Abstract
Chromosomal DNA replication is dependent on processive DNA synthesis. Across the three domains of life and in certain viruses, a toroidal sliding clamp confers processivity to replicative DNA polymerases by encircling the DNA and engaging the polymerase in protein/protein interactions. Sliding clamps are ring-shaped; therefore, they have cognate clamp loaders that open and load them onto DNA. Here we use biochemical and mutational analyses to study the structure/function of the Methanosarcina acetivorans clamp loader or replication factor C (RFC) homolog. M. acetivorans RFC (RFCMa), which represents an intermediate between the common archaeal RFC and the eukaryotic RFC, comprises two different small subunits (RFCS1 and RFCS2) and a large subunit (RFCL). Size exclusion chromatography suggested that RFCS1 exists in oligomeric states depending on protein concentration, while RFCS2 exists as a monomer. Protein complexes of RFCS1/RFCS2 formed in solution; however, they failed to stimulate DNA synthesis by a cognate DNA polymerase in the presence of its clamp. Determination of the subunit composition and previous mutational analysis allowed the prediction of the spatial distribution of subunits in this new member of the clamp loader family. Three RFCS1 subunits are flanked by an RFCS2 and an RFCL. The spatial distribution is, therefore, reminiscent of the minimal Escherichia coli clamp loader that exists in space as three γ-subunits (motor) flanked by the δ′ (stator) and the δ (wrench) subunits. Mutational analysis, however, suggested that the similarity between the two clamp loaders does not translate into the complete conservation of the functions of individual subunits within the RFCMa complex.
Processive DNA synthesis is required to gain the speed necessary for the replication of large genomes as found in organisms across the three domains of life. Therefore, in each domain of life, molecular machines that ensure rapid DNA synthesis have evolved (17). At the center of the DNA synthesis machinery are the so-called replicative DNA polymerases and their processivity factors. Cells in all three domains of life, in addition to certain viruses, have evolved ring-shaped proteins, collectively called the processivity or sliding clamps, that repress the frequent dissociation of the DNA polymerase from the DNA template (17). The bacterial sliding clamp is known as the β-subunit, the eukaryotic functional homolog is called proliferating cell nuclear antigen (PCNA), and due to its similarity to the eukaryotic protein, the archaeal sliding clamp is also termed PCNA (4, 6, 12, 15). Among the viruses, the bacteriophage T4 clamp or gp45 has been well characterized (1). In each case, the clamp contains a cavity that is wide enough to allow it to encircle and slide freely along DNA (16, 18). However, since the clamp is a ring-shaped protein, it requires another protein to open and load it onto DNA. Thus, each clamp comes with its loader, also collectively known as the clamp loaders (2, 5, 29). Whereas the bacterial clamp loader is termed the γ-complex, the eukaryotic functional homolog is known as replication factor C or RFC (3, 19).
Cullmann and coworkers demonstrated that the heteropentameric RFC complex in eukaryotes is encoded by five different genes: four different genes coding for the RFC small subunit (RFCS) proteins and the last gene coding for the RFC large subunit (RFCL) proteins (9). In a subsequent analysis of clamp loaders in archaea, it was demonstrated that Methanothermobacter thermautotrophicus (20), Sulfolobus solfataricus (29), and Pyrococcus furiosus (5) each contain only one RFCS gene and one RFCL gene. The polypeptide sequences of the RFCS of the archaea exhibit very high homology to those of the four eukaryotic RFCS (6), suggesting the relatedness of the archaeal/eukaryotic RFC proteins. Interestingly, four protomers of the archaeal RFCS protein form a complex with one large subunit protein to act as the functional clamp loader (5, 29). Thus, similar to the eukaryotic RFC, the archaea appeared to have a pentameric RFC complex that harnesses energy, derived from ATP hydrolysis, to break open the ring-shaped clamp. The loader then loads the opened clamp onto a primer-template junction, thereby facilitating the interaction between the clamp and its cognate replicative DNA polymerase to ensure processive DNA synthesis (10, 13, 14).
We recently described a new form of clamp loader in the domain Archaea (7). Unlike all previously described clamp loaders from this domain, the Methanosarcina acetivorans RFC (RFCMa) homolog has three different subunits instead of the usual two subunits, i.e., one small subunit and one large subunit (7). This new form of archaeal clamp loader may represent an intermediate stage in the evolution of the more complex RFC in eukaryotes from the less complex ones reported for archaea (7). In this report, we determined the subunit organization and propose the spatial distribution of subunits in the new form of the RFC complex. The predicted spatial distribution of the RFCMa is similar to the Escherichia coli minimal γ-complex, which is arranged as the “stator,” “motor,” and “wrench.” Mutations in the predicted motor impaired the clamp loader's capacity to stimulate clamp-dependent DNA synthesis by a cognate DNA polymerase in M. acetivorans (PolBIMa). C-terminal truncations that removed putative clamp interacting motifs in the subunit occupying the wrench position failed to abolish the capacity of the loader to stimulate clamp-dependent DNA synthesis by the cognate DNA polymerase. Our results, therefore, suggested that although the M. acetivorans and the E. coli clamp loaders likely adopt similar subunit organization/spatial distribution, the functions of the individual subunits are unlikely to be fully conserved.
MATERIALS AND METHODS
Cloning, expression, and purification of the M. acetivorans clamp loader complex.
The cloning, expression, and purification of RFCS1Ma, RFCS2Ma, RFCLMa, and RFCMa complex were described in our previous report (7).
Truncational analysis of the RFCMa complex.
In a previous report (7), we showed that the M. acetivorans clamp loader complex (RFCMa) is composed of two small subunits (RFCS1Ma and RFCS2Ma) and a large subunit (RFCLMa). The plasmid constructs for the expression of the individual subunits were designated pET28/rfcl, pET28/rfcs1, and pET28/rfcs2 for N-terminal His6-tagged RFCL, N-terminal His6-tagged RFCS1, and N-terminal His6-tagged RFCS2, respectively. To create an expression system for the RFC complex, RFCS1 and RFCS2 were placed in frame in the pACYCDuet vector (Novagen) to express the non-His6-tagged and His6-tagged proteins encoded by rfcs1 and rfcs2, respectively. This expression vector was named pACYCDuet/rfcs1/rfcs2. The pET28a vector was previously modified through the substitution of the kanamycin resistance gene with that for ampicillin resistance (4). For the coexpression of the pACYCDuet/rfcs1/rfcs2 construct, which contained a chloramphenicol resistance gene, and pET28a/rfcl, the recombinant E. coli culture was supplemented with ampicillin and chloramphenicol at 100 μg/ml and 50 μg/ml, respectively. RFCL and RFCS2 in the RFCMa complex, therefore, contained N-terminal His6 tags.
To create C-terminal truncation derivatives of RFCL, a PCR method was used. All primers are shown in Table 1. To make five truncations of decreasing length from the C-terminal region of RFCLMa, PCR amplifications with a forward primer MacRFCLF in combination with five different reverse primers—RFCLΔ1(1-550)R, RFCLΔ2(1-475)R, RFCLΔ3(1-432)R, RFCLΔ4(1-410)R, and RFCLΔ5(1-385)R—were carried out. The template was the gene coding for wild-type RFCLMa, and the RFC complexes derived from the truncated proteins were designated RFC-LΔ1, RFC-LΔ2, RFC-LΔ3, RFC-LΔ4, and RFC-LΔ5, respectively. Each truncation targeted a region predicted to form a loop in the protein structure (Predictprotein [http://www.ebi.ac.uk/∼rost/predictprotein/predictprotein.html]). Each PCR product was initially cloned into a pGEM-T Easy vector (Promega) and sequenced to confirm the integrity of the sequence. The RFCL truncated genes were then individually cloned into pET28a as described for the wild-type RFCL gene (7).
TABLE 1.
Oligonucleotides used in this studya
| Experiment | Oligonucleotide | Nucleotide sequence |
|---|---|---|
| Truncations | MacRFCS2F | AAAACATATGGAATCAATGCAGGATCTCTGGACTCT |
| MacRFCS2R | TTTTTCTCGAGTCAGGAGAATGTGGAAATCAATTTTTCG | |
| RFCS2Δ(85-110)F | GACTTTTTCGACCAGGGAATAGATATTTTTAAGGAAGTCGTT | |
| RFCS2Δ(85-110)R | AACGACTTCCTTAAAAATATCTATTCCCTGGTCGAAAAAGTC | |
| MacRFCLF | AAAAACATATGATGTCGGCAATCGAATGGGCTGAAAAAT | |
| RFCLΔ1(1-550)R | CTCGAGTTAGGTTTTCTGCTCTACAGATTCGGA | |
| RFCLΔ2(1-475)R | CTCGAGTTATTTGCCGGCAGAACCTTCCTTTTG | |
| RFCLΔ3(1-432)R | CTCGAGTTAATTCTTACCCTCTTCAAGAAGTTT | |
| RFCLΔ4(1-410)R | CTCGAGCTATGCACTTCCTGTAAGGTACATC | |
| RFCLΔ5(1-385)R | CTCGAGCTAGTCCTTCAGCATGCGGGAATAAAG | |
| Mutations | RFCS1-K18A | GAGATCTGGATTGAAGCATACAGGCCTGTCAGG |
| RFCS1-E82A | AGGGAAAACTTTACCGCACTTAATGCTTCCGAT | |
| RFCS1-E119A | ATCATTTTTCTTGATGCAGCCGATGCTCTAACA | |
| RFCS1-E136A | CTCCGCAGGACCATGGCACGGTTCAGCAGCAAC | |
| RFCS1-D205A | TACGTAGCTCAGGGAGCCATGCGAAAAGCTGTC | |
| Primer extension | (M13 6205-6234) | ATTCGTAATCATGGTCATAGCTGTTTCCTG |
The experiments in which the oligonucleotides were used are indicated. The truncation oligonucleotides were used to truncate RFCS2Ma and RFCLMa. The mutation oligonucleotides were used to create site-directed mutations in the RFC boxes of the RFCS1Ma gene. The primer extension oligonucleotide was used in DNA synthesis with M13 ssDNA as the template. Restriction sites incorporated into the oligonucleotides are underlined.
Deletion of a 26-amino-acid insertion sequence in RFC2Ma.
A 26-amino-acid insertion sequence in RFCS2Ma was deleted by a PCR overlap method described in our previous report (30). The PCR primers that were used to delete the insertion sequence, as well as fuse the flanking sequences, were MacRFCS2F/RFCS2Δ(85-110)R and RFCS2Δ(85-110)F/MacRFCS2R (Table 1).
Point mutations in five RFC boxes in RFCS1Ma.
The oligonucleotides that were used to create point mutations in RFC boxes II, IV, V, VI, and VIII of RFCS1 are listed in Table 1 as RFCS1-K18A, RFCS1-E82A, RFCS1-E119A, RFCS1-E136A, and RFCS1-D205A, respectively. The method used was as described by the manufacturer for the QuikChange site-directed mutagenesis kit (Stratagene). All DNA inserts were sequenced (W. M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign) to confirm the presence of the desired mutations.
Estimation of subunit organization by size exclusion chromatography.
To estimate the subunit organization of individual RFCSMa subunits and also their mixtures, the purified proteins were dialyzed against buffer A containing 50 mM Tris-HCl (pH 8.0) and 300 mM NaCl. Different concentrations of each protein in a 100-μl total volume were then loaded onto a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) already equilibrated with the same buffer, and the chromatography was developed at 4°C. For the mixtures of RFCS1Ma and RFCS2Ma, various amounts of the individual proteins were mixed and dialyzed against buffer A for 1 h before the gel filtration analysis. The chromatography was developed with the same buffer at a flow rate of 0.5 ml/min, and fractions of 0.5 ml were collected and aliquots were analyzed on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gel filtration column was calibrated by running a set of protein standards (thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; catalase, 58 kDa; ovalbumin, 43 kDa; and RNase A, 13.7 kDa) under the same conditions.
Stoichiometry of RFCMa complex and spatial arrangement.
The stoichiometric ratio of RFCS1, RFCS2, and RFCL in the RFCMa complex was determined through a densitometric method as in our previous report on Pyrococcus furiosus RFC (RFCPf) complex (5). The RFCMa complex was resolved through SDS-PAGE, and after being stained with Coomassie brilliant blue followed by destaining, the gels were scanned with a densitometer. Each band was then quantified from a calibration curve obtained by using each purified subunit at different concentrations to obtain a relationship between protein band intensity and the amount of protein loaded for the electrophoretic analysis.
CD scan.
The wild-type RFCMa complex and its mutants were subjected to circular dichroism (CD) scans to determine if individual mutations impacted the secondary structural elements of the protein complex. The method used was the same as described in our previous report (23). However, in this case, we used 0.5 μg/μl of each protein in a buffer composed of 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 5 mM MgCl2, 10% glycerol, and 1 mM dithiothreitol. Triplicate data sets were collected per protein under investigation, and all data were normalized against readings obtained from buffer without protein.
Cloning, expression, and purification of PCNAMa and PolBIMa.
The methods for the production of M. acetivorans PCNA or the sliding clamp and the M. acetivorans DNA PolBIMa were as described in our previous report (7).
Primer extension analysis.
To prepare the substrate per primer extension reaction, 500 ng of bacteriophage M13mp18 single-stranded DNA (ssDNA) template and 1 pmol 5′-32P-end-labeled primer (Table 1), complementary to positions 6205 to 6234 of the template, were annealed by heating to 95°C for 5 min and cooling slowly to room temperature in a buffer composed of 20 mM Tris-HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2, and 2 mM β-mercaptoethanol. Where multiple reactions were carried out, the annealing reaction mixture was scaled up by the number of reactions required. Thus, each primer extension reaction (total volume of 20 μl) contained 1 pmol of the end-labeled primer annealed to 500 ng of template, 250 μM of each deoxynucleoside triphosphate, and 20 mM ATP in the buffer used for the annealing reaction. To initiate primer extension, the cognate DNA polymerase, PolBIMa, was added to the reaction mixture at 0.24 μM. Where the effects of accessory factors were investigated, they were added at 77.5 nM of RFCMa complex or its mutants, and 0.3 μM of PCNAMa (based on the PCNA trimer) per reaction. In the case of the RFCSMa, the amounts of RFCS added ranged from 1.5 to 15 μM/reaction. The primer extension reaction was carried out at 37°C for 5 min and terminated with 6 μl of stop solution (98% formamide, 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). After being heated to 95°C for 5 min, the samples were resolved on a 1% alkali agarose gel and visualized by autoradiography.
Amino acid sequence alignments.
The amino acid sequence alignments were carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the shading was manually carried out.
RESULTS
Oligomerization state of M. acetivorans clamp loader small subunits.
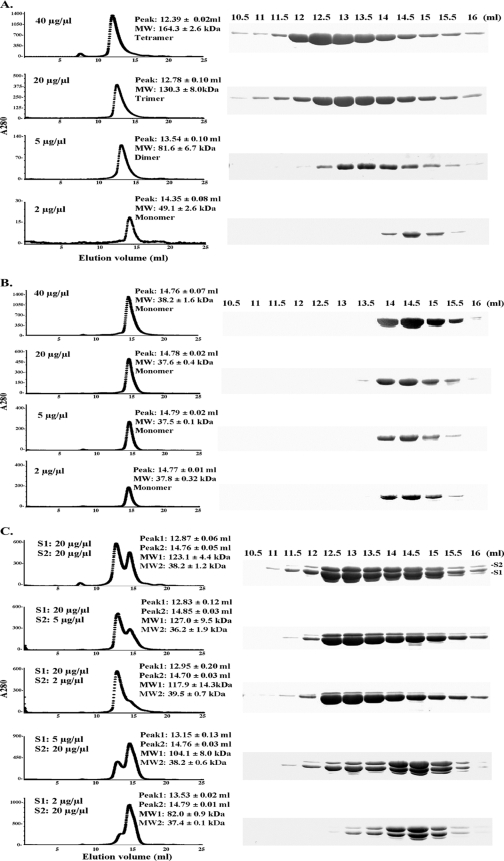
We successfully expressed both RFCS1Ma and RFCS2Ma as N-terminal His6-tagged proteins. After purification, however, the His6 tag was removed from each protein by protease (thrombin) digestion. In a previous analysis of an archaeal RFCS (Pyrococcus furiosus homolog), we reported that the protein oligomerized in solution (5). Subsequent analysis by electron microscopy (25) and X-ray crystallography (32) of the archaeal RFCS detected proteins existing as mostly homohexamers, although at different pH values, RFCSPf also existed as homopentamers and homotetramers. These oligomerization states were not observed for either of the two RFCSMa, although RFCS1Ma was estimated to exist as a dimer (7). Thus, in the present experiment, we sought to examine the oligomerization state of the two RFCSMa proteins as their concentrations were varied. RFCS1Ma and RFCS2Ma have estimated molecular masses of 37.9 and 38.3 kDa, respectively. The elution volumes in our gel filtration analysis, however, suggested that the oligomerization state of RFCS1Ma was concentration dependent. As protein concentration increased, we observed a change from monomers to homodimers, homotrimers, and finally homotetramers at 40 μg/μl (Fig. 1A). On the other hand, increasing concentrations of RFCS2Ma to 40 μg/μl did not lead to oligomerization of this protein, which was estimated to exist as a monomer in solution (Fig. 1B). It is of interest that we observed differential staining of RFCS1Ma and RFCS2Ma in our SDS-PAGE analysis (Fig. 1A and B). It is our assumption that this is due to differences in the amino acid contents, especially arginine, and to a lesser extent amino acids such as histidine, lysine, and the aromatic residues, such as tryptophan, tyrosine, and phenylalanine (8, 11). In the case of RFCLMa, it consistently eluted in the void volume, as reported earlier (7), suggesting a protein of a very large molecular mass (results not shown).
FIG. 1.
Gel filtration analysis of RFCMa subunits and mixtures of RFCS1Ma and RFCS2Ma. The native molecular masses of RFCMa subunits were estimated by gel filtration on a Superdex 200 HR 10/30 column fitted to a high-performance liquid chromatography apparatus. Indicated amounts of highly purified individual RFCSMa proteins and their mixtures were dialyzed and then injected into the column to estimate native molecular masses. The results are presented as RFCS1Ma (A), RFCS2Ma (B), and a mixture of RFCS1Ma and RFCS2Ma (C). The elution volumes are indicated, and fractions were analyzed by SDS-PAGE to visualize the proteins. RFCS1Ma tends to degrade with time. The band that appears below RFCS1Ma is known to be RFCS1Ma processed at the N terminus (7). MW, molecular weight.
In the absence of RFCS1, RFCS2 eluted at a peak elution volume around 14.8 ml (Fig. 1B). However, when the two purified proteins were dialyzed together and analyzed by size exclusion chromatography, RFCS2Ma eluted earlier with RFCS1Ma, suggesting an interaction between the two proteins (Fig. 1C). RFCS1Ma alone at 2 μg/μl was estimated as a monomer (Fig. 1A), and at a concentration of 20 μg/μl, RFCS2Ma also eluted as a monomer (Fig. 1B). However, when we mixed RFCS1Ma and RFCS2Ma at 2 μg/μl and 20 μg/μl, respectively, we observed two peaks representing dimers (82.0 ± 0.9 kDa) and monomers (37.4 ± 0.1 kDa). This suggested that the two proteins interacted and that the dimers were likely heterodimers of the two proteins. RFCS1Ma at 5 μg/μl eluted as a dimer (Fig. 1A). However, when mixed with RFCS2Ma at 20 μg/μl, there were two peaks that were estimated as 104.1 ± 8.0 kDa and 38.2 ± 0.6 kDa. This may suggest a heterotrimer of RFCS1Ma/RFCS2Ma and monomers of the individual proteins, respectively. By mixing RFCS1Ma and RFCS2Ma at concentrations of 20/2, 20/5, and 20/20 μg/μl, respectively, we also observed two peaks. The elution volume of the first peak suggested either a trimer or tetramers of the RFCS proteins (117 to 127 kDa), and the second peak suggested monomers (36.2 to 38.2 kDa).
Subunit organization and spatial distribution of the RFCMa complex.
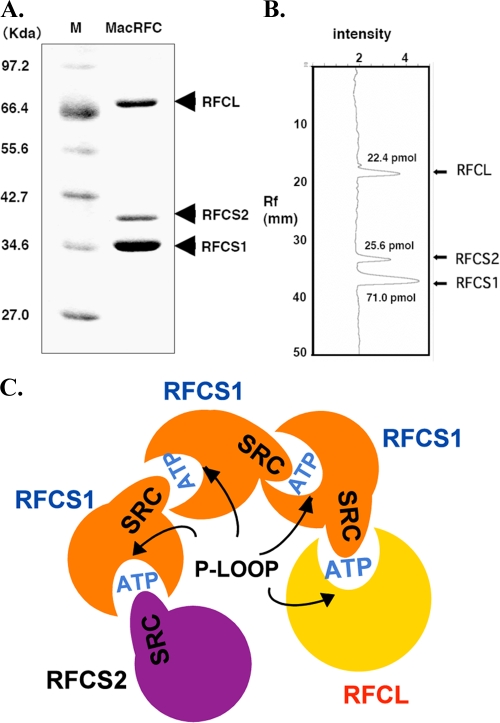
The stoichiometric ratio of RFCS1, RFCS2, and RFCL in the functional RFC complex was determined through a densitometric method as described in our previous report for the Pyrococcus furiosus RFC complex (5). The concentrations of the RFC subunits shown in Fig. 2A were calculated to be 71.0 pmol, 25.6 pmol, and 22.4 pmol for RFCS1, RFCS2, and RFCL, respectively (Fig. 2B). The results, therefore, suggested that RFCS1Ma, RFCS2Ma, and RFCLMa form a stable complex with a stoichiometric ratio of 3:1:1. Note that the differential staining of RFCS1Ma and RFCS2Ma should not impact our estimated stoichiometry, since the individual proteins were used to generate their individual prediction equation that related intensity to the amount of protein loaded on the gel. Based on the stoichiometric information and our previous data on mutagenesis in the individual subunits (7) within the RFC complex, we predicted the spatial distribution in Fig. 2C, and this is further discussed later.
FIG. 2.
Determination of the stoichiometry of the RFCMa complex. (A) RFCMa (MacRFC) complex resolved through SDS-PAGE. (B) Densitometer scan of the gel generates a relationship between protein band intensity and the amount of protein loaded for the electrophoretic analysis. Rf, relative to front value. (C) Schematic representation of the predicted spatial distribution of the RFCMa complex, which is similar to that of the E. coli clamp loader, based on stoichiometric ratio and mutational analysis at individual ATPase sites in the RFC complex (17). RFC proteins are members of AAA+ ATPases, and the ATPase sites are at the interface between two adjacent subunits. One subunit provides a Walker A motif and the other an arginine finger from its SRC motif to form a competent ATPase site. Note that in the model, an ATPase site is missing at the interface of the large subunit and RFCS2, since RFCS2 has a Walker A motif but RFCL lacks an SRC motif.
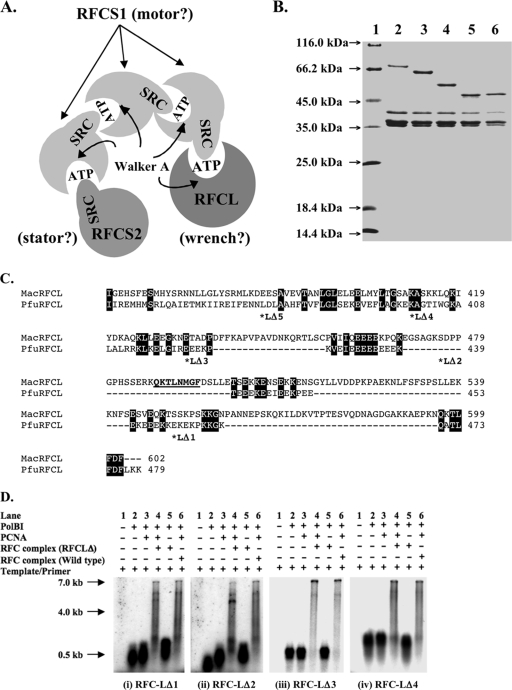
Truncational analysis of the large subunit of RFCMa complex.
The spatial arrangement of the subunits in the RFCMa complex suggested above is similar to that of the E. coli minimal clamp loader, which also has a 3:1:1 ratio and is made up of three γ subunits (motor) flanked by the stator (δ′) and the wrench (δ) (10). The wrench interacts with the E. coli sliding clamp and on its own is able to open the clamp through this interaction (17). In Fig. 3A, the M. acetivorans RFCL, which occupies the position of the wrench in the E. coli clamp loader, has two putative PCNA interacting protein (PIP) boxes at the C-terminal region: PIP box 1 (QKTLFDF; amino acids 596 to 602) and PIP box 2 (QKTLNMGF; amino acids 488 to 495) in Fig. 3C. The PIP boxes are known to mediate interactions with sliding clamps or PCNA (34). We, therefore, carried out a series of truncations from the C terminus of RFCLMa to determine whether the removal of elements in the C-terminal region, especially the PIP boxes, will affect clamp stimulation of DNA synthesis in the presence of the RFCMa complex. As shown in Fig. 3B, we successfully produced four of the five RFC complex proteins that contained truncated derivatives of RFCLMa. The last RFCL-truncated derivative, RFC-LΔ5 (which has the last 222 amino acids removed), could not be obtained (results not shown). An examination of the E. coli cells expressing this RFC complex showed that the truncated RFCL failed to express. The first and second truncation mutants, RFC-LΔ1 and RFC-LΔ2, removed PIP box 1 and both PIP box 1/PIP box 2, respectively (7). However, as shown in Fig. 3Di and ii, lanes 4, both mutant clamp loader complexes stimulated PCNAMa-dependent DNA synthesis by PolBIMa, suggesting that both RFC-LΔ1 and RFC-LΔ2 are capable of loading the sliding clamp. The results were, therefore, similar to those of the primer extension in the presence of the wild-type RFC complex (Fig. 3D, lanes 6). The subsequent deletions that removed 175 amino acids (RFC-LΔ3; Fig. 3C) and 197 amino acids (RFC-LΔ4; Fig. 3C) from the C terminus, respectively, also did not lead to the failure to load PCNAMa (Fig. 3Diii and iv, lanes 4).
FIG. 3.
Hypothetical functions of the M. acetivorans clamp loader subunits. (A) The proposed model of the RFCMa subunit arrangement is similar to that of the E. coli minimal clamp loader, the γ-complex. The RFCS2Ma, RFCS1Ma, and RFCLMa subunits occupy the positions of the stator, the motor, and the wrench, respectively, in the E. coli protein complex. (B) SDS-PAGE of the purified recombinant wild-type RFCMa complex and RFCMa complex proteins containing truncated RFCLMa. Lane 1, protein molecular mass markers (Fermentas); lane 2, wild-type RFCMa complex; lane 3, M. acetivorans RFC-LΔ1 complex; lane 4, M. acetivorans RFC-LΔ2 complex; lane 5, M. acetivorans RFC-LΔ3 complex; lane 6, M. acetivorans RFC-LΔ4 complex. (C) Alignment of the C-terminal region of RFCLPf (PfuRFCL) and RFCLMa (MacRFCL) showing the single and double PIP boxes in RFCLPf and RFCLMa, respectively. In addition, the points of the truncations in RFCLMa are denoted by asterisks and the different truncations shown as LΔX, where X represents 1, 2, 3, 4, or 5. The basic amino acid sequence in RFCLPf (KEKEKPKKGK) and the mostly acidic amino acid sequence (EEEKEEIEEKPEEEKEEE) are shown. (D) Effects of RFCMa complex proteins containing truncated RFCLMa on the primer extension capacity of PolBIMa in the presence of PCNAMa. Primer extensions were compared as follows: lanes 1, template alone or negative control; lanes 2, template/PolBIMa; lanes 3, template/PolBIMa/PCNAMa; lanes 4, template/PolBIMa/PCNAMa/M. acetivorans RFC-LΔ mutants; lanes 5, template/PolBIMa/M. acetivorans RFC-LΔ (i.e., lanes 4 without PCNAMa); lanes 6, template/PolBIMa/PCNAMa/RFCMa wild type or positive control. The substrate for DNA synthesis was 0.5 μg of singly primed M13mp18 ssDNA in DNA polymerase reaction buffer (described in Materials and Methods). The reaction mixtures were incubated at 37°C for 5 min, and the products were resolved by 1% alkali agarose gel electrophoresis and visualized by autoradiography. Note that the RFCMa complex in the absence of PCNA has no effect on DNA synthesis by PolBIMa (7).
Mutations in some conserved RFC boxes of RFCS1Ma impair RFCMa-dependent PCNAMa stimulation of DNA synthesis by PolBIMa.
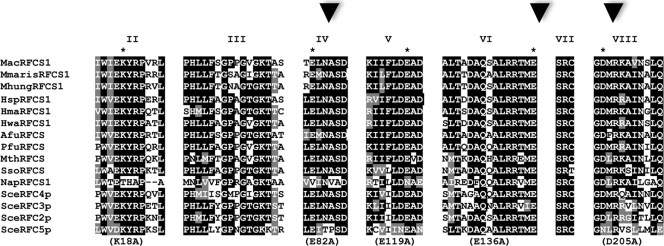
Eight conserved motifs, referred to as RFC box I to box VIII, were previously identified in RFC proteins (9). The conservation of seven of these RFC boxes (box II to box VIII) was later confirmed in archaeal RFCS proteins (6). In the present work, we hypothesized that other than RFC box III (Walker A motif or P-loop) and box VII (SRC motif), which have been shown to be critical for ATPase activity and, therefore, the clamp loading function of RFC proteins including the RFCMa complex (7), the other RFC boxes, especially those in the putative motor domain, are likely to play important roles in the function of this new clamp loader. To test our hypothesis, we made a single mutation at a conserved residue in each of RFC box II [RFCS1(K18A)Ma], box IV [RFCS1(E82A)Ma], box V [RFCS1(E119A)Ma], box VI (D136A), and box VIII (D205A) (Fig. 4). Each mutant RFCS1 was used in reconstituting an RFCMa complex that harbored that single mutation. To ensure that each mutation did not lead to gross changes in the secondary structural elements, each RFC complex containing the respective mutation was highly purified (results not shown) and subjected to CD scan. As shown in Fig. 5A, except for the RFC complex protein containing RFCS1Ma-E119A (mutation in box V), the other mutant RFC complex proteins exhibited CD spectra that were similar to that of the wild-type RFCMa complex. It should be noted, however, that the clamp loader complex containing E82A (RFC box IV) also showed some deviation from the wild type. The results, thus, suggested that at least three of the mutations (K18A, E136A, and D205A) did not severely impact the secondary structural elements of the RFCMa complex. Using primer extension analysis, we observed that three of the mutations made in RFCS1Ma (putative motor) impaired the capacity of the mutant RFC complex to stimulate PCNAMa-dependent DNA synthesis by PolBIMa. These were the mutations in RFC box IV (E82A; Fig. 5Bii, lane 4), box VI (E136A; Fig. 5Biv, lane 4), and box VIII (D205; Fig. 5Bv, lane 4). While the mutation in RFC box II (K18A) did not appear to impair the capacity of the RFCMa complex to stimulate PCNAMa-dependent DNA synthesis by PolBIMa (Fig. 5Bi, lane 4), we observed a decreased aptitude in the case of the clamp loader complex containing the E119A mutation (Fig. 5Biii, lane 4). Each protein was compared with the wild-type RFCMa complex, serving as a positive control (Fig. 5B, lanes 6).
FIG. 4.
An alignment showing the conserved RFC boxes in RFCS in both archaea and eukaryote Saccharomyces cerevisiae. The GenBank accession numbers of the proteins are as follows: Methanosarcina acetivorans C2A RFCS1 (MacRFCS1), NP_615630; Methanoculleus marisnigri JR1 RFCS1 (MmarisRFCS1), YP_001047161; Methanospirillum hungatei JF-1 RFCS1 (MhungRFCS1), YP_502463; Halobacterium sp. strain NRC-1 RFCS1 (HspRFCS1), NP_280914; Haloarcula marismortui RFCS1 (HmaRFCS1), YP_137064; Haloquadratum walsbyi DSM RFCS1 (HwaRFCS1), YP_659332; Archaeoglobus fulgidus RFCS (AfuRFCS), NP_070884; Pyrococcus furiosus RFCS (PfuRFCS), NP_577822; Methanothermobacter thermautotrophicus RFCS (MthRFCS), NP_275384; Sulfolobus solfataricus RFCS (SsoRFCS), NP_342275; Natronomonas pharaonis RFCS1 (NapRFCS1), YP_326110; Saccharomyces cerevisiae RFCS4 (SceRFC4p), EDV10522; S. cerevisiae RFCS3 (SceRFC3p), NP_014109; S. cerevisiae RFCS2 (SceRFC2p), EDV12808; and S. cerevisiae RFCS5 (SceRFC5p), NP_009644. Organisms with subunits designated -S1 are archaeal organisms with two RFCS (refer to the legend to Fig. 6). Conserved and similar amino acid residues are shaded black and gray, respectively. The asterisks denote the residue mutated to alanine in a given RFC box. Mutations in the RFC boxes with arrowheads impaired RFC-dependent PCNAMa stimulation of DNA synthesis by PolBIMa.
FIG. 5.
Mutational analysis within different conserved RFC boxes in the motor domain of the RFCMa complex. (A) The highly purified RFCMa complex and its RFC box mutant derivatives were subjected to CD scans. The CD spectra of each protein from 200 nm to 260 nm were collected in triplicate and normalized against readings from buffer without protein. The samples analyzed were the wild-type RFCMa complex (MacRFC-WT); RFCS1(K18A)Ma complex [MacRFC-S1(K18A)], mutation in RFC box II; RFCS1(E82A)Ma complex [MacRFC-S1(E82A)], mutation in RFC box IV; RFCS1(E119A)Ma complex [MacRFC-S1(E119A)], mutation in RFC box V; RFCS1(E136A)Ma complex [MacRFC-S1(E136A)], mutation in RFC box VI; and RFCS1(D205A)Ma complex [MacRFC-S1(D205A)], mutation in RFC box VIII. (B) Effect of individual mutations in the RFC boxes on the capacity of the RFCMa complex to stimulate primer extension of PolBIMa in the presence of PCNAMa. Lane 1, template alone; lane 2, template/PolBIMa; lane 3, template/PolBIMa/PCNAMa; lane 4, template/PolBIMa/PCNAMa/RFCMa mutant complex; lane 5, lane 4 without PCNAMa; lane 6, template/PolBIMa/PCNAMa/RFCMa wild type (positive control). The primer extension reaction mixtures were incubated at 37°C for 5 min, and the products were analyzed by 1% alkali agarose gel electrophoresis, followed by visualization through autoradiography. The results in panels i, ii, iii, iv, and v represent the RFC complex containing the RFCS1Ma box II, IV, V, VI, and VIII mutants, respectively.
The insertion sequence downstream of RFC box IV in RFCS2Ma is not required for clamp loading.
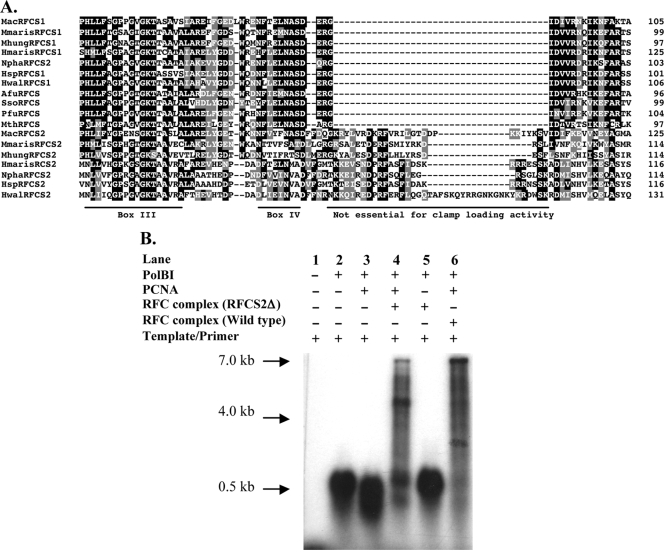
In archaeal RFCS2 polypeptides deposited in GenBank (http://www.ncbi.nlm.nih.gov/), an insertion sequence occurs downstream of RFC box IV (9) as shown in Fig. 6A. In the order Methanosarcinales, the insertion sequence comprises 26 amino acids that are highly conserved (7). To determine if the insertion sequence plays a role in PCNA loading, a PCR approach was used to delete the insertion sequence from RFCS2Ma, and an RFC complex containing the mutant RFCS2Ma, instead of the wild-type protein, was highly purified (results not shown). The RFCMa complex containing the mutant RFCS2 (RFCS2Δ; Fig. 6B) was tested for its capacity to enhance primer extension by PolBIMa in the presence of the cognate clamp or PCNAMa. As shown in Fig. 6B, lane 4, deletion of the insertion sequence in RFCS2Ma, although it appears to have slightly impaired the capacity of the protein complex to stimulate PCNAMa-dependent DNA synthesis by PolBIMa, did not abolish the clamp loading activity. We look forward to characterizing the mutant in detail. It should also be noted that the deleted sequence may play a role in a different function, for example, in interactions with other proteins.
FIG. 6.
Effect of an insertion sequence in RFCS2Ma on PCNAMa-dependent DNA synthesis by PolBIMa. (A) Alignment depicting insertion sequences downstream of RFC box IV regions in RFCS2Ma and its homologs. The GenBank accession numbers of the proteins are as follows: M. acetivorans RFCS1 (MacRFCS1), NP_615630; M. marisnigri RFCS1 (MmarisRFCS1), YP_001047161; M. hungatei JF-1 RFCS1 (MhungRFCS1), YP_502463; H. marismortui RFCS1 (HmarisRFCS1), YP_137064; N. pharaonis RFCS1 (NphaRFCS1), YP_326110; Halobacterium sp. strain NRC-1 RFCS1 (HspRFCS1), NP_280914; Haloquadratum walsbyi DSM RFCS1 (HwalRFCS1), intein-containing, YP_659332; A. fulgidus RFCS (AfuRFCS), NP_070884; S. solfataricus RFCS (SsoRFCS), NP_342275; RFCSPf (PfuRFCS), intein-containing, NP_577822; M. thermautotrophicus RFCS (MthRFCS), NP_275384; M. acetivorans RFCS2 (MacRFCS2), NP_615114; M. marisnigri RFCS2 (MmarisRFCS2), YP_001047146; M. hungatei JF-1 RFCS2 (MhungRFCS2), YP_502304; H. marismortui RFCS2 (HmarisRFCS2), YP_136998; N. pharaonis RFCS2 (NphaRFCS2), YP_326194; Halobacterium sp. strain NRC-1 RFCS2 (HspRFCS2), NP_280882; and H. walsbyi DSM RFCS2 (HwalRFCS2), YP_659090. Conserved and similar amino acid residues are black and gray, respectively. (B) Deletion of the insertion sequence in RFCS2Ma does not abolish the stimulation of DNA synthesis by the RFCMa complex. Lane 1, template alone; lane 2, template/PolBIMa; lane 3, template/PolBIMa/PCNAMa; lane 4, template/PolBIMa/PCNAMa/RFCMa-RFCS2Δ complex; lane 5, lane 4 without PCNAMa; lane 6, template/PolBIMa/PCNAMa/RFCMa wild type or positive control. The substrate for DNA synthesis was 0.5 μg of singly primed M13mp18 ssDNA in DNA polymerase reaction buffer. The reaction mixtures were incubated at 37°C for 5 min, the products were resolved by 1% alkali agarose gel electrophoresis, and signals were detected by autoradiography.
The RFCS1Ma/RFCS2Ma complex is unable to stimulate PCNAMa-dependent DNA synthesis by PolBIMa.
As demonstrated above, we detected interactions that suggested heterodimer formation and either heterotrimer or heterotetramer formation between RFCS1Ma and RFCS2Ma (Fig. 1C). The peak fraction from each subunit organization was used, in a primer extension analysis, to determine whether the complex of the M. acetivorans RFCS can stimulate PCNAMa-dependent DNA synthesis by PolBIMa. In each case, the complex of the small subunits rather inhibited DNA synthesis, suggesting that they cannot load PCNA (see Fig. S1 in the supplemental material). The results were, therefore, similar to our previous findings that the individual subunits of RFC inhibit DNA synthesis by PolBIMa (7). A similar finding showing the inhibition of a cognate DNA polymerase (Pol δ) by human RFCS has been reported elsewhere (28).
DISCUSSION
Pyrococcus furiosus RFCS (5, 27) and Archaeoglobus fulgidus RFCS (32) were shown to exist in oligomeric forms. Unlike the usual archaeal clamp loader proteins, the RFCMa complex has two small subunits (7). In the present report, we show that RFCS1Ma can exist as monomers and also as oligomers, ranging from homodimers to homotetramers, and that the oligomerization state is dependent on the protein concentration. In contrast, RFCS2Ma only exists as monomers in solution, irrespective of the protein concentration. The amino acid sequence alignment of RFCS1Ma and RFCS2Ma with other archaeal RFCS proteins (results not shown) suggests that RFCS1Ma is more similar to the known archaeal RFCS proteins. Thus, the RFCS1Ma polypeptide exhibits 59%, 50%, 49%, and 63% identities to the RFCS proteins found, respectively, in P. furiosus, Methanothermobacter thermautotrophicus, Sulfolobus solfataricus, and A. fulgidus (5, 20, 29, 31). RFCS2Ma, on the other hand shows 29 to 38% identities to these proteins. A mixture of RFCS1Ma/RFCS2Ma coeluted during size exclusion chromatography (Fig. 1C), suggesting that the two proteins interacted in solution. Although the protein complexes are likely to contain both RFCS1Ma and RFCS2Ma, they may also contain protein complexes made up of only RFCS1, since this protein can undergo self-oligomerization. Samples from the peak fractions of the oligomerized RFCS1Ma/RFCS2Ma failed to stimulate PCNAMa-dependent DNA synthesis by PolBIMa, suggesting that the complexes of the RFCSMa proteins are incapable of loading the sliding clamp.
The stoichiometry and spatial distribution of RFCMa subunits in the functional clamp loader were also determined in this report. In our model, three RFCS1Ma protomers are flanked by an RFCS2Ma on one side and by an RFCLMa on the other side (Fig. 2C). This arrangement can be inferred from the estimated stoichiometry and our previous mutational analysis that showed that a critical mutation in the Walker A motif of RFCS2 has no effect on clamp-loading activity (7). The observation implied that the Walker A motif of RFCS2Ma does not participate in ATP binding/hydrolysis. RFC proteins are AAA+ ATPases (17), and the ATPase sites are formed by a Walker A motif (RFC box III) from one subunit and an arginine finger (R in the SRC motif or RFC box VII) from an adjacent subunit. In the spatial arrangement shown in Fig. 2C, the Walker A (P-loop) of RFCS2 does not participate in ATP binding, and therefore, it does not influence ATP hydrolysis. A mutation in the Walker A motif of RFCLMa (one ATPase site) also did not impair clamp-loading activity by RFCLMa. On the other hand, mutations in either the SRC or Walker A motif of RFCS1Ma, which should affect three ATP binding sites according to our predicted spatial distribution (Fig. 2C), abolished clamp-loading activity (7). Both ATP binding and hydrolysis are required for the conformational changes that lead to clamp loading (17), and our previous mutational data (7) therefore support our proposed spatial distribution. The spatial arrangement of the RFCMa complex is, therefore, likely to be similar to the E. coli minimal clamp loader (10, 17).
Subunits of the E. coli γ-complex are capable of binding to the sliding clamp or β-subunit. The δ wrench exhibits the strongest interaction with the β-clamp, and by itself, it can load and unload the clamp (17, 21, 33). The γ3, on the other hand, binds less tightly and is also less competent in unloading the β-clamp (22). The δ′ is known as the stator or the stationary part of the γ-complex upon which the other subunits move, and its arginine finger together with the P-loop or Walker A motif of the adjacent γ-protomer forms the first ATPase site (site 1) in the γ-complex (35), as also shown in Fig. 3A for RFCMa. In archaea, RFCL is known to interact with the sliding clamp or PCNA (4). The C-terminal region of RFCLMa, which occupies the position analogous to the E. coli wrench, harbors two putative PIP boxes (Fig. 3C), although previously described RFCL proteins in archaea have only one PIP box (4, 6). This interaction between PCNA and the PIP box, by analogy, is similar to the interaction between the wrench (clamp-opening subunit) and the sliding clamp. Each of our four deletions from the C terminus of RFCLMa (Fig. 3B), however, failed to abolish the stimulation of PCNA-dependent enhancement of DNA synthesis by PolBIMa. Thus, it is likely that although the subunit organization and spatial distribution of the RFCMa complex are similar to the minimal E. coli clamp loader, interaction through the PIP boxes in the large subunit (wrench position) is not required for clamp loading. Note that in Methanosarcina acetivorans, instead of an octapeptide PIP box, several PCNA-interacting proteins have rather a conserved heptapeptide PIP box (7; I. K. O. Cann et al., unpublished data).
It was demonstrated previously that the RFCLPf contains 10 amino acids that form a basic cluster (motif) immediately upstream of the PIP box, and upstream of the basic cluster is another amino acid cluster mainly composed of acidic residues (26). These clusters are present in RFCLMa (Fig. 3C). However, unlike the RFCLPf and its homologs in other thermophilic archaea, such as Aeropyrum pernix, Methanocaldococcus jannaschii, and M. thermautotrophicus, the RFCLMa has >30 amino acids separating the basic residues from the most C-terminal PIP box. Furthermore, the acidic-residue-rich (glutamic acid-rich) region is split by several amino acids in the case of RFCLMa (Fig. 3C). The basic amino acid cluster, and not the acidic cluster, was shown to be required for the formation of a ternary complex of RFCPf/PCNAPf/DNA (26). Furthermore, although the PIP box in RFCLPf was shown to bind tightly to the sliding clamp (24), in support of our data presented here, the PIP box was shown to be nonessential for the RFC-dependent enhancement of DNA synthesis by PolBIPf in the presence of its cognate PCNA (24).
The analysis of the polypeptides constituting the eukaryotic RFC proteins clearly showed that they contain conserved motifs that were designated RFC boxes (9). These motifs were also conserved in archaeal RFC proteins (6, 7). Although RFC box 1 is unique to the eukaryotic RFCL, box II to box VIII are common to all RFCS proteins (6). The functions of RFC box III (Walker A motif) and RFC box VII (SRC motif) in the archaeal and eukaryotic proteins are well described since the two motifs fold to form the competent ATPase sites in the protein complex (3, 10). To our knowledge, the other RFC boxes have not been investigated for their contribution to the structure/function of an RFC complex. Thus, as a first step to understanding the importance of these motifs in the RFCMa complex, we targeted conserved amino acids in the RFC boxes of RFCS1Ma, which may serve as the motor in the clamp loader complex. The importance of RFCS1 was demonstrated in our previous report, where we showed that single mutations in a conserved lysine in the Walker A motif or the conserved arginine in the SRC motif in this subunit abolished RFC-dependent stimulation of PolBIMa during DNA synthesis in the presence of PCNA. This observation suggested failure to load the clamp by the mutant RFCMa complex (7). Interestingly, E119A slightly stimulated PCNAMa-dependent DNA synthesis by PolBIMa. Whereas the mutation in RFC box II (K18A) did not appear to impact the clamp loader's capacity to stimulate PCNA-dependent DNA synthesis by PolBIMa (Fig. 5B), the three remaining mutations completely abolished the capacity of the clamp loader to stimulate DNA synthesis by the cognate DNA polymerase in the presence of its clamp.
Hence, we demonstrate that, in addition to box III (Walker A motif) and box VII (SRC motif), the integrity of three other RFC boxes in the motor domain of the RFCMa complex is critical to the clamp-loading reaction. These data agree with our recent biophysical analysis that captures the loading of a dye-labeled clamp to a dye-labeled primer/template junction of a DNA template (C. Liu et al., unpublished data). Similar assays have been used to observe clamp loading in other systems (36).
The RFCMa complex represents the only biochemically characterized member of this new family of archaeal/eukaryotic RFC. This type of RFC is expected in most haloarchaea, since the available genome sequences of representatives, such as Halobacterium sp. strain NRC-1, Haloarcula marismortui, Haloquadratum walsbyi, and Natronomonas pharaonis, have the three subunits in their genomes. In addition, several mesophilic methanogens, such as Methanospirillum hungatei and Methanoculleus marisnigri, have the genes encoding orthologs of RFCMa subunits. It is anticipated that the subunit organization, spatial distribution, and mutational data described for RFCMa will be applicable to the clamp loader complex proteins in these organisms and other relatives yet to be described.
Supplementary Material
Acknowledgments
We thank William Metcalf (University of Illinois) for providing M. acetivorans genomic DNA and Taekjip Ha and Chen Liu (University of Illinois) for insightful discussions.
This research was supported by a National Science Foundation grant, MCB-0238451, to I.K.O.C. Also, Y.-H.C. and Y.L. were supported by National Science Foundation grant MCB-0238451.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Benkovic, S. J., A. M. Valentine, and F. Salinas. 2001. Replisome-mediated DNA replication. Annu. Rev. Biochem. 70:181-208. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, G. D., E. R. Goedken, S. L. Kazmirski, M. O'Donnell, and J. Kuriyan. 2005. DNA polymerase clamp loaders and DNA recognition. FEBS Lett. 579:863-867. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, G. D., M. O'Donnell, and J. Kuriyan. 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429:724-730. [DOI] [PubMed] [Google Scholar]
- 4.Cann, I. K., S. Ishino, I. Hayashi, K. Komori, H. Toh, K. Morikawa, and Y. Ishino. 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 181:6591-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cann, I. K., S. Ishino, M. Yuasa, H. Daiyasu, H. Toh, and Y. Ishino. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:2614-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann, I. K., and Y. Ishino. 1999. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics 152:1249-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y. H., S. A. Kocherginskaya, Y. Lin, B. Sriratana, A. M. Lagunas, J. B. Robbins, R. I. Mackie, and I. K. Cann. 2005. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J. Biol. Chem. 280:41852-41863. [DOI] [PubMed] [Google Scholar]
- 8.Compton, S. J., and C. G. Jones. 1985. Mechanism of dye response and interference in the Bradford protein assay. Anal. Biochem. 151:369-374. [DOI] [PubMed] [Google Scholar]
- 9.Cullmann, G., K. Fien, R. Kobayashi, and B. Stillman. 1995. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4661-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey, M. J., D. Jeruzalmi, J. Kuriyan, and M. O'Donnell. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3:826-835. [DOI] [PubMed] [Google Scholar]
- 11.De Moreno, M. R., J. F. Smith, and R. V. Smith. 1986. Mechanism studies of Coomassie blue and silver staining of proteins. J. Pharm. Sci. 75:907-911. [DOI] [PubMed] [Google Scholar]
- 12.Dionne, I., N. P. Robinson, A. T. McGeoch, V. L. Marsh, A. Reddish, and S. D. Bell. 2003. DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus. Biochem. Soc. Trans. 31:674-676. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, X. V., and P. M. Burgers. 2001. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem. 276:34768-34775. [DOI] [PubMed] [Google Scholar]
- 14.Gomes, X. V., S. L. Schmidt, and P. M. Burgers. 2001. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J. Biol. Chem. 276:34776-34783. [DOI] [PubMed] [Google Scholar]
- 15.Grabowski, B., and Z. Kelman. 2003. Archaeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 57:487-516. [DOI] [PubMed] [Google Scholar]
- 16.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 17.Indiani, C., and M. O'Donnell. 2006. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7:751-761. [DOI] [PubMed] [Google Scholar]
- 18.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2002. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 12:217-224. [DOI] [PubMed] [Google Scholar]
- 19.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106:429-441. [DOI] [PubMed] [Google Scholar]
- 20.Kelman, Z., and J. Hurwitz. 2000. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem. 275:7327-7336. [DOI] [PubMed] [Google Scholar]
- 21.Leu, F. P., M. M. Hingorani, J. Turner, and M. O'Donnell. 2000. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem. 275:34609-34618. [DOI] [PubMed] [Google Scholar]
- 22.Leu, F. P., and M. O'Donnell. 2001. Interplay of clamp loader subunits in opening the beta sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 276:47185-47194. [DOI] [PubMed] [Google Scholar]
- 23.Lin, Y., C. E. Guzman, M. C. McKinney, S. K. Nair, T. Ha, and I. K. Cann. 2006. Methanosarcina acetivorans flap endonuclease 1 activity is inhibited by a cognate single-stranded-DNA-binding protein. J. Bacteriol. 188:6153-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumiya, S., S. Ishino, Y. Ishino, and K. Morikawa. 2002. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells 7:911-922. [DOI] [PubMed] [Google Scholar]
- 25.Mayanagi, K., T. Miyata, T. Oyama, Y. Ishino, and K. Morikawa. 2001. Three-dimensional electron microscopy of the clamp loader small subunit from Pyrococcus furiosus. J. Struct. Biol. 134:35-45. [DOI] [PubMed] [Google Scholar]
- 26.Nishida, H., S. Ishino, T. Miyata, K. Morikawa, and Y. Morikawa. 2005. Identification of the critical region in replication factor C from Pyrococcus furiosus for the stable complex formation with proliferating cell nuclear antigen and DNA. Genes Genet. Syst. 80:83-93. [DOI] [PubMed] [Google Scholar]
- 27.Oyama, T., Y. Ishino, I. K. Cann, S. Ishino, and K. Morikawa. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8:455-463. [DOI] [PubMed] [Google Scholar]
- 28.Pan, Z. Q., M. Chen, and J. Hurwitz. 1993. The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis. Proc. Natl. Acad. Sci. USA 90:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisani, F. M., M. De Felice, F. Carpentieri, and M. Rossi. 2000. Biochemical characterization of a clamp-loader complex homologous to eukaryotic replication factor C from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 301:61-73. [DOI] [PubMed] [Google Scholar]
- 30.Robbins, J. B., M. C. McKinney, C. E. Guzman, B. Sriratana, S. Fitz-Gibbon, T. Ha, and I. K. Cann. 2005. The Euryarchaeota: nature's medium for engineering of single-stranded DNA binding proteins. J. Biol. Chem. 280:15325-15339. [DOI] [PubMed] [Google Scholar]
- 31.Seybert, A., D. J. Scott, S. Scaife, M. R. Singleton, and D. B. Wigley. 2002. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res. 30:4329-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seybert, A., M. R. Singleton, N. Cook, D. R. Hall, and D. B. Wigley. 2006. Communication between subunits within an archaeal clamp-loader complex. EMBO J. 25:2209-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner, J., M. M. Hingorani, Z. Kelman, and M. O'Donnell. 1999. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 18:771-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warbrick, E. 1998. PCNA binding through a conserved motif. Bioessays 20:195-199. [DOI] [PubMed] [Google Scholar]
- 35.Yao, N., L. Coryell, D. Zhang, R. E. Georgescu, J. Finkelstein, M. M. Coman, M. M. Hingorani, and M. O'Donnell. 2003. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 278:50744-50753. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang, Z., B. L. Yoder, P. M. Burgers, and S. J. Benkovic. 2006. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc. Natl. Acad. Sci. USA 103:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.