Data are the mean ± s.e.mean of at least three separate experiments. Results were analysed using a one-site binding hyperbola for each data set. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; IL-8, interleukin-8.
Investigating allosteric interactions using kinetic binding studies
The most sensitive method for detecting allosteric effects on ligand affinity is to use kinetic radioligand binding assays, as an altered dissociation rate is indicative of an allosteric action (Christopoulos and Kenakin, 2002). Specifically, the disintegration characteristics of a radioligand-receptor complex can only be modified by the binding of a compound to a site distinct from the radioligand binding site. Consequently, the rate of dissociation of [3H]-SB265610 was measured as before, using a saturating concentration of SB265610, but also in the presence of a saturating concentration of IL-8. The rate of dissociation of [3H]-SB265610 was unchanged when IL-8 was included with SB265610 (Figure 6A). Interestingly, IL-8 alone was unable to compete with [3H]-SB265610 binding, as no displacement was observed at an IL-8 concentration of 100 nmol·L−1. This, as previously described, may be because the CXCR2 receptor population exists almost entirely in the inactive R conformation so that a small degree of agonist binding (∼2%) would not cause any significant degree of displacement of bound SB265610.
Figure 6.
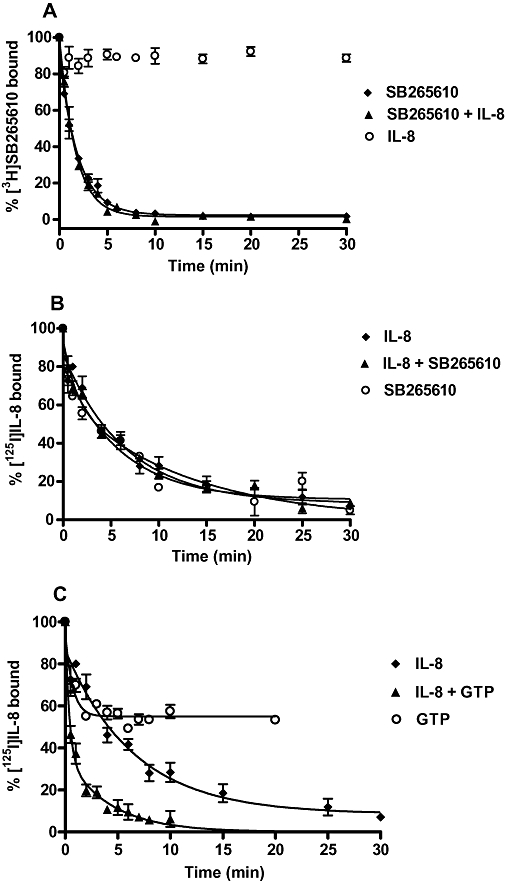
(A) Dissociation of [3H]-SB265610 (2 nmol·L−1) from the CXCR2 receptor in CHO membranes in the presence of 1 µmol·L−1 SB265610, 1 µmol·L−1 SB265610 and 100 nmol·L−1 IL-8 and 100 nmol·L−1 IL-8 alone. (B) Dissociation of [125I]-IL-8 from the CXCR2 receptor in CHO membranes in the presence of 10 nmol·L−1 IL-8, 10 nmol·L−1 IL-8 and 1 µmol·L−1 SB265610 and 1 µmol·L−1 SB265610 alone. (C) Dissociation of [125I]-IL-8 from the CXCR2 receptor in CHO membranes in the presence of 10 nmol·L−1 IL-8, 10 nmol·L−1 IL-8 and 100 µmol·L−1 GTP and 100 µmol·L−1 GTP alone. Data shown are representative of ≥3 separate experiments performed in duplicate. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; IL-8, interleukin-8.
The influence of SB265610 on the dissociation kinetics of [125I]-IL-8 was then examined. The rate of [125I]-IL-8 dissociation was unchanged when SB265610 was included with IL-8. In addition, SB265610 alone produced an [125I]-IL-8 dissociation curve that was identical to the dissociation curve produced in the presence of IL-8 (Figure 6B). In contrast, when GTP was included with IL-8, a two-phase [125I]-IL-8 dissociation curve was observed (Figure 6C, Table 2). The kinetics of the first, faster phase were equivalent to the rate of [125I]-IL-8 dissociation with GTP alone and the second phase displayed the same kinetics as observed with IL-8 alone (Table 2). Interestingly, GTP (100 µmol·L−1) was only able to displace [125I]-IL-8 from ∼50% of the receptor sites. These data are consistent with the presence of two populations of high-affinity receptor conformations.
Table 2.
Dissociation constants koff and t1/2 for [125I]-IL-8 binding in CHO membranes expressing human CXCR2 receptor in the presence of IL-8, GTP and SB265610
|
Dissociation constant (koff) (min−1) |
Half-life (t1/2) (min) |
n | |||
|---|---|---|---|---|---|
| k1 | k2 | (t1/2)1 | (t1/2)2 | ||
| IL-8 | 0.167 ± 0.022 | – | 4.672 ± 0.687 | – | 7 |
| IL-8 + SB265610 | 0.150 ± 0.019 | – | 4.793 ± 0.635 | – | 3 |
| IL-8 + GTPa | 0.165 ± 0.027 | 3.428 ± 1.088 | 4.581 ± 0.796 | 0.251 ± 0.053 | 4 |
| SB265610 | 0.148 ± 0.016 | – | 4.779 ± 0.490 | – | 3 |
| GTPb | 1.129 ± 0.411 | – | 0.791 ± 0.257 | – | 3 |
Data are mean ± s.e.mean for the indicated number of experiments.
Indicates that two-phase exponential decay was the preferred fit when compared with one phase, with a P-value of <0.0001 for three experiments and 0.0002 for the fourth experiment.
Indicates that the dissociation curve did not reach zero, instead GTP alone could only cause the dissociation of 45.22 ± 5.66% [125I]-IL-8.
CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; IL-8, interleukin-8.
Functional studies using [35S]-GTPγS binding assays
To investigate the influence of SB265610 on the function of the CXCR2 receptor, stimulation of [35S]-GTPγS binding by IL-8 and GROα in CHO-CXCR2 membranes was examined. Concentration-response curves to both IL-8 and GROα were determined in the presence of a range of concentrations of SB265610, the effects of which are shown in Figure 7A and B. At the lowest concentration of SB265610, parallel rightward shifts of the agonist concentration-response curve were observed with no significant diminution of Emax. At higher concentrations of SB265610, however, there were significant reductions in the maximum response of each agonist. Table 3 shows that in the presence of 1 µmol·L−1 SB265610, the Emax values for IL-8 and GROα were significantly reduced by 71.06 and 59.50% respectively.
Figure 7.
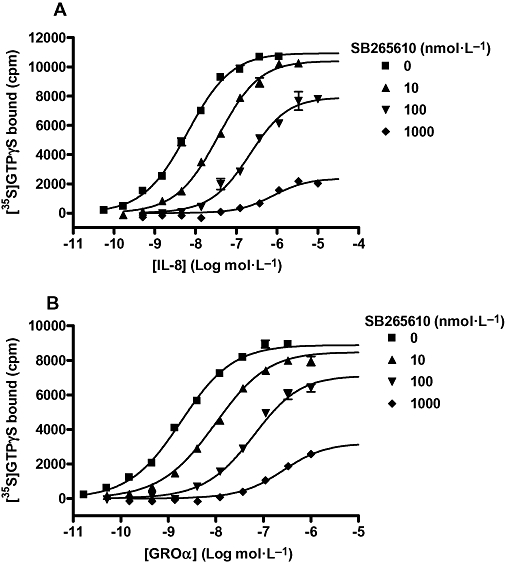
Stimulation of [35S]-GTPγS binding in CHO-CXCR2 membranes by IL-8 (A) and GROα (B) in the absence and presence of a range of concentrations of SB265610 (10, 100 and 1000 nmol·L−1). The data were globally analysed using the operational allosteric ternary complex model (Equation 6). The best-fit parameter values for the IL-8 data were: KB=−8.71, KA=−6.02 and τ= 147.5, and for the GROα data; KB=−8.66, KA=−6.34 and τ= 239.5. Data shown are representative of three separate experiments, performed in duplicate. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; GRO, growth related oncogene; IL-8, interleukin-8.
Table 3.
pEC50 values and % inhibition of Emax from IL-8 and GROα stimulated [35S]-GTPγS binding in CHO membranes expressing human CXCR2 receptor, in the presence and absence of SB265610
| SB265610 (nmol·L−1) |
IL-8 |
GROα |
||
|---|---|---|---|---|
| Inhibition of Emax % | pEC50 | Inhibition of Emax % | pEC50 | |
| 0 | n/a | 8.15 ± 0.04 | n/a | 8.52 ± 0.13 |
| 10 | 5.90 ± 2.20 | 7.48 ± 0.01 | 12.20 ± 2.01 | 7.94 ± 0.08 |
| 100 | 27.57 ± 4.05 | 6.71 ± 0.03 | 28.07 ± 3.27 | 7.23 ± 0.09 |
| 1000 | 71.06 ± 1.61 | 6.12 ± 0.13 | 59.50 ± 1.20 | 6.49 ± 0.13 |
Data were obtained by fitting unconstrained four parameter logistic curves and are presented as means ± s.e.mean from three separate experiments.
CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; GRO, growth related oncogene; IL-8, interleukin-8; n/a, not available.
The data in Figure 7A and B were globally analysed using Equation 6, based on an operational extension of the ATCM shown in Figure 2. As SB265610 was shown to have no effect on the binding affinity of IL-8, the term α was fixed to 1. Best-fit values for KB, KA, τ and β are shown in Table 4. The mean KA values for IL-8 and GROα show that they are both predicted to have very low affinity for the inactive, uncoupled state of the receptor. The mean KB values for SB265610 were −8.74 ± 0.04 and −8.61 ± 0.04 versus IL-8 and GROα respectively; these values are in close agreement with Kd values determined using [3H]-SB265610 (−8.93). The τ values were similar for both IL-8 and GROα, indicating they have a similar intrinsic efficacy. Finally, the β values for SB265610 calculated against IL-8 and GROα were very low, 0.0018 ± 0.0018 and 0.0029 ± 0.0003 respectively. This suggests that SB265610 very efficiently inhibits receptor activation. Indeed, for many of the global fits of β, zero was within the 95% confidence limits, so it is possible that SB265610 acts to completely inhibit receptor activation once bound. This is consistent with the ability of SB265610 to inhibit constitutive activity of the CXCR2 receptor. Figure 8 shows that SB265610 inhibits basal levels of [35S]-GTPγS binding in a concentration-dependent manner with a mean pIC50 value of 7.83 ± 0.12 (n= 3).
Table 4.
KA, KB and τ and β values determined by fitting the data in Figure 7 to the operational allosteric ternary complex model (Equation 6)
| Parameter |
[35S]-GTPγS binding |
|
|---|---|---|
| IL-8 | GROα | |
| pKA | −6.13 ± 0.07 | −6.40 ± 0.08 |
| pKB | −8.72 ± 0.01 | −8.63 ± 0.04 |
| τ | 110.5 ± 22.2 | 161.6 ± 29.5 |
| β | 0.0018 ± 0.0018 | 0.0029 ± 0.0003 |
For this analysis, α was fixed to 1.0. Data are mean ± s.e.mean from at least three separate experiments.
GRO, growth related oncogene; IL-8, interleukin-8.
Figure 8.
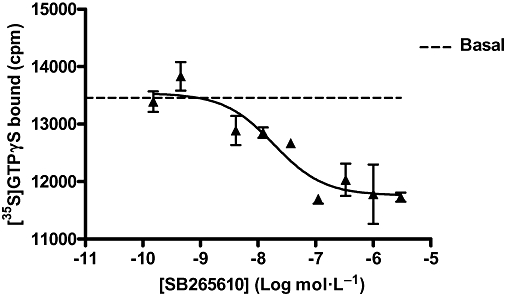
Effect of SB265610 on basal [35S]-GTPγS binding in CHO-CXCR2 membranes, in the absence of GROα. Data shown are representative of three separate experiments. CHO, chinese hamster ovary; CXCR, C-X-C chemokine receptor; GRO, growth related oncogene.
Human neutrophil chemotaxis
The effect of a range of concentrations of SB265610 on GROα-stimulated human neutrophil chemotaxis was investigated (Figure 9). All concentrations of SB265610 produced parallel rightward shifts of the GROα concentration-response curve with no effect on the Emax, the expected pattern of a competitive antagonist, as suggested by Auten et al. (2001). As there was no diminution of maximal agonist response, the data could not be reliably fitted to the operational ATCM. It is possible, however, that there is larger receptor reserve in the neutrophils, which would conceal the insurmountable nature of this antagonist. The notion of increased spare receptors in neutrophils is supported by the increased potency of GROα in this system.
Figure 9.
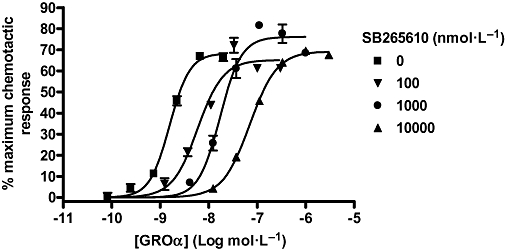
Effect of SB265610 on GROα stimulated chemotaxis in human neutrophils. Data shown are representative of three separate experiments, each performed in duplicate. GRO, growth related oncogene.
Discussion
Unlike most other chemokine receptor antagonists, SB265610 and analogues have been described previously as competitive antagonists at the CXCR2 receptor. The aim of this study was to further investigate the nature of antagonism of SB265610 to determine whether it interacts with the CXCR2 receptor at the IL-8 and GROα binding site, thereby displaying a unique mechanism of action.
A key feature of competitive antagonists is that they can be overcome by increasing the concentration of agonist, that is, the agonist Bmax should not change. In our hands, however, SB265610 reduced the Bmax in [125I]-IL-8 saturation binding experiments with no change in [125I]-IL-8 affinity. In addition, further [125I]-IL-8 binding studies demonstrated that the IC50 of SB265610 did not change with elevations in [125I]-IL-8 concentration. We were able to show that it was possible to detect competitive action in this system by performing the same experiments with IL-8 and producing a linear relationship between IC50 and radioligand concentration (data not shown). A reduction in agonist binding sites typically arises from covalent binding to the receptor, irreversibly removing receptor sites without affecting the affinity of the agonist (Furchgott, 1966). However, studies with [3H]-SB265610 showed that it was fully reversible, with a rapid rate of dissociation from the CXCR2 receptor (t1/2 of ≈2 min).
In functional studies SB265610 produced concentration-dependent reductions in the Emax of both IL-8 and GROα stimulated [35S]-GTPγS binding. Insurmountable antagonism is commonly observed with competitive/orthosteric antagonists as an artefact of slow dissociation kinetics from the receptor (discussed by Kenakin et al., 2006). As many of the assays commonly used to attribute compound mechanism of action utilize rapid readouts such as second messenger generation (e.g. calcium transients), they are seldom, if ever, at equilibrium (Lew et al., 2000). This can be compounded if the system studied has a low receptor reserve (i.e. number of receptors versus efficacy of agonist, see Rang (1966). In the present study, we were careful to use an assay format that included an agonist-antagonist-receptor pre-incubation period prior to addition of the substrate to ensure the system was at, or close to, equilibrium. In addition to this, we were also able to establish that SB265610 has rapid dissociation kinetics. It is therefore unlikely that the reduction in maximal agonist response observed with SB265610 is due to hemi-equilibrium conditions.
When the functional GTPγS data were globally fit to an operational ATCM (Figure 2) with α fixed to 1, the β values determined were not significantly different from zero, indicating that once bound, SB265610 very effectively inhibits receptor signalling. This pattern of allosteric behaviour is equivalent to non-competitive enzyme inhibition as described by Dixon (1972), where the ligand blocks enzyme function without affecting substrate binding.
In chemotaxis assays performed with human neutrophils, however, SB265610 caused concentration-dependent rightward shifts in the agonist response curves, without causing a diminution of maximal agonist response. These data are consistent with previous reports on human neutrophils (Widdowson et al., 2004). It is likely, however, that human neutrophils are a more efficiently coupled system with a larger receptor reserve, masking the allosteric nature of SB265610. This is supported by the higher potency of GROα in the neutrophil chemotaxis experiment over the GTPγS assay. It is possible, therefore, that allosteric antagonists may, in some systems, appear competitive and this highlights the importance of testing ligands in range of different assay systems.
How, then, does a reversible antagonist remove agonist binding sites? Agonists preferentially bind to the active conformation of the receptor, which is stabilized when coupled to guanine nucleotide-free G proteins. Uncoupling the receptor-G protein complex (e.g. with guanine nucleotides) reduces the number of high-affinity binding sites, normally resulting in a shift in apparent agonist affinity towards the inactive conformation without a reduction in Bmax. In some cases, however, where the agonist has very low affinity at the inactive conformation, a decrease in Bmax may be observed as high-affinity sites are removed (discussed by Keen, 1997), presumably because these low-affinity interactions are washed away during the filtration process. This was observed in our system when a range of concentrations of GTP was included in [125I]-IL-8 saturation binding experiments, where GTP behaved in a similar manner to SB265610, reducing the Bmax of [125I]-IL-8 with no change in Kd. This supports the notion that IL-8 has a very low affinity for the uncoupled form of the receptor and suggests that SB265610 may behave in a similar way as GTP to uncouple the receptor-G protein complex. In contrast to SB265610, however, GTP was not able to fully inhibit [125I]-IL-8 binding, demonstrating the presence of guanine nucleotide sensitive and insensitive populations of high-affinity sites. The guanine nucleotide insensitive population could possibly comprise G protein-coupled receptors that are poorly accessible to GTP (Cohen et al., 1996), but more likely represent receptor that is stabilized in its high-affinity conformation by an alternative binding partner, as observed for the NK1 receptor fused to β-arrestin 1 (Martini et al., 2002) and the glucagon-like peptide-1 receptor fused to β-arrestin 2 (Jorgensen et al., 2005). Importantly, SB265610 removed all high-affinity sites, most likely acting as an inverse agonist to stabilize the inactive conformation of the receptor (Costa and Herz, 1989). This is supported by experiments showing that SB265610 was able to inhibit basal [35S]-GTPγS binding in the absence of agonist.
If SB265610 was able to bind simultaneously with IL-8 at the CXCR2 receptor, the dissociation rate of IL-8 should be increased as SB265610 converts high-affinity sites to lower-affinity sites either directly or by uncoupling receptor from G protein in a similar manner to GTP. To investigate this, both GTP and SB265610 were examined in kinetic experiments. As expected, GTP increased the dissociation rate of IL-8, confirming that the G protein behaves as an allosteric modulator of agonist affinity. In contrast, SB265610 did not alter the dissociation kinetics of [125I]-IL-8. This may be because IL-8 blocks the access of SB265610 to its binding site in the receptor; that is, it can only bind once IL-8 has dissociated.
In summary, SB265610 exhibits elements of an inverse agonist that stabilizes the inactive conformation of the CXCR2 receptor. It does not compete directly with the agonist, but rather binds to an allosteric region to prevent receptor activation without directly affecting agonist affinity. The molecular mechanism of such antagonism at the receptor level has not been adequately described to date, but may arise if the binding site of SB265610 is located close to the site of G protein coupling or to a region of the receptor that is responsible for the transduction of the activation signal following agonist binding. Further work using mutagenesis approaches will be required to locate the precise site of interaction between SB265610 and the CXCR2 receptor.
There are a number of other examples in the chemokine field of similar mechanisms of antagonism. Watson et al. (2005) demonstrated a reduction in [125I]-MIP-1α binding sites by 873140, a low molecular weight antagonist of the CCR5 receptor. In functional calcium assays, 873140 also caused diminution of the maximum MIP-1α response. During the course of this study, Gonsiorek et al. (2007) published results on the dual CXCR1/CXCR2 antagonist Sch527123, which they classify as a ‘non-competitive allosteric antagonist’. As with SB265610, Sch527123 causes concentration-dependent reductions in the maximal response to agonist in [35S]-GTPγS and FLIPR assays, in a transfected cell line. In contrast to our results, however, high concentrations of Sch527123 (>60-fold Kd) completely abrogated the agonist response in human neutrophil chemotaxis. However, in both of these studies and in contrast to SB265610, the antagonists 873140 and Sch527123 show slow dissociation from the receptor, which could have contributed to the insurmountable effects on agonist efficacy (Kenakin et al., 2006).
In conclusion, we have demonstrated that SB265610, like many other chemokine receptor antagonists, is able to inhibit receptor function by binding to an allosteric site on the CXCR2 receptor. Indeed, it may be postulated that in order to successfully inhibit the action of a large protein agonist, small molecular weight compounds must prevent receptor activation rather than interfere with the orthosteric binding site directly. Moreover, this mechanism of action may be more desirable than competitive antagonism. When a receptor bound by competitive antagonist is exposed to high concentrations of agonist, the system will re-equilibrate according to the relative concentrations of ligand, possibly resulting in the majority of receptors bound by agonist. In contrast, an antagonist that directly interferes with receptor activation will inhibit receptor function irrespective of the concentration of agonist present. This insurmountability will result in a more complete block of receptor signalling and potentially more efficacious drugs.
Acknowledgments
Neil Press and Joe Fullerton for synthesizing SB265610. Matthias Frommherz (DMPK Isotope laboratories in Basel) for making [3H]-SB265610. Mark Dowling for reviewing this manuscript and for the useful discussions we have had throughout the course of this study.
Glossary
Abbreviations:
- ATCM
allosteric ternary complex model
- CCR
CC chemokine receptor
- CHO
Chinese hamster ovary
- COPD
chronic obstructive pulmonary disease
- CXCR
C-X-C chemokine receptor
- GRO
growth-related oncogene
- IL-8
interleukin-8
- SB265610
1-(2-bromo-phenyl)-3-(7-cyano-3H-benzotriazol-4-yl)-urea
- SPA
scintillation proximity assay
- WGA
wheatgerm agglutinin
Conflicts of interest
None.
References
- Auten RL, Richardson RM, White JR, Mason SN, Vozzelli MA, Whorton MH. Nonpeptide CXCR2 antagonist prevents neutrophil accumulation in hyperoxia-exposed newborn rats. J Pharmacol Exp Ther. 2001;299:90–95. [PubMed] [Google Scholar]
- Bertini R, Allegretti M, Bizzarri C, Moriconi A, Locati M, Zampella G, et al. Noncompetitive allosteric inhibitors of the inflammatory chemokine receptors CXCR1 and CXCR2: prevention of reperfusion injury. Proc Natl Acad Sci USA. 2004;101:11791–11796. doi: 10.1073/pnas.0402090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzarri C, Beccari AR, Bertini R, Cavicchia MR, Giorgini S, Allegretti M. ELR+ CXC chemokines and their receptors (CXC chemokine recptor 1 and CXC chmokine receptor 2) as new therapeutic targets. Pharmacol Ther. 2006;112:139–149. doi: 10.1016/j.pharmthera.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Black JW, Leff P. Operational models of pharmacological agonists. Proc R Soc Lond B Biol Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cerretti DP, Kozlosky CJ, Vanden Bos T, Nelson N, Gearing DP, Beckmann MP. Molecular characterisation of receptors for human interleukin-8, GRO/melanoma growth-stimulatory activity and neutrophil activating peptide-2. Mol Immunol. 1993;30:359–367. doi: 10.1016/0161-5890(93)90065-j. [DOI] [PubMed] [Google Scholar]
- Christopoulos A. Quantification of allosteric interactions at G protein-coupled receptors using radioligand binding assays. In: Enna SJ, editor. Current Protocols in Pharmacology. New York: John Wiley & Sons; 2000. pp. 1.21.1–1.22.40. [DOI] [PubMed] [Google Scholar]
- Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- Clark RA. Isolation and functional analysis of neutrophils. In: Coligan JE, editor. Current Protocols in Immunology. New York: John Wiley & Sons; 1996. pp. 7.23.1–7.23.3. [DOI] [PubMed] [Google Scholar]
- Cohen FR, Lazareno S, Birdsall NJ. The effects of saponin on the binding and functional properties of the human adenosine A1 receptor. Br J Pharmacol. 1996;117:1521–1529. doi: 10.1111/j.1476-5381.1996.tb15316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T, Herz A. Antagonists with negative intrinsic activity at delta-opioid receptors coupled to GTP-binding proteins. Proc Nati Acad Sci USA. 1989;86:7321–7325. doi: 10.1073/pnas.86.19.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. The graphical determination of Km and Ki. Biochem J. 1972;129:197–202. doi: 10.1042/bj1290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert FJ. Estimation of the affinities of allosteric ligands using radioligand binding and pharmacological null methods. Mol Pharmacol. 1988;33:187–194. [PubMed] [Google Scholar]
- Furchgott RF. The use of -haloalkylamines in the differentiation of receptors and in the determination of dissociation constants of receptor-agonist complexes. In: Harper NJ, Simmonds AB, editors. Advances in Drug Research. New York: Academic Press; 1966. pp. 21–55. Vol 3. [Google Scholar]
- Geiser T, Dewald B, Ehrengruber MU, Clark-Lewis I, Bagglioni M. The interleukin-8-related chemotactic cytokines GROα, GROβ and GROγ activate human neutrophil and basophil leukocytes. J Biol Chem. 1993;268:15419–15424. [PubMed] [Google Scholar]
- Gonsiorek W, Fan X, Hesk D, Fossetta J, Qiu H, Jakway J, et al. Pharmacological characterization of Sch527123, a potent allosteric CXCR1/CXCR2 antagonist. J Pharmacol Exp Ther. 2007;322:477–485. doi: 10.1124/jpet.106.118927. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Martini L, Schwartz TW, Elling CE. Characterization of Glucagon-Like Peptide-1 Receptor {beta}-Arrestin 2 Interaction: a high-affinity receptor phenotype. Mol Endocrinol. 2005;19:812–823. doi: 10.1210/me.2004-0312. [DOI] [PubMed] [Google Scholar]
- Katancik JA, Sharma A, de Nardin E. Interleukin 8, neutrophil-activating peptide-2 and GROalpha bind to and elicit cell activation via specific and different amino acid residues of CXCR2. Cytokine. 2000;12:1480–1488. doi: 10.1006/cyto.2000.0742. [DOI] [PubMed] [Google Scholar]
- Keatings VM, Collins PD, Scott DM, Barnes PJ. Differences in interleukin-8 and tumor necrosis factor-α in induced sputum from patients with chronic obstructive pulmonary disease or asthma. Am J Respir Crit Care Med. 1996;153:530–534. doi: 10.1164/ajrccm.153.2.8564092. [DOI] [PubMed] [Google Scholar]
- Keen M. Radioligand-binding methods. In: Challis RAJ, editor. Methods in Molecular Biology, Vol. 83: Receptor Signal Transduction Protocols. Totowa, NJ: Humana Press Inc; 1997. pp. 1–24. [Google Scholar]
- Kenakin T. A Pharmacology Primer: Theory, Application, and Methods. London: Elsevier Academic Press; 2004. [Google Scholar]
- Kenakin T. A Pharmacology Primer: Theory, Application, and Methods. 2nd edn. London: Elsevier Academic Press; 2006. [Google Scholar]
- Kenakin T, Jenkinson S, Watson C. Determining the potency and molecular mechanism of action of insurmountable antagonists. J Pharmacol Exp Ther. 2006;319(2):710–723. doi: 10.1124/jpet.106.107375. [DOI] [PubMed] [Google Scholar]
- Koller DY, Nething I, Otto J, Urbanek R, Eichler I. Cytokine concentrations in sputum from patients with cystic fibrosis and their relation to eosinophil activity. Am J Respir Crit Care Med. 1997;155:1050–1054. doi: 10.1164/ajrccm.155.3.9116985. [DOI] [PubMed] [Google Scholar]
- Lew MJ, Ziogas J, Christopoulos A. Dynamic mechanisms of non-classical antagonism by competitive AT(1) receptor antagonists. Trends Pharmacol Sci. 2000;21:376–381. doi: 10.1016/s0165-6147(00)01523-6. [DOI] [PubMed] [Google Scholar]
- Martini L, Hastrup H, Holst B, Fraile-Ramos A, Marsh M, Schwartz TW. NK1 receptor fused to beta-arrestin displays a single-component, high-affinity molecular phenotype. Mol Pharmacol. 2002;62:30–37. doi: 10.1124/mol.62.1.30. [DOI] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- Motulsky HJ, Christopoulos A. Analysing kinetic binding data. In: Motulsky HJ, Christopoulos A, editors. Fitting Models to Biological Data using Linear and Nonlinear Regression. New York: Oxford University Press; 2003. pp. 245–251. [Google Scholar]
- Price MR, Baillie GL, Thomas A, Stevenson LA, Easson M, Goodwin R, et al. Allosteric Modulation of the Cannabinoid CB1 Receptor. Mol Pharmacol. 2005;68:1484–1495. doi: 10.1124/mol.105.016162. [DOI] [PubMed] [Google Scholar]
- Rang HP. The kinetics of action of acetylcholine antagonists in smooth muscle. Proc R Soc Lond B Biol Sci. 1966;164:488–510. doi: 10.1098/rspb.1966.0045. [DOI] [PubMed] [Google Scholar]
- Sabroe I, Peck MJ, Van Keulen BJ, Jorritsma A, Simmons G, Clapham PR, et al. A small molecule antagonist of CCR1 and CCR3. Potent inhibition of eosinophil function and CCR3-mediated HIV-1 entry. J Biol Chem. 2000;275:25985–25992. doi: 10.1074/jbc.M908864199. [DOI] [PubMed] [Google Scholar]
- Sarau HM, Widdowson K, Palovich MR, White JR, Underwood DC, Griswold DE. Interleukin-8 receptor (CXCR2) antagonists. Prog Respir Res. 2001;31:293–296. [Google Scholar]
- Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharm. 2005;67:1268–1282. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- Widdowson KL, Elliott JD, Veber DF, Nie H, Rutledge MC, McCleland BW, et al. Evaluation of potent and selective small-molecule antagonists for the CXCR2 chemokine receptor. J Med Chem. 2004;47:1319–1321. doi: 10.1021/jm034248l. [DOI] [PubMed] [Google Scholar]


