Abstract
To cause infection, Salmonella enterica serovar Typhimurium uses type III secretion systems, which are encoded on two Salmonella pathogenicity islands, SPI-1 and SPI-2, the latter of which is thought to play a crucial role in bacterial proliferation in Salmonella-containing vacuoles (SCVs) after invading cells. S. Typhimurium SrfJ, located outside SPI-2, is also known to be involved in Salmonella pathogenicity and has high amino acid sequence homology with human lysosomal glucosylceramidase (GlcCerase). We present the first crystal structure of SrfJ at a resolution of 1.8 Å. The overall fold of SrfJ shares high structure similarities with that of human GlcCerase, comprising two distinctive domains: a (β/α)8-barrel catalytic domain and a β-sandwich domain. As in human GlcCerase, the pocket-shaped active site of SrfJ is located on the C-terminal side of the barrel, and two conserved glutamic acid residues are used for the enzyme catalysis. Moreover, a glycerol-bound form of SrfJ reveals that the glucose ring moiety of the substrate might similarly bind to the enzyme as to human GlcCerase, suggesting that SrfJ might function as a glycoside hydrolase. Although some structural differences are observed between SrfJ and human GlcCerase in the substrate entrance of the active site, we speculate that, based on the high structural similarities to human GlcCerase in the overall fold and the active-site environment, SrfJ might have a GlcCerase activity and use the activity to enhance Salmonella virulence by modifying SCV membrane lipids.
Gram-negative bacterial pathogens deliver effector proteins into host cells through type III secretion systems (TTSS). The TTSS apparatus is a molecular syringe which spans the inner and outer membranes of pathogens and secretes translocon and effector proteins. Translocon proteins locate at the tip of the needle structure and are involved in the translocation of effector proteins by forming pores in the host cell membrane (3). The translocated effector proteins function to manipulate diverse host cellular processes such as cytoskeleton assembly, vesicle transport, and signal transduction, thereby promoting bacterial virulence (9).
Salmonella enteric serovar Typhimurium (S. Typhimurium) causes a systemic infection in mice and is an intensively studied model of typhoid fever. This gram-negative bacterium can invade host cells and then survive by replicating within a membrane-bound compartment known as the Salmonella-containing vacuole (SCV) (16). Both invasion and intracellular survival are mediated by numerous virulence genes, which are clustered within the pathogenicity islands, SPI-1 and SPI-2 (18). The regulation of virulence proteins encoded by each pathogenicity island depends on the different stages of infection. While most of the genes within SPI-1 are required for the invading host cells and the early stages of SCV development (8), the genes in SPI-2 play a crucial role in bacterial proliferation in SCVs after cell invasion (23). The SsrA-SsrB two-component regulatory system is known to regulate the expression of genes within SPI-2 for bacterial virulence (4). Recent works have shown that several genes located outside of SPI-2 are under the control of the SsrA-SsrB regulator as well, and these have been proposed as putative virulence factors (10, 25).
S. Typhimurium SrfJ was initially identified as a gene that is strongly activated by SsrB outside SPI-2 (25). Furthermore, a mutation on srfJ leads to mild attenuation of virulence in mice (22). Interestingly, SrfJ shares high amino acid sequence similarity with human lysosomal glucosylceramidase (GlcCerase) (25), which is a peripheral membrane protein catalyzing the hydrolysis of glucosylceramide (GlcCer) to β-glucose and ceramide in the presence of the modulator protein saposin C and lipid (11). Inherited defects in GlcCerase result in lysosomal GlcCer accumulation and, as a consequence, Gaucher disease, the most common lysosomal storage disease (19). Both human GlcCerase and SrfJ have been grouped into glycoside hydrolase (GH) family 30 containing GlcCerase (EC 3.2.1.45), β-1,6-glucanse (EC 3.2.1.75), and β-xylosidase (EC 3.2.1.37) of the GH-A clan in the CAZy database (http://afmb.cnrs-mrs.fr/CAZY). Among the members of GH family 30, structural information is available only on the human enzyme. The biochemical function of SrfJ and its role in Salmonella virulence remain to be elucidated. In order to better understand the function of SrfJ, we have determined the crystal structure of SrfJ from S. Typhimurium at a resolution of 1.8 Å.
MATERIALS AND METHODS
Cloning, expression, and purification of SrfJ.
The SrfJ gene was amplified by PCR using S. Typhimurium chromosomal DNA as a template. The PCR product was then subcloned into the pET30a (Novagen) vector that is designed to express a C-terminal His6-tagged protein. The resulting expression vector pET30a::SrfJ was transformed in the Escherichia coli BL21(DE3) strain, and cells were grown in LB medium supplemented with an appropriate amount of kanamycin. After induction with 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for a further 20 h at 22°C, the culture was harvested by centrifugation at 5,000 × g at 22°C. The cell pellet was resuspended in ice-cold buffer A (50 mM Tris-HCl, pH 8.0, 5 mM β-mercaptoethanol) and disrupted by ultrasonication. The cell debris was removed by centrifugation at 11,000 × g for 1 h, and the lysate was bound to Ni-nitrilotriacetic acid agarose (Qiagen). After a washing step with buffer A containing 10 mM imidazole, the bound proteins were eluted with 300 mM imidazole in buffer A. A trace amount of contaminants was removed by applying HiTrap Q anion-exchange and Superdex 75 size exclusion chromatography. The purified SrfJ protein showed ∼95% purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown) and was concentrated up to 30 mg/ml in buffer A and used for crystallization.
Crystallization and structure determination of SrfJ.
Crystals of the SrfJ protein were obtained by the hanging-drop vapor diffusion method at 22°C by mixing and equilibrating 2 μl of protein and 2 μl of precipitant solution against a precipitant solution containing 0.1 M phosphate citrate, pH 4.2, 1.6 M NaH2PO4, and 0.4 M K2HPO4. For data collection, 20% (wt/vol) glycerol was added to the crystallizing precipitant as a cryoprotectant, and the crystals were immediately placed in a 100 K nitrogen gas stream. X-ray diffraction data of the crystals were collected at a resolution of 1.7 Å at the 6C1 beamline of the Pohang Accelerator Laboratory (South Korea) using a Quantum 210 charge-coupled-device detector (San Diego, CA). The data were then indexed, integrated, and scaled using the HKL2000 suite (21). The crystals belonged to the space group C2221 with unit cell dimensions of a = 92.6 Å, b = 162.3 Å, and c = 169.6 Å. The structure was determined by molecular replacement with the CCP4 version of MOLREP (24) using the structure of human GlcCerase (Protein Data Bank [PDB] code 2v3e) as a search model. The model building was performed using the program Coot (6), and the refinement was performed with CCP4 Refmac 5 (20) and CNS (2). The x-ray diffraction and structure refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics of SrfJ
| Statistics | Resulta |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions (Å) | a = 92.6, b = 162.3, c = 169.6 |
| Resolution (Å) | 30.0-1.7 |
| Rsymb | 8.1 (38.3) |
| I/σ(I) | 19.3 (1.7) |
| Completeness (%) | 90.2 (45.9) |
| Redundancy | 2.7 |
| Structure refinement | |
| Resolution (Å) | 30.0-1.8 |
| No. of reflections | 117,878 |
| Rworkc/Rfree | 19.9/21.5 |
| No. of atoms | |
| Protein | 7,136 |
| Water | 364 |
| No. of molecules | |
| Glycerol | 4 |
| PO4 | 4 |
| RMS deviationsd | |
| Bond lengths (Å) | 0.0053 |
| Bond angles (°) | 1.3627 |
| B factor (Å2) | 27.01 |
The values in parentheses are statistics from the highest resolution shell.
Rsym = Σ |Iobs − Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is average over symmetry equivalents.
Rwork = Σ ||Fo| − |Fc||/Σ |Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 5% of the data.
RMS, root mean square.
Protein structure accession number.
The atomic coordinate and structure factor have been deposited in the PDB under accession number 2wnw.
RESULTS AND DISCUSSION
Overall fold of SrfJ.
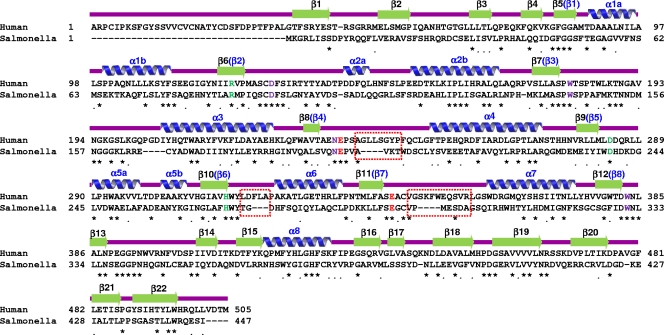
The 1.8-Å resolution crystal structure of SrfJ from S. Typhimurium was determined by molecular replacement using human GlcCerase (PDB code 2v3e) as a search model, which shares 30% amino acid sequence identity with SrfJ over 447 residues (Fig. 1). In the crystal two SrfJ molecules are present per asymmetric unit with contact areas of 1,292 Å2 corresponding to 7.5% of the total solvent-accessible surface area. However, a size exclusion chromatography analysis suggests that SrfJ exists as a monomer in solution (data not shown), which is consistent with the monomeric state of human GlcCerase (5).
FIG. 1.
Alignment of amino acid sequences of SrfJ (Salmonella) and human GlcCerase (Human). Secondary structure elements are shown based on the crystal structure of SrfJ and labeled in black above the sequences. Secondary structure elements for the (β/α)8-barrel catalytic domain are labeled in blue above the sequences. Identical and highly conserved residues are indicated with asterisks and dots, respectively. Residues used for enzyme catalysis are shown in red. Residues involved in the binding of the glucose ring moiety of the substrate and those involved in the hydrogen bond network with the catalytic residues are shown in purple and light green, respectively. Three loops (β8-α4, β10-α6, and β11-α7) showing considerable structural differences between the two proteins are indicated by red rectangles.
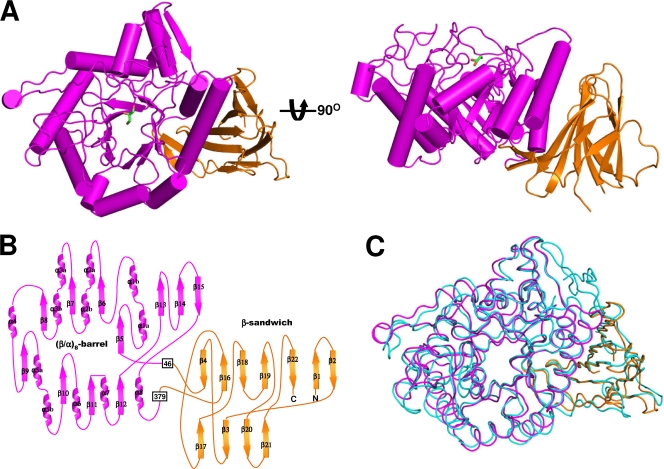
The SrfJ structure is composed of eight α-helices and 22 β-strands and folds into two distinct domains: a catalytic domain (residues 46 to 379) and a β-sandwich domain (residues 1 to 45 and 380 to 447). The catalytic domain consists of the common (β/α)8-barrel fold, a structural fold common to many GHs, and a three-stranded mixed β-sheet (Fig. 2A and B). The barrel is composed of eight parallel β-strands (β5 to β12) at the center and eight surrounding parallel α-helices (α1 to α8). The mixed β-sheet (β13 to β15) is inserted between the eighth strand (β12) and the eighth helix (α8) on the C-terminal side of the barrel (Fig. 2B). The connecting loops on the C-terminal side of the barrel are generally longer than those on the opposite side of the barrel, and they take part in the formation of the active-site pocket. The β-sandwich domain consists of two twisted β-sheets: an antiparallel seven-stranded β-sheet and a mixed four-stranded β-sheet (Fig. 2B). The domain organization of SrfJ is that generally observed in GHs and consists of a (β/α)8-barrel catalytic domain and one or more auxiliary modules. The overall structure of SrfJ can be well superposed on that of human GlcCerase, with a root mean square deviation of 1.32 Å over 428 Cα-carbon atoms (Fig. 2C). The β-sandwich domain of human GlcCerase was proposed to play an important regulatory function based on the pathogenic mutations (5). We speculate that the domain of SrfJ plays the same regulatory roles as in human GlcCerase.
FIG. 2.
Overall shape of SrfJ. (A) A cartoon of the overall fold of SrfJ. The figure at right is rotated 90° in a vertical direction relative to the left-hand figure. The (β/α)8-barrel and the β-sandwich domains are shown in magenta and orange, respectively. A glycerol molecule bound in SrfJ is shown in a stick model (green). (B) Two-dimensional topology of SrfJ. Secondary structure elements of SrfJ are labeled appropriately and shown with the same color scheme as in panel A. (C) Structural superposition of SrfJ with human GlcCerase (PDB code 2v3e). The (β/α)8-barrel and the β-sandwich domains of SrfJ are shown in the same color scheme as in panel A, and human GlcCerase is depicted in light cyan.
Compared with human GlcCerase, a 34-residue amino-terminal region is missing in SrfJ (Fig. 1 and 2C). In human GlcCerase, the region is known to be required for glycosylation and disulfide bond formation during protein targeting in human cells (5). Mutations of the region showed reduced enzymatic activity, which was speculated to be caused by structural instability (15). However, SrfJ does not contain the region because it does not need to undergo the mammalian protein targeting process but, rather, is produced in S. Typhimurium and secreted to the host.
Active site of SrfJ.
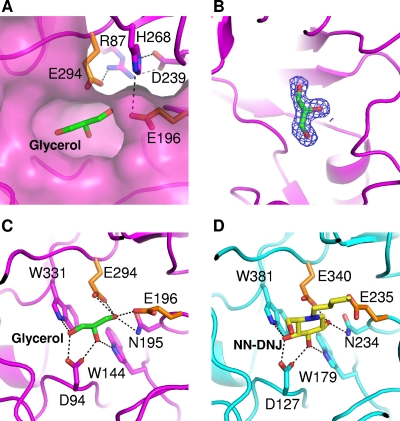
A pocket-shaped active site of SrfJ contains several conserved polar and aromatic residues from the connecting loops on the C-terminal side of the barrel. In SrfJ, residues Glu196 and Glu294 are located at the same positions as the catalytic residues Glu235 and Glu340 of human GlcCerase, in which these residues are used for the catalytic acid/base and the catalytic nucleophile, respectively (Fig. 3A). Moreover, the catalytic residues Glu196 and Glu294 of SrfJ form hydrogen bonds with Arg87, Asp239, and His268 as in human GlcCerase, where residues Glu235 and Glu340 form hydrogen bonds with Arg120, Asp282, and His311, contributing to the activation of the catalytic residues (Fig. 3A).
FIG. 3.
The active site and substrate binding of SrfJ. (A) The active site of SrfJ. A pocket-shaped active site is presented in a surface-fill model. Two catalytic residues are presented in a stick model (orange). Residues involved in the hydrogen bond network with the catalytic residues are shown in a stick model in magenta. (B) Experimental electron density map showing the bound glycerol molecule. The 2Fo − Fc electron density (blue mesh) is contoured at 2.0 σ. (C) The mode of glycerol binding to SrfJ. A bound glycerol molecule is shown in a stick model in green. Two catalytic residues are presented as in panel A. Residues involved in glycerol binding are shown in a stick model in magenta. (D) The binding mode of NN-DNJ to human GlcCerase. A bound NN-DNJ molecule is shown in a stick model in yellow. The residues for enzyme catalysis and those involved in NN-DNJ binding are shown in a stick model in orange and cyan, respectively.
In the present crystal structure of SrfJ, a glycerol molecule used for cryo-protection of the crystal was found to occupy the active site, which might mimic the substrate binding mode of the enzyme (Fig. 3B). The hydroxyl groups of the glycerol molecule are hydrogen bonded with the side chains of polar residues such as Asp94, Trp144, Asn195, Glu196, Glu294, and Trp331 (Fig. 3C). The binding mode of the glycerol molecule to SrfJ is extremely similar to that of N-nolyl-deoxynojirimycin (NN-DNJ) to human GlcCerase, in which hydroxyl groups of the glucose moiety are hydrogen bonded with side chains of polar residues such as Asp127, Trp179, Asn234, Glu235, Glu340, and Trp381 (1) (Fig. 3D). Notably, the positions not only of polar residues involved in the substrate recognition but also those of hydroxyl groups of glycerol and NN-DNJ are almost identical between these two proteins. These structural similarities observed in the active-site environment and the substrate binding mode lead us to suggest that SrfJ might act as a GH by recognizing a glucose-containing molecule as a substrate and that it has an enzymatic mechanism identical to that of human GlcCerase (1, 5).
Structural differences with human GlcCerase.
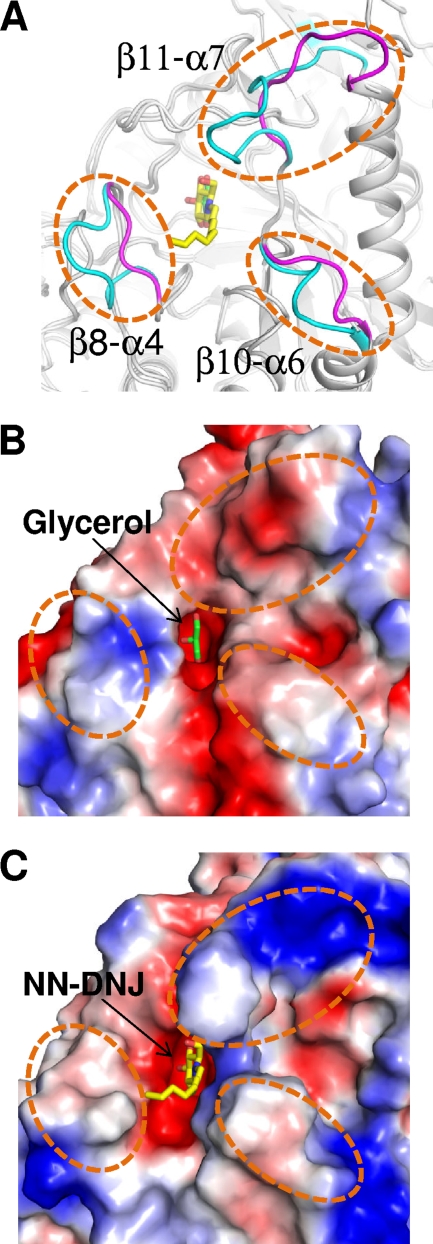
Despite the structural similarities between SrfJ and human GlcCerase in the overall fold and the active-site environment, considerable structural differences are observed in the connecting loops on the C-terminal side of the catalytic barrel. In particular, three loops (β8-α4, β10-α6, and β11-α7) that are known to contribute to the formation of the substrate entrance in human GlcCerase (1, 17) are observed to be shorter in SrfJ by deletion of several hydrophobic residues of the regions present in human GlcCerase (Fig. 1 and 4A). Because of the structural differences in the loop regions, the substrate entrance of SrfJ is more acidic than that of human GlcCerase, in which a relatively hydrophobic environment is observed (Fig. 4B and C). In human GlcCerase, the substrate entrance plays a crucial role in the stabilization of the substrate alkyl chain. In spite of the deletion in the loops, the substrate entrance is well constituted in SrfJ also (Fig. 4B). Moreover, residues Ala199 and Tyr270, whose corresponding residues Ala238 and Tyr313 in human GlcCerase are involved in stabilization of alkyl chain of NN-DNJ, are conserved in SrfJ. In fact, the alkyl chains of the GlcCer are known to be recognized with low specificity by human GlcCerase, unlike the specific binding recognition of the glucose ring to the enzyme (1, 5). Based on these observations, we speculate that, given the structural similarities in the active-site environment with human GlcCerase, SrfJ might recognize alkyl chain-containing glycosides as a substrate and act as a GlcCerase.
FIG. 4.
Structural differences between SrfJ and human GlcCerase. (A) SrfJ and human GlcCerase are superposed and presented in cartoon diagrams (gray). Three loops (β8-α4, β10-α6, and β11-α7) showing considerable structural differences between the two proteins are represented in magenta and cyan for SrfJ and human GlcCerase, respectively, and highlighted with dashed circles. (B and C) Surface models of SrfJ (B) and human GlcCerase (C) showing differences of electrostatic potentials at the substrate entrance regions. The figures are presented in the same direction and dimension as in panel A.
Recent studies revealed that ceramides play important roles in cellular infection of some pathogens including Pseudomonas aeruginosa (13), Staphylococcus aureus (7), and Neisseria gonorrhoeae (12), and the generation of ceramides results in the formation of a large ceramide-enriched membrane platform that mediates the internalization of pathogens (14). Considering the structural similarities between SrfJ and human GlcCerase and the involvement of ceramides in cellular infection, we speculate that SrfJ might act, using the GlcCerase activity, as an effector by modifying SCV membrane lipids to enhance bacterial virulence. Further investigations should be addressed to elucidating a detailed enzymatic mechanism of SrfJ and its role in Salmonella pathogenicity.
Summary.
We determined the crystal structure of S. Typhimurium SrfJ, a protein involved in Salmonella virulence (22). It is the first structure among the members of bacterial GH family 30. A 1.8-Å resolution crystal structure of SrfJ reveals that the protein comprises two distinctive domains, a (β/α)8-barrel catalytic domain and a β-sandwich domain. The structure of SrfJ is highly similar to that of human GlcCerase, whose mutations are known to cause Gaucher disease by lysosomal GlcCer accumulation (11). Despite considerable structural differences in the substrate entrance, high structural similarities are observed in the active-site environment and the substrate binding mode between SrfJ and human GlcCerase. Based on the observed structural similarities between these two proteins, we speculate that SrfJ, using its GlcCerase activity, might contribute to virulence during the infection of Salmonella.
Acknowledgments
This work was funded by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science and Technology, Republic of Korea, and was also supported by a grant from the Gyeongbuk Sea Grant Program, funded by the Ministry of Land, Transport and Maritime Affairs of the Korean government.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Brumshtein, B., H. M. Greenblatt, T. D. Butters, Y. Shaaltiel, D. Aviezer, I. Silman, A. H. Futerman, and J. L. Sussman. 2007. Crystal structures of complexes of N-butyl- and N-nonyl-deoxynojirimycin bound to acid beta-glucosidase: insights into the mechanism of chemical chaperone action in Gaucher disease. J. Biol. Chem. 282:29052-29058. [DOI] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 4.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 5.Dvir, H., M. Harel, A. A. McCarthy, L. Toker, I. Silman, A. H. Futerman, and J. L. Sussman. 2003. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 4:704-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 7.Esen, M., B. Schreiner, V. Jendrossek, F. Lang, K. Fassbender, H. Grassme, and E. Gulbins. 2001. Mechanisms of Staphylococcus aureus induced apoptosis of human endothelial cells. Apoptosis 6:431-439. [DOI] [PubMed] [Google Scholar]
- 8.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 9.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 10.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385-2396. [DOI] [PubMed] [Google Scholar]
- 11.Grabowski, G. A., S. Gatt, and M. Horowitz. 1990. Acid beta-glucosidase: enzymology and molecular biology of Gaucher disease. Crit. Rev. Biochem. Mol. Biol. 25:385-414. [DOI] [PubMed] [Google Scholar]
- 12.Grassme, H., E. Gulbins, B. Brenner, K. Ferlinz, K. Sandhoff, K. Harzer, F. Lang, and T. F. Meyer. 1997. Acidic sphingomyelinase mediates entry of N. gonorrhoeae into nonphagocytic cells. Cell 91:605-615. [DOI] [PubMed] [Google Scholar]
- 13.Grassme, H., V. Jendrossek, A. Riehle, G. von Kurthy, J. Berger, H. Schwarz, M. Weller, R. Kolesnick, and E. Gulbins. 2003. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat. Med. 9:322-330. [DOI] [PubMed] [Google Scholar]
- 14.Gulbins, E., and P. L. Li. 2006. Physiological and pathophysiological aspects of ceramide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290:R11-R26. [DOI] [PubMed] [Google Scholar]
- 15.Kacher, Y., B. Brumshtein, S. Boldin-Adamsky, L. Toker, A. Shainskaya, I. Silman, J. L. Sussman, and A. H. Futerman. 2008. Acid beta-glucosidase: insights from structural analysis and relevance to Gaucher disease therapy. Biol. Chem. 389:1361-1369. [DOI] [PubMed] [Google Scholar]
- 16.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 17.Lieberman, R. L., B. A. Wustman, P. Huertas, A. C. Powe, Jr., C. W. Pine, R. Khanna, M. G. Schlossmacher, D. Ringe, and G. A. Petsko. 2007. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat. Chem. Biol. 3:101-107. [DOI] [PubMed] [Google Scholar]
- 18.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145-156. [DOI] [PubMed] [Google Scholar]
- 19.Meikle, P. J., J. J. Hopwood, A. E. Clague, and W. F. Carey. 1999. Prevalence of lysosomal storage disorders. JAMA 281:249-254. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 23.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 24.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 25.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]