Abstract
Energy-coupling factor (ECF) transporters, a recently discovered class of importers of micronutrients, are composed of a substrate-specific transmembrane component (S component) and a conserved energy-coupling module consisting of a transmembrane protein (T component) and pairs of ABC ATPases (A proteins). Based on utilization of a dedicated (subclass I) or shared (subclass II) energy-coupling module, ECF systems fall into two subclasses. The T components are the least-characterized proteins of ECF importers, and their function is essentially unknown. Using RcBioN and LmEcfT, the T units of the subclass I biotin transporter (RcBioMNY) of a gram-negative bacterium and of the subclass II folate, pantothenate, and riboflavin transporters of a lactic acid bacterium, respectively, we analyzed the role of two strongly conserved short motifs, each containing an arginine residue. Individual replacement of the two Arg residues in RcBioN reduced ATPase activity, an indicator of the transporter function, by two-thirds without affecting the modular assembly of the RcBioMNY complex. A double Arg-to-Glu replacement destroyed the complex and abolished ATPase activity. The corresponding single mutation in motif II of LmEcfT, as well as a double mutation, led to loss of the T unit from the subclass II ECF transporters and inactivated these systems. A single Arg-to-Glu replacement in motif I, however, abolished vitamin uptake activity without affecting assembly of the modules. Our results indicate that the conserved motif I in T components is essential for intramolecular signaling and, in cooperation with motif II, for subunit assembly of modular ECF transporters.
Energy-coupling factor (ECF) transporters are a recently discovered novel class of importers of micronutrients in prokaryotes (9, 12, 13, 20). They are composed of a conserved energy-coupling module consisting of a transmembrane protein (T component) and pairs of ATP-binding cassette-containing proteins (A proteins), as well as an S unit (S component) through which substrate specificity is conveyed. S components represent a group of highly diverse small integral membrane proteins with predicted or experimentally established individual specificity for transition metal ions, B vitamins or their precursors, biotin, lipoate, and intermediates of salvage pathways. A link between substrate specificity and S components has been experimentally demonstrated in the case of CbiMN (Co2+) and NikMN (Ni2+) (13), RibU (riboflavin) (4, 6, 19), BioY (biotin) (9), FolT (folate) (8, 12), and ThiT (thiamine) (8, 12, 14). Based on utilization of a dedicated or shared AAT module, ECF transporters fall into two subclasses. Members of subclass I are encoded by operons containing one or two ABC ATPase genes, a T-component gene, and an S-component gene (12). RcBioMNY, the biotin transporter of Rhodobacter capsulatus, is the prototype of these systems. Previous work has established that the solitary BioY (S component) can function as a low-affinity transporter. It is converted into a high-affinity system in the presence of the AAT module BioMN (9). Notably, the majority of ECF transporters belong to subclass II. These promiscuous systems are widespread in the Firmicutes and also occur in members of the Thermotogales lineage and in archaea (12). In general, the cells contain a single ecfA1A2T operon and a number of genes for S units that are scattered around the genome. Cooccurrence of subclass I and subclass II ECF transporters is found in archaeal and bacterial species. The bioinformatic prediction for subclass II systems suggesting that several diverse S components interact with the same EcfA1A2T module has recently been confirmed experimentally for folate, riboflavin, and thiamine transporters of Bacillus subtilis, Lactobacillus casei, and Leuconostoc mesenteroides and for the hypothetical pantothenate transporter of L. mesenteroides (12).
The modular composition of ECF transporters poses questions about their oligomeric structure, the specificity of subunit recognition, and the intersubunit signaling that couples substrate uptake to ATP hydrolysis by the ABC ATPase domains. These issues are essentially unsolved for any ECF transporter.
In the present study, we confirm the predicted role of L. mesenteroides PanT (LmPanT) as a pantothenate-specific S component that functionally interacts with the LmEcfA1A2T module. The main focus is on the role of the T components, which are the least-characterized proteins of ECF importers. T proteins have moderately similar primary structures. Two 3-amino-acid signatures with Ala-Arg-Gly as the consensus sequence in the C-terminal part are the most conserved feature in the T units. Using RcBioMNY as a member of subclass I and LmEcfA1A2T plus LmFolT, LmPanT, or LmRibU as representatives of subclass II ECF transporters, we obtained evidence that replacement of either of the Arg residues in the T proteins strongly reduces or abolishes activity of the systems. While double mutations interfered with complex stability in each case, single replacements of the Arg residue in motif I did not have a marked impact on stability of the complexes, suggesting that this residue is important primarily for intramolecular signaling in both subclasses of ECF transporters.
MATERIALS AND METHODS
Bacterial strains, plasmids, and purification of transporter complexes.
Escherichia coli UT5600 with plasmids pFDX500 (lacIq) and pRcBioMNY encoding an N-terminally deca-His-tagged BioM, an untagged BioN, and a C-terminally FLAG-tagged BioY was used for production and purification of BioMNY complexes using a recently described protocol (9), with slight modifications. Briefly, 800 ml of a cell suspension was harvested, washed, and concentrated. The cells were broken by three passages through a French pressure cell. Membranes were isolated by ultracentrifugation and solubilized in the presence of 2% dodecyl-β-d-maltoside. Four milliliters of the solubilized material was mixed with 500 μl Ni-nitrilotriacetic acid (NTA) matrix (Invitrogen) and incubated overnight at 4°C. After transfer to an empty column, the matrix was washed with 80 ml buffer containing 0.05% dodecyl-β-d-maltoside, 300 mM NaCl, 15% glycerol, and imidazole at concentrations up to 130 mM. RcBioMNY was eluted with 1 ml of buffer containing 300 mM imidazole. Imidazole was subsequently removed by filtration through a PD-10 column (GE Healthcare), and samples were removed and used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (2 μg) and ATPase assays (1 μg).
For production of subclass II ECF transporters, E. coli BL21 cells containing plasmids pLacI-RARE2 (encoding several tRNAs to compensate for the differences in codon bias between L. mesenteroides and E. coli genes), pLmEcfA1A2T (with a wild-type or mutant ecfT allele [see below]), and pLmFolT, pLmPanT, or pLmRibU were used. The resulting transporter complexes contained N-terminally deca-His-tagged LmEcfA1, untagged LmEcfA2, a C-terminally FLAG-tagged LmEcfT variant, and C-terminally FLAG-tagged LmFolT, LmPanT, or LmRibU. Complexes were purified by using a slightly modified version of a previous protocol (12) from 2 liters of cell suspension. Solubilized membrane protein was incubated with 1 ml Ni-NTA matrix. The matrix was washed with 40 ml buffer containing imidazole at a concentration up to 75 mM, and the protein was eluted with 12 ml buffer containing 500 mM imidazole. After 40-fold concentration with Amicon concentrators (30-kDa cutoff) and then 2-fold concentration with Amicon Ultra concentrators (10-kDa cutoff), 5 to 10 μg of protein was removed and used for SDS-PAGE. Western blotting was performed by standard protocols using monoclonal anti-FLAG and monoclonal anti-oligo-His alkaline phosphatase conjugates (Sigma).
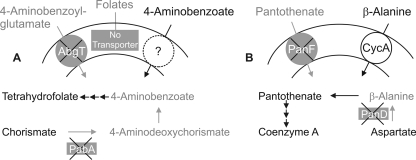
An E. coli K-12 pabA abgT mutant (kindly provided by A. Eudes and A. D. Hanson, University of Florida, Gainesville) and E. coli strain DV1 (with panD2 panF1 metB1 as relevant markers) (16) were used as test strains to analyze folate and pantothenate transporter activities, respectively. Strain phenotypes are shown in Fig. 1.
FIG. 1.
Phenotypes of vitamin synthesis- and vitamin transport-deficient E. coli strains. (A) The pabA abgT strain (kindly provided by A. Eudes and A. D. Hanson, Gainesville, FL) lacks the transporter for the folate catabolite 4-aminobenzoyl-glutamate and cannot produce the folate precursors 4-aminodeoxychorismate and 4-aminobenzoate. It can take up exogenous 4-aminobenzoate by an unknown transporter. E. coli K-12 naturally lacks transport systems for folates. (B) E. coli DV1 (panD panF) lacks the sodium/pantothenate symporter and is unable to produce the pantothenate precursor β-alanine. It can utilize exogenous β-alanine by means of the CycA amino acid transporter (15).
Growth assays.
Plate assays with E. coli DV1 containing plasmid pLacI-RARE2 plus an empty vector, an ampicillin resistance-conferring variant of pLmEcfA1A2T, pLmPanT, or pLmEcfA1A2T plus pLmPanT were performed on mineral salts agar (12) supplemented with Casamino Acids (0.04%), ampicillin, 1 mM isopropyl-β-d-galactopyranoside (IPTG), and either no additional supplement, 1 μM β-alanine, or 1 μM filter-sterilized Ca2+-d-(+)-pantothenate. Prior to spotting on the plates, liquid cultures were grown in medium containing β-alanine (1 μM) overnight. Cells were washed twice, resuspended in 35 mM Na-K phosphate buffer (pH 7.0), incubated for 5 h at 37°C with shaking, and spotted (10 μl) after serial 10-fold dilution on the agar plates. The plates were incubated for about 48 h at 37°C.
The consequences of amino acid replacement in EcfT for vitamin uptake were tested by performing growth assays with liquid cultures. For analysis of pantothenate uptake activity, precultures were grown in mineral salts medium supplemented with 134 μM l-methionine, 6 μM thiamine, and the appropriate antibiotics (basic medium) plus 1 μM β-alanine for 18 h at 37°C. Cells were diluted 1:50 in basic medium containing 0.5 mM IPTG and either no additional supplement, 1 μM β-alanine, or 1 μM Ca2+-pantothenate. Growth was analyzed with duplicate samples (200 μl per well) using a microtiter plate reader (SpectraMax M2; Molecular Devices) at 37°C. Tests for folate uptake activity were done with the E. coli pabA abgT mutant containing the tRNA-encoding plasmid pACYC-RIL and expressing LmFolT and LmEcfA1A2T variants as indicated below. Precultures of the recombinant strains were grown at 37°C overnight in basic medium supplemented with 2 μM 4-aminobenzoate, diluted 1:50 in basic medium, cultivated for 24 h, diluted again 1:50 in basic medium, and incubated for another 6 h. After addition of IPTG (0.5 mM) plus either 4-aminobenzoate (10 μM), (6R,S)-5-formyl-5,6,7,8-tetrahydrofolic acid (10 μM) (purchased as the calcium salt from Schircks Laboratories, Jona, Switzerland), or no additional supplement, growth was monitored with the microtiter plate reader.
Construction of mutant plasmids.
Mutants with Arg codons replaced by Glu codons in T-component genes were constructed by PCR as follows, and all mutations were verified by nucleotide sequencing. A plasmid-borne copy of bioN was used as the template for CGG164-to-GAG164 codon replacement with two overlapping primers using a protocol analogous to Stratagene's QuikChange protocol. A 253-bp NdeI/MscI fragment was used to replace the corresponding fragment of pRcBioMNY. A CGC195-to-GAA195 exchange was generated with a mutagenic primer that overlapped with the NdeI site and an inverse primer located in bioY. A 533-bp NdeI/KasI fragment of pRcBioMNY was replaced by the corresponding synthetic DNA. A BioNR164/195E-encoding plasmid was obtained by insertion of the 253-bp NdeI/MscI fragment containing the BioNR164E mutation mentioned above into NdeI/MscI-treated pRcBioMNR195EY. Plasmid pLmEcfA1A2T was used as the template for mutagenesis of ecfT. The R184E mutation was created by a single round of PCR using a mutagenic (CGT184-to-GAA184) forward primer that overlaps with the recognition site for the Mva1269I restriction endonuclease and a reverse primer located in the vector. The PCR product was treated with Mva1269I and NotI, and the resulting 305-bp fragment was inserted into Mva1269I/NotI-digested pLmEcfA1A2T. Two rounds of PCR were used for isolation of the R225E and R184/225E mutants. The mutagenic forward primer mentioned above and a mutagenic (CGA225-to-GAA225) reverse primer were used during the first round of PCR for construction of the double mutant. The resulting 155-bp product and the reverse vector primer were the primers used in the second PCR round. Treatment of the product with Mva1269I and NotI gave the 305-bp fragment that was used to produce pLmEcfA1A2TR184/225E. The same procedure was used to isolate the R225E single mutation, except that an Mva1269I site-overlapping primer with the wild-type sequence was used.
Peptide mass fingerprinting.
Coomassie blue-stained protein was excised from acrylamide gels and digested with trypsin (Promega, Mannheim, Germany) according to the manufacturer's protocol. Peptides were separated with an Agilent 1200 high-performance liquid chromatograph using a Zorbrax 300SB C18 column. Buffer A was 0.1% formate in water, and buffer B was acetonitrile containing 0.1% formate. A gradient from 5 to 60% buffer B over 60 min with a flow rate of 0.2 ml/min was applied, followed by 80% acetonitrile, 0.1% formate for 5 min and reequilibration for 15 min with 5% acetonitrile containing 0.1% formate. The high-performance liquid chromatograph was coupled with an Agilent MSD electrospray ionization-time of flight mass spectrometer. Nitrogen was used as the sheath gas at a flow rate of 12 liters/min and a temperature of 350°C. The cone voltage was 225 V. Spectral data were collected over a range from m/z 200 to 2800. Masses were resolved using the molecular feature extractor of Agilent MASSHUNTER software. The masses of all ions eluting between 20 and 50 min with a signal-to-noise ratio greater than 3 and a peptide-like abundance spectrum were determined. PEPTIDEMAP of the software suite PROWL (http://prowl.rockefeller.edu) was used to compare experimentally determined masses with those of LmEcfA1 and LmEcfA2 in silico digests (allowing one missed cleavage site) with a minimal matching accuracy of 0.05 Da.
ATPase assays.
The ATPase activity of BioMNY complexes was determined by quantitation of the amount of inorganic phosphate released from ATP as previously described (9). The values shown in Fig. 6 represent the means of duplicate determinations.
FIG. 6.
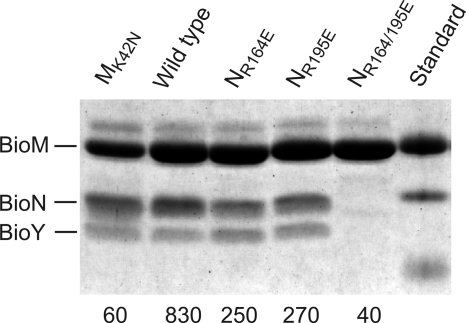
Consequences of replacements in BioN for complex formation and ATPase activity. BioMNY complexes were purified using the His tag on BioM. Two micrograms of protein was subjected to SDS-PAGE, and the gel was stained with Coomassie blue. The molecular masses of the standard proteins are 25, 20, and 15 kDa. The number below each lane represents the ATPase activity of the purified sample, expressed in nmol Pi min−1 (mg protein)−1. The BioMK42NNY complex (left lane) with inactivated ATPase domains served as a negative control.
RESULTS
T components RcBioN and LmEcfT.
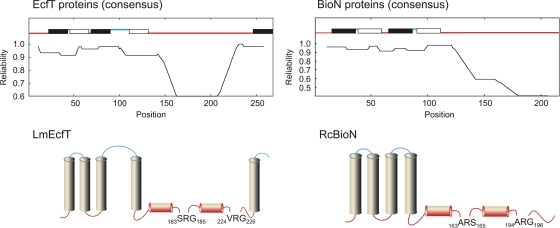
In contrast to the A and S units that catalyze ATP hydrolysis and confer substrate specificity, respectively, the distinct role of T proteins is elusive. Previous work on the R. capsulatus biotin transporter established that BioM (A protein) and BioN (T component) form stable complexes in the absence of BioY (S component). The bipartite BioMN complex, however, lacks ATPase and biotin-transport activities, which are found only in the presence of BioY. Biotin transport and biochemical analyses have shown that upon overproduction of BioM and BioY in the absence of BioN, the two former proteins form functional but instable complexes (9). The L. mesenteroides EcfA1, EcfA2, and EcfT proteins form complexes in the presence as well as in the absence of an S unit (12). Uncertainties about the functions of these proteins and the fact that T components are part of several hundred ECF transporters prompted us to take a closer look at these proteins. In silico topological analyses of a multitude of T proteins of ECF transporters for various substrates did not produce a conclusive general picture of the number and orientation of transmembrane segments. Likewise, amino acid sequence comparisons identified only moderate overall similarity. Closer inspection of the aligned amino acid sequences (not shown), however, revealed that two short motifs in the C-terminal region with Ala-Arg-Gly as the consensus sequence are the feature most strongly conserved among the various T units. To analyze the significance of these signatures for the structure and function of ECF transporters, we chose the RcBioN and LmEcfT proteins as representatives of subclass I and subclass II systems, respectively, for experimental analysis. First, we attempted to localize the two motifs in the cytoplasmic, extracytoplasmic, or transmembrane region. In order to obtain more reliable predictions of the membrane topology, RcBioN and 22 BioN proteins from gram-negative bacteria were aligned. Likewise, the LmEcfT sequence was compared with the sequences of 22 EcfT proteins from lactobacteria. The two derived consensus sequences were subjected to topological analysis using the SCAMPI (2), PRO-TMHMM, PRODIV-TMHMM (17), OCTOPUS (18), and TOPCONS (3) algorithms, and the results are shown in Fig. 2. EcfT proteins from lactobacteria are predicted to contain four transmembrane helices in the N-terminal region and a fifth membrane-spanning domain close to the C terminus. The conserved Arg-containing motifs (183SRG185 and 224VRG226 in LmEcfT) are located in a cytoplasmic domain. BioN proteins in gram-negative bacteria have four membrane-spanning regions resulting in cytoplasmic localization of the Arg motifs (163ARS165 and 194ARG196 in RcBioN). The model for BioN with inside orientation of the C terminus is consistent with previous results obtained with a bacterial two-hybrid system. These experiments indicated that the C terminus of RcBioN is in direct contact with the cytoplasmic RcBioM ABC ATPase (P. Schulz and T. Eitinger, unpublished results).
FIG. 2.
Topology of T components. (Upper panels) The consensus amino acid sequences of 23 EcfT proteins from lactobacteria (left panel) and 23 BioN proteins from gram-negative bacteria (right panel) were calculated with CLUSTALW. The consensus sequences were analyzed by TOPCONS based on the SCAMPI, PRO-TMHMM, PRODIV-TMHMM, and OCTOPUS algorithms. Predicted transmembrane helices are indicated by solid (in-to-out orientation) and open (out-to-in orientation) bars. Only the helices with a TOPCONS reliability score greater than 0.6 were considered. Red and blue indicate cytoplasmic and extracytoplasmic locations, respectively. (Lower panels) Topological models of the L. mesenteroides EcfT (LmEcfT) and R. capsulatus BioN (RcBioN) proteins based on the consensus profiles. The conserved signatures with “ARG” as the consensus sequence are located adjacent to helical regions with various hydrophobicities in individual T components. The colors indicate cytoplasmic and extracytoplasmic locations as described above.
PanT is a pantothenate-specific S component.
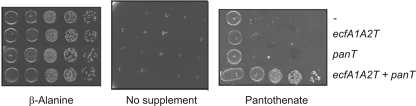
Previous experiments in our lab have shown that LmFolT, LmPanT, and LmRibU physically interact with the LmEcfA1A2T module, and we were curious about whether the Arg-containing signatures in LmEcfT mentioned above are involved in these interactions. Data on vitamin uptake activity were available only for the LmFolT-LmEcfA1A2T combination, and they demonstrated that FolT has specificity for folates (12). In these experiments, the L. mesenteroides proteins were produced in a folate-deficient mutant of E. coli K-12 (Fig. 1A) and tested for complementation activity in the presence of the vitamin, a method originally described by Klaus et al. (10). Although the efficiency of heterologous production of lactobacterial membrane proteins in E. coli is low, this organism is a suitable host. E. coli K-12 derivatives, in contrast to members of the Firmicutes, do not contain any ECF transporter, and thus, there is not interference with endogenous ECF components. In order to have another functional assay for vitamin uptake activity, we used a β-alanine-auxotrophic, pantothenate transport-deficient E. coli strain (Fig. 1B) to test the hypothetical function of LmPanT as a pantothenate-specific S component. The results are shown in Fig. 3. Recombinant mutant strains containing an empty vector and producing LmEcfA1A2T, LmPanT, or a combination of these two proteins were able to grow on mineral medium supplemented with β-alanine, but they could not grow in unsupplemented medium. In the presence of pantothenate, only the cells that contained both LmEcfA1A2T and LmPanT grew. These results clearly indicate that LmPanT has specificity for pantothenate and that its activity relies on the ECF. Analysis of the riboflavin uptake activity of LmRibU using a similar strategy was not possible, because mutants of E. coli K-12 with defined lesions in riboflavin synthesis genes are not available and these genes are considered essential (1).
FIG. 3.
Assay of pantothenate uptake activity. E. coli strain DV1 (see the text and Fig. 1B for the genotype and phenotype, respectively) containing an empty vector (−) or expressing the L. mesenteroides genes indicated on the right was pregrown in β-alanine-containing mineral salts medium. Cultures were harvested and washed in sterile buffer. Aliquots of 10-fold serial dilutions were spotted onto mineral salts agar plates containing 1 mM IPTG and either no additional supplement, 1 μM β-alanine, or 1 μM Ca2+-pantothenate.
Critical role of two conserved Arg residues in LmEcfT for activity of subclass II ECF transporters.
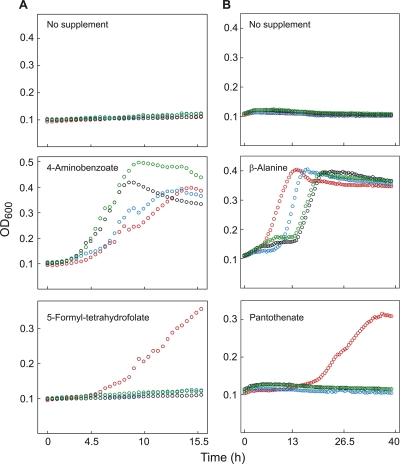
The Arg residues in the 183SRG185 and 224VRG226 motifs of LmEcfT were replaced individually and in combination by Glu since we expected that an opposite charge would have strong consequences. The mutant LmEcfA1A2T proteins were produced in combination with LmFolT in the folate-deficient E. coli strain and in combination with LmPanT in the pantothenate-deficient strain, and growth of the recombinants was monitored in liquid medium (Fig. 4). As expected, none of the strains grew in the absence of an appropriate vitamin precursor, and all strains grew when such a precursor was present. Notably, only the wild-type ecfT allele allowed utilization of folate and pantothenate, indicating that each of the single mutations, as well as the double mutations, in LmEcfT inactivated the folate and pantothenate transporters.
FIG. 4.
Mutations in EcfT abolish folate and pantothenate uptake. (A) The E. coli pabA abgT strain containing pACYC-RIL, pLmFolT, and a variant of pLmEcfAAT with a wild-type (red circles), R184E (blue circles), R225E (black circles), or R184/225R (green circles) ecfT allele were grown in minimal medium containing no supplement, 10 μM 4-aminobenzoate, or 10 μM 5-formyl-tetrahydrofolate. (B) E. coli DV1 (panD panF) harboring pLacI-RARE2, pLmPanT, and variants of pLmEcfAAT as indicated above was grown in unsupplemented minimal medium or in medium containing 1 μM β-alanine or 1 μM Ca2+-pantothenate. All cultures contained 0.5 mM IPTG and were grown in duplicate in microtiter plates at 37°C. Growth was monitored by determining the optical density at 600 nm (OD600) and is expressed as the mean of duplicate results. See Fig. 1 for the strain phenotypes.
Consequences of Arg residue replacement for stability of LmEcf transporter complexes.
To investigate whether loss of activity in mutant transporters was the result of diminished complex stability, wild-type and mutant L. mesenteroides ECF transporters were produced in E. coli BL21. Solubilized membrane protein was chromatographed on immobilized Ni-NTA, taking advantage of the His tag on LmEcfA1, and the preparations were subjected to SDS-PAGE and subsequently to Western blotting and peptide mass fingerprinting. As indicated in Fig. 5, Western blotting resulted in comparable subunit patterns for wild-type and mutant transporters harboring the LmEcfTR184E variant. A different picture was obtained for transporters with the LmEcfTR225E and LmEcfTR184/225E variants. In these cases the S unit copurified with LmEcfA1 (although to a lesser extent in the case of the pantothenate and riboflavin transporters). Clearly, the modified T components, which were detectable in membranes of recombinant E. coli cells (not shown), were lost. LmEcfA2 was untagged and thus could not be detected by Western blotting. Since tagged LmEcfA1 (31.8 kDa) and untagged LmEcfA2 (31.2 kDa) have similar molar masses, the corresponding size ranges were excised from the Coomassie blue-stained acrylamide gels and analyzed by peptide mass fingerprinting. Masses corresponding to the masses of LmEcfA1- and LmEcfA2-derived peptides were detected in each case (see Fig. S1 in the supplemental material), suggesting that LmEcfA2 was present in the complexes irrespective of the nature of the LmEcfT variant. In conclusion, replacement of one of the strictly conserved Arg residues in LmEcfT abolished transport activity but differentially affected complex stability. The subunit composition of transporters with the LmEcfTR184E variant resembled that of their wild-type counterparts. In contrast, the R225E replacement in LmEcfT destabilized the complexes and led to loss of the T unit.
FIG. 5.
Complexes of EcfA1A2T with FolT, PanT, and RibU. E. coli BL21 containing pLacI-RARE2 was used as the host. The EcfA1A2T module with wild-type (WT) and mutant (R184E, R225E, R184/225E) ecfT genes was produced from a plasmid (resulting in deca-His-tagged EcfA1, untagged EcfA2, and FLAG-tagged EcfT) in the presence of another plasmid encoding FLAG-tagged FolT, PanT, or RibU. Membranes of the recombinants were isolated and solubilized with dodecyl-β-d-maltoside. Solubilized material was chromatographed on immobilized Ni-NTA. Enriched proteins were separated by SDS-PAGE and blotted onto nitrocellulose membranes. The membranes were probed with monoclonal anti-oligo-His or anti-FLAG antibody-alkaline phosphatase conjugates. The marker proteins (left lane in each panel) had molecular masses of (from top to bottom) 37, 25, and 20 kDa.
Differential effects of Arg replacements in BioN for the RcBioMNY transporter.
In contrast to the L. mesenteroides transporters, RcBioMNY can be produced in large quantities in E. coli. To investigate whether the conserved Arg residues in the T component are also important for subclass I ECF transporters, we introduced Arg-to-Glu replacements into the 163ARS165 and 194ARG196 signatures of RcBioN. Wild-type and mutant transporters were solubilized from membranes of recombinant E. coli and purified by Ni-chelate affinity chromatography, and the subunit content was analyzed by SDS-PAGE and Coomassie blue staining. Figure 6 shows the results. Single exchanges of R164 or R195 in RcBioN had no detectable effect on the amount of the RcBioMNY biotin transporter and its subunit composition. In the case of the double mutant, however, RcBioM (containing the His tag) purified as the single unit, and the RcBioN and RcBioY proteins were lost. It was previously shown that the tripartite RcBioMNY complex displays ATPase activity, in contrast to the solitary nucleotide-binding protein (RcBioM), the ECF (RcBioMN), and the tripartite RcBioMK42NNY complex with replacement of the essential K42 residue in the Walker A site (9). Thus, ATPase activity is a suitable indicator of complex integrity and function. ATPase assays of the purified mutant complexes revealed that both the R164E and R195E replacements reduced this activity by two-thirds. As expected, the double mutant showed only background activity due to the lack of RcBioN and RcBioY. These results suggest that the single Arg replacements in RcBioN affect intramolecular signaling rather than complex stability and that the double exchange destabilizes the RcBioMNY complex.
DISCUSSION
ECF transporters are a widespread class of import systems that have highly conserved nucleotide-binding domains (or ATP-binding cassette-containing domains [A domains]) in common with canonical ABC importers (see reference 5 for a recent review of bacterial ATP-binding cassette systems). The fundamental differences include the fact that ECF systems are independent of extracytoplasmic solute-binding proteins, the utilization of an ECF that contains a conserved transmembrane (T) protein, and the fact that the shared use of the energy-coupling module by numerous substrate-specific (S) components is the rule rather than exception in the subclass II ECF systems. As noted above, the substrate specificity of several S components has been analyzed. These experiments characterized the S proteins as high-affinity substrate capture proteins with equilibrium dissociation constants in the nanomolar range (binding of folates to L. casei FolT [8]) or subnanomolar range (binding of thiamine to L. casei ThiT [8] and binding of riboflavin to Lactococcus lactis RibU [6]). Experiments with RcCbiMN and RcBioY, the S units of the ECF-type cobalt and biotin transporters of R. capsulatus, respectively, demonstrated that these proteins not only bind their substrates but also act as transporters in the absence of their energy-coupling modules under certain conditions (9, 13). Whether other solitary S components can function as transporters needs to be established.
Whereas distinct functions can be assigned to the A and S units of ECF transporters, the biochemical role of their T components remains to be defined. T proteins occur in bacteria, archaea, algae, and plants and form a family of moderately conserved integral membrane proteins. Due to their similarity to CbiQ, the T unit of prokaryotic cobalt transporters, many T proteins are misannotated in databases as cobalt transporters or as components of cobalt transporters. In most cases T proteins are encoded by operons containing genes for S, A, and T units (subclass I ECF transporters) or A and T units (subclass II). The exceptions include a special group of T components in cyanobacteria and their plant homologs. Since the genome context in cyanobacteria does not point to a specific role and experimental data are not available, the function of these T proteins is essentially unknown. Considering the presence of potential plastid-specific transit peptides at their N termini, plant T proteins (e.g., the T proteins encoded by the locus AT3G21580 in the Arabidopsis thaliana genome and by the gene Os05g0400600 of Oryza sativa) may be localized in chloroplasts.
In the present study we attempted to obtain insight into the function of RcBioN and LmEcfT, the T components of a subclass I biotin transporter and of the vitamin transporters of a member of the Firmicutes. Close inspection of the amino acid sequences of more than 400 T proteins revealed that two arginine-containing short motifs are the most conserved signatures, suggesting that they have a critical role. This suggestion was confirmed by physiological and biochemical assays when Arg residues were replaced. Individual exchanges in RcBioN did not obviously change the subunit composition of the RcBioMNY complex but led to curtailed activity. Likewise, replacement within motif I of LmEcfT did not result in destruction of ECF transporter complexes but nonetheless abolished vitamin uptake activity. These results correspond in part to the findings of previous studies of the role of the so-called EAA loop, an intracellular loop in the transmembrane proteins of canonical ABC importers that is in direct contact with the ABC proteins (for a review, see reference 5). It has long been known that distinct amino acid replacements in the EAA loops of MalF and MalG of the maltose ABC transporter abolish sugar uptake but do not prevent complex formation with the MalK ABC ATPase (11). Whereas single exchanges in RcBioN and the replacement in motif I of LmEcfT did not disrupt complex assembly, double mutations and the replacement in motif II of LmEcfT had strong destabilizing consequences. These results show that the conserved Arg-containing motifs in T components are important determinants of complex stability and/or intramolecular signaling in ECF transporters.
Supplementary Material
Acknowledgments
We thank Aymerick Eudes and Andrew D. Hanson (University of Florida, Gainesville) for kindly providing the E. coli pabA abgT mutant strain and Philipp Schulz (Berlin, Germany) for assistance with BioN mutant construction.
This work was funded by grant EI 374/3-1 from the Deutsche Forschungsgemeinschaft to T.E.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernsel, A., H. Viklund, J. Falk, E. Lindahl, G. von Heijne, and A. Elofsson. 2008. Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. USA 105:7177-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernsel, A., H. Viklund, A. Hennerdal, and A. Elofsson. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465-W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess, C. M., D. J. Slotboom, E. R. Geertsma, R. H. Duurkens, B. Poolman, and D. van Sinderen. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 188:2752-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duurkens, R. H., M. B. Tol, R. Geertsma, H. P. Permentier, and D. J. Slotboom. 2007. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 282:10380-10386. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Eudes, A., G. B. Erkens, D. J. Slotboom, D. A. Rodionov, V. Naponelli, and A. D. Hanson. 2008. Identification of genes encoding the folate- and thiamine-binding membrane proteins of Lactobacillus casei and other Firmicutes. J. Bacteriol. 190:7591-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebbeln, P., D. A. Rodionov, A. Alfandega, and T. Eitinger. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. USA 104:2909-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaus, S. M., E. R. Kunji, G. G. Bozzo, A. Noiriel, R. D. de la Garza, G. J. Basset, S. Ravanel, F. Rebeille, J. F. Gregory III, and A. D. Hanson. 2005. Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 280:38457-38463. [DOI] [PubMed] [Google Scholar]
- 11.Mourez, M., M. Hofnung, and E. Dassa. 1997. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 16:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodionov, D. A., P. Hebbeln, A. Eudes, J. ter Beck, I. A. Rodionova, G. B. Erkens, D. J. Slotboom, M. S. Gelfand, A. L. Osterman, A. D. Hanson, and T. Eitinger. 2009. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191:42-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodionov, D. A., P. Hebbeln, M. S. Gelfand, and T. Eitinger. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schauer, K., J. Stolz, S. Scherer, and T. M. Fuchs. 2009. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J. Bacteriol. 191:2218-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider, F., R. Krämer, and A. Burkovski. 2004. Identification and characterization of the main β-alanine uptake system of Escherichia coli. Appl. Microbiol. Biotechnol. 65:576-582. [DOI] [PubMed] [Google Scholar]
- 16.Vallari, D. S., and C. O. Rock. 1985. Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J. Bacteriol. 164:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viklund, H., and E. Elofsson. 2004. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 13:1908-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viklund, H., and A. Elofsson. 2008. A method that improves topology prediction for transmembrane proteins by using two-track ANN-based preference scores and an improved topological grammar. Bioinformatics 24:1662-1668. [DOI] [PubMed] [Google Scholar]
- 19.Vogl, C., S. Grill, O. Schilling, J. Stülke, M. Mack, and J. Stolz. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhulin, I. B. 2009. It's computation time for bacteriology! J. Bacteriol. 191:20-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.