Abstract
FlhF proteins are putative GTPases that are often necessary for one or more steps in flagellar organelle development in polarly flagellated bacteria. In Campylobacter jejuni, FlhF is required for σ54-dependent flagellar gene expression and flagellar biosynthesis, but how FlhF influences these processes is unknown. Furthermore, the GTPase activity of any FlhF protein and the requirement of this speculated activity for steps in flagellar biosynthesis remain uncharacterized. We show here that C. jejuni FlhF hydrolyzes GTP, indicating that these proteins are GTPases. C. jejuni mutants producing FlhF proteins with reduced GTPase activity were not severely defective for σ54-dependent flagellar gene expression, unlike a mutant lacking FlhF. Instead, these mutants had a propensity to lack flagella or produce flagella in improper numbers or at nonpolar locations, indicating that GTP hydrolysis by FlhF is required for proper flagellar biosynthesis. Additional studies focused on elucidating a possible role for FlhF in σ54-dependent flagellar gene expression were conducted. These studies revealed that FlhF does not influence production of or signaling between the flagellar export apparatus and the FlgSR two-component regulatory system to activate σ54. Instead, our data suggest that FlhF functions in an independent pathway that converges with or works downstream of the flagellar export apparatus-FlgSR pathway to influence σ54-dependent gene expression. This study provides corroborative biochemical and genetic analyses suggesting that different activities of the C. jejuni FlhF GTPase are required for distinct steps in flagellar gene expression and biosynthesis. Our findings are likely applicable to many polarly flagellated bacteria that utilize FlhF in flagellar biosynthesis processes.
Flagellar biosynthesis in bacteria is a complex process that requires expression of more than 50 genes in a sequential manner to ensure that the encoded proteins are secreted and interact in a proper order to construct a flagellar organelle (8). Formation of a flagellum to impart swimming motility is often an essential determinant for many bacteria to infect hosts or reside in an environmental niche. As such, flagella and flagellar motility are required for Campylobacter jejuni to initiate and maintain a harmless intestinal colonization in many wild and agriculturally important animals (16, 17, 19, 35, 47, 49), which leads to large reservoirs of the bacterium in the environment and the human food supply (13). In addition, flagellar motility is essential for the bacterium to infect human hosts to cause a diarrheal disease, which can range from a mild, watery enteritis to a severe, bloody diarrheal syndrome (4). Due to its prevalence in nature and in the food supply, C. jejuni is a leading cause of enteritis in humans throughout the world (7).
C. jejuni belongs to a subset of motile bacteria that produce polarly localized flagella, which includes important pathogens of humans, such as Helicobacter, Vibrio, and Pseudomonas species. These bacteria have some commonalities in mechanisms for flagellar gene expression and biosynthesis, such as using both alternative σ factors, σ28 and σ54, for expression of distinct sets of flagellar genes (1, 6, 9, 11, 18, 20-22, 26, 36, 40, 44, 45, 49). In addition, these bacteria produce the putative FlhF GTPase, which is required in each bacterium for at least one of the following: expression of a subset of flagellar genes, biosynthesis of flagella, or the polar placement of the flagella. For instance, FlhF is required for expression of some σ54- and σ28-dependent flagellar genes and for production of flagella in the classical biotype of Vibrio cholerae (10). However, V. cholerae flhF mutants of another biotype can produce a flagellum in a minority of cells, but the flagellum is at a lateral site (14). Similar lateral flagella were found in flhF mutants of Pseudomonas aeruginosa and Pseudomonas putida (34, 37). FlhF of Vibrio alginolyticus may also be involved in the polar formation of flagella and may possibly influence the number of flagella produced (28, 29). Demonstration that FlhF is polarly localized in some of these species and the fact that FlhF has been observed to assist the early flagellar MS ring protein, FliF, in localizing to the old pole in one biotype of V. cholerae give credence that FlhF may be involved in the polar placement of flagella in the respective organisms (14, 29, 34).
Bioinformatic analysis indicates that the FlhF proteins belong to the SIMIBI class of NTP-binding proteins (30). More specifically, the GTPase domains of FlhF proteins are most similar to those of the signal recognition particle (SRP) pathway GTPases, such as Ffh and FtsY. Because of the homology of the GTPase domains, these three proteins may form a unique subset within the SIMIBI proteins. Whereas the GTPase activities of the interacting Ffh and FtsY proteins have been extensively characterized (32, 38, 39, 42), little is known about the GTP hydrolysis activity of FlhF. Structural determination of FlhF of Bacillus subtilis indicates that the potential GTPase activity of FlhF is likely varied relative to those of Ffh and FtsY (2). However, no biochemical analysis has been performed to verify or characterize the ability of an FlhF protein to hydrolyze GTP. As such, no studies have correlated the biochemical activity of FlhF in relation to GTP hydrolysis with the role that FlhF performs in flagellar gene expression or biosynthesis.
Through previous work, we have delineated the regulatory cascades governing flagellar gene expression in C. jejuni. We have found that formation of the flagellar export apparatus (FEA), a multiprotein inner membrane complex (consisting of the proteins FlhA, FlhB, FliF, FliO, FliP, FliQ, and FliR) that secretes most of the flagellar proteins out of the cytoplasm to form the flagellum, is required to activate the FlgS sensor kinase to begin a phosphorelay to the cognate FlgR response regulator (23, 24). Once activated by phosphorylation, FlgR likely interacts with σ54 in RNA polymerase to initiate expression of many flagellar genes encoding components of the flagellar basal body, rod, and hook (20, 24). After formation of the hook, flaA, encoding the major flagellin, is expressed via σ28 and RNA polymerase to generate the flagellar filament and complete flagellar biosynthesis (6, 18, 20, 21, 49). In two separate genetic analyses, we found that flhF mutants of C. jejuni are nonmotile and show a more than 10-fold reduction in expression of σ54-dependent flagellar genes, indicating that FlhF is required for both flagellar gene expression and biosynthesis (20). However, it is unclear how FlhF influences expression of σ54-dependent flagellar genes. Furthermore, it is unknown if the GTPase activity of FlhF is required for flagellar gene expression or biosynthesis in C. jejuni.
We have performed experiments to determine that C. jejuni FlhF specifically hydrolyzes GTP, confirming that FlhF is a GTPase. Whereas the FlhF protein is required for motility, flagellar biosynthesis, and expression of σ54-dependent flagellar genes, the GTPase activity of the protein significantly influences only proper biosynthesis of flagella. These results suggest that multiple biochemical activities of FlhF (including GTPase activity and likely other, as yet uncharacterized activities mediated by other domains) are required at distinct steps in flagellar gene expression and biosynthesis. In addition, we provide biochemical and genetic evidence that FlhF likely functions in a pathway separate from the FEA-FlgSR pathway in C. jejuni to influence expression of σ54-dependent flagellar genes. This study provides corroborative genetic and biochemical analysis of FlhF to indicate that FlhF has multiple inherent activities that function at different steps in development of the flagellar organelle, which may be applicable to many polarly flagellated bacteria.
MATERIALS AND METHODS
Bacterial strains and growth.
All C. jejuni strains used in this study are derivatives of strain 81-176, a clinical isolate from an individual with gastroenteritis that has subsequently been shown to promote diarrheal disease in human volunteers and commensal colonization of the chick gastrointestinal tract (4, 19, 27). C. jejuni was grown under microaerobic conditions on Mueller-Hinton (MH) agar with antibiotics as appropriate, as previously described (44). Escherichia coli strains DH5α, XL1-Blue, and BL21(DE3) were cultured with Luria-Bertani (LB) agar or broth with antibiotics as appropriate, as previously described (16).
Construction of mutants.
Mutants of C. jejuni were constructed by previously described methods (18). pDRH416 was used in PCR-mediated mutagenesis to provide plasmids containing various mutations and deletions of flhF, including (20, 31): flhFΔG (which lacks amino acids 274 to 476, removing the GTPase domain; pSNJ132), flhF(K295A) (pDRH1275), flhF(D321A) (pMB620), and flhF(R324A) (pMB619). These plasmids were electroporated into DRH1054 (81-176 Smr flhF::cat-rpsL) (20) to replace flhF::cat-rpsL with the flhF mutant alleles on the chromosome. To delete astA in these genetic backgrounds and in DRH1056 (81-176 Smr ΔflhF) (20), the strains were electroporated first with pDRH424 to create cat::rpsL insertions in astA and then with pDRH449 to remove the astA::cat-rpsL mutation (and thus astA) from the chromosome (20).
Replacement of flgR with flgR alleles encoding deletions of the N-terminal receiver domain (FlgRΔreceiver) or the C-terminal domain (FlgRΔCTD) was achieved by first electroporating DRH1438 (81-176 Smr ΔastA ΔflhF) with pDRH443 to create a kan::rpsL insertion in flgR (SNJ170) (20). This strain was then electroporated with pDRH1855 or pDRH1856 to replace flgR::kan-rpsL with the flgR alleles encoding the domain deletions (24).
To create a ΔastA ΔflhF ΔflhB mutant of C. jejuni, DRH1438 (81-176 Smr ΔastA ΔflhF) was electroporated with pDRH781 to create a cat::rpsL insertion in flhB (MB511) (20). This strain was then electroporated with pDRH742 to remove flhB::cat-rpsL (and thus flhB) from the chromosome of C. jejuni to generate MB532 (81-176 Smr ΔastA ΔflhF ΔflhB) (20).
For construction of a ΔastA ΔflgG mutant, the flgFG locus of C. jejuni 81-176 was amplified by PCR with primers containing 5′ BamHI sites. After cloning of the DNA into BamHI-digested pUC19 to generate pDRH1349, a cat-rpsL cassette from pDRH265 was inserted into the BglII site within flgG to create pDRH2567 (18). DRH461 (81-176 Smr ΔastA) (20) was electroporated with pDRH2567 to create DRH2623 (81-176 Smr ΔastA flgG::cat-rpsL). pDRH1349 was subjected to PCR-mediated deletion mutagenesis to delete the entire coding sequence of flgG from the plasmid (creating pDRH2425). This plasmid was electroporated into DRH2623 to remove flgG::cat-rpsL (and thus flgG) from the chromosome to generate SNJ925 (81-176 Smr ΔastA ΔflgG).
Deletion of fliF from C. jejuni was performed by first amplifying the fliF locus from C. jejuni 81-176 by PCR with primers containing 5′ BamHI sites. After cloning of the DNA into BamHI-digested pUC19 to create pDRH1777, a cat-rpsL cassette from pDRH265 was inserted into the MfeI site within fliF to create pDRH1814 (18). This plasmid was electroporated into DRH212 (81-176 Smr) (18) to create DRH2067 (81-176 Smr fliF::cat-rpsL). pDRH1777 was used in PCR-mediated deletion mutagenesis to delete codons 2 through 548 of the coding sequence of fliF, fusing the start codon to the last 13 codons of fliF. This plasmid was electroporated into DRH2067 to remove fliF::cat-rpsL (and thus fliF) from the chromosome to create DRH2074 (81-176 Smr ΔfliF).
Purification of FlhF proteins.
Wild-type flhF, flhF(K295A), flhF(D321A), and flhF(R324A) from codon 2 to the stop codon and flhFΔG from codons 2 through 273 were PCR amplified with primers containing in-frame 5′ BamHI sites. The DNA fragments were digested with BamHI and ligated into BamHI-digested pQE30 to create N-terminal His6-tagged fusions to wild-type FlhF (pDRH2270), FlhF(K295A) (pMB174), FlhF(D321A) (pMB640), FlhF(R324A) (pMB681), and FlhFΔG (pMB176). These constructs were then transformed into E. coli XL-1 Blue. Bacteria were grown in 500 ml LB broth (for purification of wild-type FlhF, FlhF(D321A), FlhF(R324A), and FlhFΔG) or 2 liters of LB broth [for purification of FlhF(K295A)] at 37°C to an optical density at 600 nm (OD600) of 0.8 and induced with 1 mM (final concentration) of isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 1 h. The bacteria were washed once with 350 mM NaCl, 10 mM MgCl2, and 10 mM KCl (pH 8.0) and then resuspended in 40 ml of the same buffer containing 1% Nonidet P-40 and 1 tablet of Complete protease inhibitor cocktail (Roche). The resuspended bacteria were passaged four times through an EmulsiFlex-C5 cell disrupter (Avesin) at 15,000 to 20,000 lb/in.2. Nickel-nitrilotriacetic acid agarose beads (Qiagen) were used for purification of proteins under native conditions according to the manufacturer's instructions. Eluted proteins after chromatography were analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fractions with the highest purity of FlhF proteins were combined, and glycerol was added to a final concentration of 10%. Protein preparations were frozen at −80°C. The concentrations of protein samples were measured by Bradford assay prior to use.
Nucleotide hydrolysis assays.
The nucleotide hydrolysis activity of purified proteins over time was performed by using a modified version of the protocol published by Weiss et al. (48). Reactions consisted of 500 nM purified wild-type and mutant FlhF proteins, 6 μCi [γ-32P]GTP or [γ-32P]ATP (100 nM, final concentration), 50 mM sodium acetate, 40 mM KCl, 5.4 mM MgCl2, 0.1 M EDTA, 3% glycerol, 1 mM dithiothreitol, and 0.1 mg/ml acetylated bovine serum albumin in a final volume of 55 μl. At time points of 0, 5, 10, 20, and 30 min, 10 μl was removed and added to 990 μl of 1 N formic acid to stop the reaction. Released phosphate in 5 μl of each stopped reaction was separated from the nucleoside by thin-layer chromatography using Cellulose PEI plates (J.T. Baker) in a buffer containing 0.4 M K2HPO4 and 0.7 M boric acid. The plates were air dried and autoradiographed by using a Storm 820 phosphorimager (Amersham Biosciences). The data were analyzed by the manufacturer's software.
Generation of polyclonal antiserum against C. jejuni proteins.
For purification of wild-type FlhF for generation of antisera, XL1-Blue/pDRH2270 was grown in 500 ml LB containing 100 μg/ml ampicillin to an OD600 of 0.6 at 37°C. IPTG was added to a final concentration of 0.1 mM, and the culture was further incubated for 4 h. The bacteria were lysed as described above, and His6-FlhF was purified from the insoluble material after solubilization in 8 M urea with nickel-nitrilotriacetic acid agarose according to the manufacturer's instructions.
The coding sequence of flgG and fliF was cloned into pGEX-4T-2 and transformed into BL21(DE3) to create pJMB515 (encoding the glutathione S-transferase [GST]-FlgG fusion protein) and pDRH2266 (encoding the GST-FliF fusion protein). For induction of protein production, 500 ml and 3 liters of LB broth with 100 μg/ml of ampicillin was inoculated with overnight cultures of BL21(DE3)/pJMB515 or BL21(DE3)/pDRH2266, respectively, and grown to an OD600 of 0.6. The bacteria were induced for 4 h with 0.1 mM IPTG at 37°C, and the bacteria were lysed as described above. After recovery of the soluble fraction after centrifugation, the fusion proteins were purified by standard procedures using glutathione Sepharose 4B.
The purified His6-FlhF, GST-FlgG, and GST-FliF proteins were used to immunize mice by standard procedures for antiserum generation by a commercial vendor (Cocalico Biologicals).
Arylsulfatase assays to monitor expression of flagellar genes.
C. jejuni strains in the ΔastA background were electroporated with pDRH532, pDRH608, or pDRH610 to create promoterless astA transcriptional fusions to flgDE2, flaA, and flaB, respectively (20). Arylsulfatase production from the transcriptional fusions in these strains was measured by previously published methods (15, 20, 50).
Fractionation of C. jejuni strains for protein localization studies and immunoblotting analysis.
Total proteins from whole-cell lysates (WCL) and total membrane of C. jejuni strains were fractionated by a protocol that was described previously (3). For whole-cell lysates, protein samples were loaded to represent the proteins recovered from 200-μl aliquots of bacterial cultures which had been equilibrated to the same density. For fractions representing total membrane, the amounts loaded represented protein samples obtained from 500 μl of bacterial culture (for detection of FliF, AtpF, or RpoA) or 5 ml of bacterial culture (for detection of FlhB).
Immunoblotting analysis was performed with specific primary antiserum at the following dilutions: anti-FlhF M1, 1:3,000; anti-RpoA M59, 1:3,500 (23); anti-FlhB Rab476, 1:1,000 (23); anti-FliF M1, 1:1,000; anti-FlgS Rab11, 1:3,500 (16); anti-FlgR Rab13, 1:5,000 (16); anti-AtpF M3, 1:1,000; and anti-FlgG M69, 1:1,000. Secondary antibodies were used at 1:5,000 to 1:10,000 dilutions.
Motility assays and transmission electron microscopy.
Motility phenotypes of C. jejuni wild-type and mutant strains were assessed as previously described (23). Briefly, strains from 16-h growth plates were suspended in MH broth to an OD600 of 0.8 and stabbed into semisolid MH motility agar by using a sterilized inoculating needle. The plates were incubated for 24 h at 37°C in microaerobic conditions and then visualized for motility.
For electron microscopy analysis of flagellar biosynthesis, the strains were grown and prepared as described above for motility assays. After dilution to an OD600 of 0.8, 1 ml of each strain was pelleted, resuspended in a 2% (vol/vol) glutaraldehyde solution, and incubated for 1 h on ice. Samples were stained with 1% (wt/vol) uranyl acetate and visualized with a FEI Technai G2 Spirit BioTWIN transmission electron microscope.
Real-time RT-PCR.
C. jejuni strains DRH212 (81-176 Smr), DRH1056 (81-176 Smr ΔflhF), MB630 [81-176 Smr flhF(D321A)], and MB628 [81-176 Smr flhF(R324A)] were grown from frozen stocks on MH agar containing appropriate antibiotics at 37°C for 48 h and restreaked 16 h prior to use. Bacteria were suspended from the agar plates in MH broth, and total RNA was extracted from the bacteria with Trizol reagent (Invitrogen). The RNA was then treated with DNase prior to analysis. A final concentration of 50 ng/μl of RNA was used in a Sybr green PCR master mix. Real-time reverse transcription-PCR (RT-PCR) was performed using a 7500 real-time PCR system (Applied Biosystems). Detection of mRNA for 16S rRNA served as an endogenous control, and the transcript levels of rpoN, flgG, flhA, flhB, and fliF in flhF mutants were compared relative to those of the wild-type strain (DRH212). The following primer pairs were used for real-time RT-PCR analysis: rpoN RT F, 5′-TTAGCACTTGATTTAGAACGCAATG-3′, and rpoN R, 5′-GGGATAAGTCCTCTTTCACAACTTAAATA-3′; flgG RT F, 5′-AATTCTGCGCGACTTTTCTTAAA-3′, and flgG RT R, 5′-AGCGCAGCAAACACAAATTG-3′; flhA RT R, 5′-AATACAATCACGCCAATGACCAT-3′, and flhA RT F, 5′-GCCCTGAAGCTGTGAGTGAGA-3′; flhB RT F, 5′-CGACGCATTATGCCGTAGCTA-3′, and flhB RT R, 5′-TGTTTTATGCGAAGAGCGAGAAA-3′; fliF RT F2, 5′-TTTAAACGAGGAAAGAACAGGTAGAAA-3′, and fliF RT R2, 5′-GTCAAGTCCTTCAACAGGACCTATATTA-3′; and 16S rRNA F, 5′-CCGGAATCGCTAGTAATCGTAGA-3′, and 16S rRNA R, 5′-ACGGGCGGTGAGTACAAGAC-3′.
RESULTS
FlhF is a GTPase.
Based on previous annotation of domains of the FlhF protein of B. subtilis (2), C. jejuni FlhF can be divided into three domains: an N-terminal basic (B) domain (amino acids 1 to 78), a middle N domain (amino acids 79 to 272), and a C-terminal G domain containing the GTPase domain (amino acids 283 to 484) (Fig. 1A). Bioinformatic analysis revealed that the B and N domains of the proteins share some homology (21 to 27.9% identity; 46.5 to 51.1% similarity), but FlhF of C. jejuni contains an additional 107 amino acids in the putative B and N domains not found in B. subtilis FlhF. Similarly, the B and N domains of other FlhF proteins from P. putida, P. aeruginosa, Helicobacter pylori, V. cholerae, and V. alginolyticus also contain additional amino acids, suggesting that these domains may confer functional and structural differences to their respective FlhF proteins compared to that of B. subtilis. More homology was noted between the G domain of the C. jejuni and B. subtilis proteins (35.6% identity and 56.5% similarity), with most of the homology occurring within the putative GTPase domain. Two subdomains of the G domain, G1 (containing the P-loop of GTPases) and G2, demonstrate conservation of residues predicted to be required for GTPase activity of the FlhF proteins (41) (Fig. 1B). Despite conservation of GTPase domains, no studies have analyzed if the proteins are able to hydrolyze GTP. We performed in vitro and in vivo biochemical and genetic analysis to determine if FlhF of C. jejuni is a GTPase and if this biochemical activity of FlhF is required for distinct steps in flagellar gene regulation or flagellar biosynthesis.
FIG. 1.
Domain organization and sequence alignments of FlhF proteins. (A) The FlhF proteins of C. jejuni 81-176 (GenBank accession number YP_999790) and B. subtilis strain 168 (GenBank accession number CAA47062) can be divided into three domains: an N-terminal basic domain (B; black rectangles), a central N domain (N; white rectangles), and a C-terminal GTPase domain (G; gray rectangles). Within the GTPase domain, multiple conserved subdomains are evident, including G1 containing the P-loop domain (striped box) and G2 containing the DXXR motif (dotted box). Numbers above proteins indicate boundaries of domains. (B) ClustalW alignment of the G1 and G2 domains of the FlhF proteins from C. jejuni 81-176 (Cj), H. pylori 26695 (Hp) (GenBank accession number NP_207825), V. cholerae O395 (Vc) (GenBank accession number YP_001217595), V. alginolyticus12G01 (Va) (GenBank accession number ZP_01258825), P. aeruginosa PAO1 (Pa) (GenBank accession number AAG04842), P. putida MK1 (Pp) (GenBank accession number AF67042), and B. subtilis (Bs). Numbers flanking each sequence indicate positions of amino acids in the respective proteins. Arrowheads indicate conserved amino acids changed to alanine residues in the C. jejuni FlhF(K295A), FlhF(D321A), and FlhF(R324A) mutant proteins analyzed in this study.
Wild-type FlhF and FlhF mutant proteins predicted to be defective in GTP hydrolysis were purified. FlhFΔG lacks amino acids 274 to 476, which removes most of the GTPase domain and presumably eliminates the hydrolysis activity of the protein. FlhF(K295A) contains an alanine substitution at a conserved lysine in the G1 (P-loop) region of the GTPase domain that is usually essential for GTP hydrolysis (41) (Fig. 1B). FlhF(D321A) and FlhF(R324A) contain alanine instead of an aspartic acid or arginine at a conserved DXXR motif of the G2 region of the GTPase domain, which has been proposed to assist in GTP hydrolysis in the SIMIBI class of GTPases (30) (Fig. 1B). However, verification of the requirement of the DXXR motif for the GTPase activity of an FlhF protein has yet to be performed.
The FlhF proteins were analyzed for GTPase activity by monitoring release of 32P from [γ-32P]GTP over time. Wild-type FlhF hydrolyzed GTP, with more than 80% of the 32P detected as free phosphate by the end of the assay (Fig. 2). In contrast, FlhF(R324A), FlhF(D321A), and FlhF(K295A) showed a more than 60 to 75% reduction in GTP hydrolysis (Fig. 2). As expected, FlhFΔG was the most defective for GTP hydrolysis, with a more than 90% reduction in activity. In comparison to GTP, wild-type FlhF demonstrated only limited hydrolysis of ATP, suggesting that FlhF is more efficient at hydrolyzing GTP than other nucleotides.
FIG. 2.
Nucleotide hydrolysis activity of wild-type (WT) and mutant FlhF proteins. Released phosphate from [γ-32P]GTP or [γ-32P]ATP was monitored in reactions with purified wild-type and FlhF mutant proteins over 30 min. FlhF proteins and nucleotides were used at final concentrations of 0.5 μM and 0.1 μM, respectively. Hydrolysis activity is expressed as the percentage of labeled phosphate released compared to the amount of labeled phosphate in nonhydrolyzed nucleotides remaining at each time point. Data presented are from a representative assay. Closed symbols indicate phosphate released from GTP with addition of WT FlhF (♦), FlhF(K295A) (▴), FlhF(D321A) (×), FlhF(R324A) (▪), and FlhFΔG (•). Open symbols indicate phosphate released from ATP with addition of WT FlhF (⋄).
GTPase activity of FlhF is specifically required for proper biosynthesis of flagella.
Wild-type flhF of C. jejuni was replaced with flhF alleles encoding the FlhF mutant proteins described above to determine if defects in GTPase activity impact flagellar gene expression, biosynthesis, or motility. FlhF(D321A) and FlhF(R324A) were detected as stable proteins in C. jejuni, produced at levels similar to those of wild-type FlhF (Fig. 3A). However, FlhF(K295A) and FlhFΔG were produced at less than 25% of the levels of wild-type FlhF. Because the reduced levels or stability of FlhF(K295A) and FlhFΔG may have caused difficulty in interpreting results of potential defects in flagellar gene expression and biosynthesis that may be unrelated to defects in GTP hydrolysis, these respective C. jejuni mutants were not further characterized.
FIG. 3.
Analysis of flhF mutants for flagellar gene expression, motility, and protein production. (A) Immunoblot analysis of production of wild-type (WT) and FlhF mutant proteins in C. jejuni. FlhF proteins from WCL were detected with murine anti-FlhF M1 antiserum (top panel), and RpoA from WCL was detected with murine anti-RpoA M59 antiserum to verify equal loading of samples (bottom panel). Arrows indicate FlhF or FlhFΔG. Strains include DRH212 (wild-type 81-176 Smr), DRH1056 (81-176 Smr ΔflhF), MB630 [81-176 Smr flhF(D321A)], MB628 [81-176 Smr flhF(R324A)], DRH1302 [81-176 Smr flhF(K295A)], and SNJ206 [81-176 Smr flhFΔG]. (B) Motility phenotypes of wild-type (WT) and mutant C. jejuni strains in MH semisolid agar. Strains include DRH212 (wild-type 81-176 Smr), DRH1056 (81-176 Smr ΔflhF), MB630 [81-176 Smr flhF(D321A)], and MB628 [81-176 Smr flhF(R324A)]. (C) Arylsulfatase assays measuring expression of the σ54-dependent transcriptional fusions, flgDE2::astA and flaB::astA, in C. jejuni strains. Results are from a typical assay with each strain tested in triplicate. Values reported for each strain are average arylsulfatase activity ± standard deviation relative to the amount of expression of each transcriptional fusion in wild-type 81-176 Smr ΔastA, which was set to 100 arylsulfatase units. For expression of flgDE2::astA (black bars), strains include wild-type DRH533 (WT), SNJ150, MB657, and MB662. For expression of flaB::astA (white bars), strains include wild-type DRH665, SNJ155, MB659, and MB666. (D) Immunoblot analysis of production of FlgG in C. jejuni strains. Anti-FlgG M69 murine antiserum was used to detect FlgG in WCL in the top panel. The lower panel shows an immunoblot for RpoA detected with anti-RpoA M59 murine antiserum to verify equal loading of samples. Strains include DRH212 (wild-type 81-176 Smr [WT]), SNJ925 (81-176 Smr ΔastA ΔflgG), DRH1056 (81-176 Smr ΔflhF), MB630 [81-176 Smr flhF(D321A)], and MB628 [81-176 Smr flhF(R324A)]. (E) Arylsulfatase assays measuring expression of flaA::astA in C. jejuni strains. Results are from a typical assay with each strain tested in triplicate. Values reported for each strain are average arylsulfatase activity ± standard deviation relative to the amount of expression of each transcriptional fusion in wild-type 81-176 Smr ΔastA, which was set to 100 arylsulfatase units. Strains include DRH655, SNJ154, MB704, and MB706.
Unlike wild-type C. jejuni, which was fully motile, a ΔflhF mutant (generated by removing codons 2 to 476 of flhF from the chromosome [20]) was nonmotile (Fig. 3B). The flhF(D321A) and flhF(R324A) mutants displayed an intermediate phenotype with a level of motility sharply reduced compared to that of the wild-type strain (Fig. 3B). These data suggest that a complete lack of FlhF is more detrimental to motility than producing an FlhF mutant protein hindered for GTPase activity. Attempts to recover transconjugants with wild-type flhF expressed from a constitutive promoter in trans to complement any flhF mutant were unsuccessful, suggesting that this method of complementation of flhF mutants may be lethal to the bacterium (data not shown). However, interruption of the two genes downstream of flhF did not result in nonmotile phenotypes like that caused by deletion of flhF (data not shown). These results confirm that any motility defects associated with the ΔflhF mutant or flhF point mutations were directly due to mutation of flhF and not to any possible polar defects in expression of downstream genes that could be essential for flagellar motility.
We next analyzed if reduced motility of the flhF(D321A) and flhF(R324A) mutants was due to reduced expression of σ54-dependent flagellar genes. Expression of two σ54-dependent flagellar genes was measured by examining the levels of arylsulfatase produced from a transcriptional fusion of flgDE2 and flaB to a promoterless astA construct. Compared to the wild-type strain, the ΔflhF mutant of C. jejuni demonstrated a 12- to 15-fold reduction in expression of σ54-dependent flagellar genes (Fig. 3C). In contrast, expression of flgDE2 was not affected in the flhF(D321A) and flhF(R324A) mutants. We did detect a minor reduction (less than twofold) in expression of flaB in the flhF(D321A) mutant but not in the flhF(R324A) mutant. These results suggest that impairing the GTPase activity of FlhF does not significantly influence the ability of C. jejuni to express flagellar genes. We also assessed the ability of the flhF(D321A) and flhF(R324A) mutants to produce proteins encoded by σ54-dependent transcripts. For this analysis, we analyzed production of FlgG, which forms the periplasmic distal rod structure of the flagellum. Like flgDE2 and flaB, flgG is predicted to be dependent on σ54 and FlhF for expression, but this analysis has never been performed. By real-time RT-PCR analysis, we confirmed that the ΔflhF mutant demonstrated a fivefold reduction in expression of flgG relative to the wild-type strain, which resulted in a very minimal level of FlgG production (Fig. 3D and data not shown). Similar to our analysis of flgDE2 and flaB expression, we detected only a slight reduction of expression of flgG in the flhF(D321A) and flhF(R324A) mutants of about 25% (data not shown). In addition, the flhF(D321A) and flhF(R324A) mutants produced wild-type levels of FlgG (Fig. 3D), indicating that production of proteins from σ54-dependent transcripts was likely not affected in FlhF mutants hindered for GTPase activity. Analysis of expression of the σ28-dependent flagellar gene flaA revealed that the flhF(D321A) and flhF(R324A) mutants expressed approximately 70% more flaA than the wild-type strain whereas the ΔflhF mutant expressed about 50% less. Whereas the significance of increased expression of flaA in these mutants is unknown as far as whether this increase would negatively impact flagellar motility, the fact that these mutants do not have decreases in flaA expression indicates that these mutants are not defective in regulating pathways for controlling activity of σ28 for expression of essential flagellar genes.
By transmission electron microscopy, we found that the reduced motility phenotypes of the flhF(D321A) and flhF(R324A) mutants were due to defects in proper flagellar biosynthesis. The production of a single flagellum at one or both poles of the bacterium was the normal flagellar biosynthesis phenotype produced by more than 92% of individual wild-type C. jejuni (Fig. 4A and Table 1). In contrast, the ΔflhF mutant rarely produced a single flagellum at one pole (Fig. 4B and Table 1). The flhF(D321A) and flhF(R324A) mutants produced a variety of flagellar biosynthesis phenotypes, with only approximately one-third producing a single flagellum at one or both bacterial poles (part of the normal phenotype) (Fig. 4C and Table 1). Another one-third of the mutants produced no flagella, compared to only 7.2% of wild-type bacteria with no flagella (Fig. 4C, D, G, and J and Table 1). The remaining 30 to 37% of the flhF(D321A) and flhF(R324A) mutants produced a variety of improper flagellar biosynthesis phenotypes not detected in wild-type C. jejuni, which included the production of two or more flagella at one pole (Fig. 4E, F, I, and J), a flagellum at a nonpolar site (flagella produced either slightly off the polar ends or at a more lateral site on the bacterium) (Fig. 4D, E, G, and H), or a significantly shorter flagellum (Fig. 4C, G, and J). The increase in the aflagellated or improper flagellar phenotypes likely contributed to the reduction in flagellar motility observed in these mutants (Fig. 3A). These results indicate that the GTPase activity of FlhF is specifically required for proper flagellar biosynthesis and may perform only a minor role, if any, in flagellar gene expression or protein production. Since the ΔflhF mutant is defective for σ54-dependent flagellar gene expression, we hypothesize that some other, as yet uncharacterized activity of FlhF is required by C. jejuni for expression of wild-type levels of σ54-dependent flagellar genes. Thus, inherent biochemical activities are required for distinct steps in flagellar gene expression and biosynthesis.
FIG. 4.
Flagellar biosynthesis phenotypes of C. jejuni wild-type (WT) and flhF mutant strains. Transmission electron microscopy of negatively stained bacteria was performed. All micrographs are between magnification ×11,500 and ×20,500. Bars = 1 μm. (A) DRH212 (WT 81-176 Smr); (B) DRH1056 (81-176 Smr ΔflhF); (C to F) MB630 [81-176 Smr flhF(D321A)]; (G to J) MB628 [81-176 Smr flhF(R324A)]. Arrowheads indicate truncated flagella produced by some bacteria in panels C, G, and J.
TABLE 1.
Proper flagellar number and placement for wild-type and C. jejuni mutant strains
| Relevant genotypea | % of bacteria withb: |
|||
|---|---|---|---|---|
| 2 flagella | 1 flagellum | 0 flagella | Other forms | |
| Wild type | 73.9 | 18.9 | 7.2 | 0 |
| ΔflhF | 0 | 1.6 | 98.4 | 0 |
| flhF(D321A) | 1.6 | 29.0 | 32.3 | 37.1 |
| flhF(R324A) | 4.1 | 29.3 | 35.8 | 30.9 |
Strains used include DRH212 (wild-type 81-176 Smr), DRH1056 (81-176 Smr ΔflhF), MB630 [81-176 Smr flhF(D321A)], and MB628 [81-176 Smr flhF(R324A)].
More than 110 individual bacteria for each strain were examined to determine the percentage of bacteria producing each phenotype. Phenotypes included bacteria that produced a single flagellum at both poles (2 flagella), a single flagellum at only one pole (1 flagellum), or no flagella (0 flagella). Bacteria producing other forms included those that produced more than one flagellum at a single pole, a flagellum at a nonpolar or lateral site, or a significantly shortened flagellum.
FlhF likely functions in pathway separate from the FEA-FlgSR pathway to activate expression of σ54-dependent flagellar genes.
Previous work has established that the formation of the FEA, rather than its secretory activity, contributes to a signal that is sensed by the cytoplasmic FlgS sensor kinase to result in autophosphorylation and phosphorelay to the cognate response regulator FlgR (23, 24). Activation of FlgR stimulates σ54 in RNA polymerase for expression of flagellar genes. Elimination of any component of the FEA, FlgS, or FlgR results in 12- to 800-fold decreases in σ54-dependent flagellar gene expression (20). To provide a better understanding of where FlhF may function in regulatory pathways for expression of σ54-dependent flagellar genes, we analyzed if FlhF is required for production of the components of the FEA-FlgSR pathway.
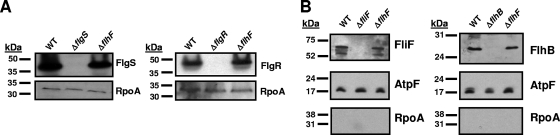
As shown in Fig. 5A, no differences in the levels of the FlgS and FlgR proteins were observed in whole-cell lysates (WCL) of wild-type and ΔflhF mutant strains. Comparing wild-type and ΔflhF mutant strains by real-time RT-PCR revealed no differences in the levels of expression of fliF, flhA, and flaB, encoding three components of the FEA (data not shown). In addition, we did not detect significant differences in the production or membrane localization of two components of the flagellar export apparatus, FlhB and FliF, between wild-type and ΔflhF mutant strains (Fig. 5B). Purity of membrane preparations from C. jejuni strains was verified by monitoring the presence of the inner membrane protein AtpF, a component of ATP synthase, and the absence of the cytoplasmic RpoA protein, a component of RNA polymerase (Fig. 5B). Whereas we could detect FlhB and FliF localized to the membrane of C. jejuni, generation of antisera specific for the other FEA components was not successful. Thus, the immunoblotting analysis was incomplete in ensuring that all components of the FEA were produced and localized to the membrane properly. The observation that a small minority (less than 2%) of individual ΔflhF bacteria produce a single flagellum is indirect and secondary evidence that FlhF is not absolutely required for FEA formation since the FEA is required to secrete proteins to build the flagellum (Table 1). Because of these combined results, we conclude that FlhF is likely not required for production of components of the FEA-FlgSR signaling pathway.
FIG. 5.
Formation of the FEA-FlgSR signaling pathway components and activity of the FEA in C. jejuni wild-type (WT) and flhF mutant strains. (A) Immunoblot analysis of FlgS and FlgR production in WCL of C. jejuni strains. Immunoblots were performed with anti-FlgS Rab11 rabbit antiserum and anti-FlgR Rab13 rabbit antiserum. Control immunoblots were performed with anti-RpoA M59 antisera to verify equal loading of protein samples. Strains include DRH212 (WT 81-176 Smr), DRH460 (81-176 Smr ΔflgS) (20), DRH737 (81-176 Smr ΔflgR) (20), and DRH1056 (81-176 Smr ΔflhF). (B) Immunoblot analysis of FliF and FlhB production and localization to the membrane fraction in C. jejuni strains. Total membranes were isolated from wild-type and mutant C. jejuni strains and then analyzed by immunoblot analysis. Immunoblots were performed with anti-FliF M1 murine antiserum and anti-FlhB Rab476 rabbit antiserum. Control immunoblots were performed with anti-AtpF M3 and anti-RpoA M59 murine antisera to verify the presence of AtpF and absence of RpoA in the membrane preparations. The membrane proteins from 500 μl of bacterial culture at equivalent densities were analyzed for FliF, AtpF, and RpoA, and those from 5 ml of bacterial culture at equivalent densities was analyzed for FlhB. Strains include DRH212 (wild-type 81-176 Smr), DRH2074 (81-176 Smr ΔfliF), SNJ471 (81-176 Smr ΔflhB), and DRH1056 (81-176 Smr ΔflhF).
We conducted additional experiments to determine if FlhF functions within the FEA-FlgSR signaling pathway or in a separate pathway that converges with the FEA-FlgSR pathway to activate expression of σ54-dependent flagellar genes. We first ensured that flhF was not required for expression of rpoN encoding σ54. Quantitative real-time RT-PCR did not reveal any differences in expression of rpoN between wild-type and the ΔflhF mutant strains (data not shown). We were unable to monitor the levels of σ54 since antisera generated against a portion of the protein were not able to detect native σ54 in C. jejuni.
In previous work, partially constitutively active forms of FlgR lacking the N-terminal receiver domain (FlgRΔreceiver) or the C-terminal domain (FlgRΔCTD) of the protein were shown to suppress the phenotypes of an FEA or flgS mutant for expression of σ54-dependent flagellar genes (23, 24). These results indicated that FlgR functioned downstream of the FEA and FlgS in the signaling cascade for expression of σ54-dependent flagellar genes. A similar suppressor analysis was performed to determine if FlgRΔreceiver or FlgRΔCTD could restore flagellar gene expression to a ΔflhF mutant. However, these partially constitutively active FlgR mutant proteins did not suppress the ΔflhF mutant for expression of σ54-dependent flagellar genes (data not shown). These results suggest that FlhF does not function upstream of the FlgSR in the FEA-FlgSR signaling cascade.
We then compared the levels of expression of one σ54-dependent flagellar gene, flaB, in mutants lacking flhF or flhB (encoding a component of the FEA) or both flhF and flhB. Our analysis revealed that the ΔflhF or ΔflhB mutants displayed, respectively, an 18- or 29-fold decease in expression of flaB::astA, but the ΔflhF ΔflhB mutant showed a 65-fold decrease in expression of this transcriptional reporter (data not shown). Because the defects of disrupting both the FEA-FlgSR pathway and FlhF were additive, these data provide further evidence that FlhF likely functions in a separate pathway that either converges with or functions downstream of the FEA-FlgSR pathway to activate σ54-dependent flagellar gene expression.
DISCUSSION
Peritrichous bacteria, which produce multiple flagella over the bacterial surface, have historically served as the model system for analyzing regulation of flagellar genes and flagellar biosynthesis. Investigations into polarly flagellated bacteria have uncovered variations in these pathways, indicating that bacteria have evolved diverse mechanisms to ensure proper expression of flagellar genes and biosynthesis of flagella. One of these differences has been the inclusion of FlhF in flagellar gene regulation and biosynthesis pathways in many polarly flagellated bacteria. With the studies conducted so far, it is evident that FlhF functions at different steps in flagellar organelle development in these bacteria.
Before this study, no experimental proof existed regarding whether FlhF proteins hydrolyze GTP or whether the GTPase activity is required for specific steps in production of flagella. By analyzing mutants of C. jejuni that either lacked FlhF or produced FlhF proteins with a reduced ability to hydrolyze GTP, we identified distinct GTPase-dependent and GTPase-independent steps in flagellar gene regulation and biosynthesis (Fig. 6). We discovered that a mutant lacking FlhF expressed approximately 5- to 15-fold-lower levels of three σ54-dependent flagellar genes and more than 98% of the bacteria did not produce a flagellum, which contributed to the nonmotile phenotype. However, mutants producing FlhF(D321A) and FlhF(R324A), which in in vitro analysis demonstrated only 30 to 40% of the GTPase activity of wild-type FlhF, were not severely defective for σ54-dependent flagellar gene expression. These mutants were significantly defective only for proper flagellar biosynthesis, which is defined for C. jejuni as the production of a single flagellum at one or both bacterial poles. Whereas we feel that our results give credence to our conclusion that the GTPase activity is required only for proper flagellar biosynthesis, the possibility remains that the residual GTPase activity we observed with the FlhF(D321A) and FlhF(R324A) proteins was sufficient for expression of σ54-dependent flagellar genes analyzed in this study. Conclusive analysis that the GTPase activity of FlhF is not required for σ54-dependent flagellar gene expression will require the in vivo production of a stable FlhF mutant protein in C. jejuni that completely lacks GTPase activity and analysis of multiple σ54-dependent promoters. As was shown by this study, this approach may be difficult, since we were unable to generate such a stable, GTPase-deficient FlhF mutant protein, perhaps due to a requirement of GTP interactions for the proper folding or stability of FlhF. Nonetheless, our results demonstrate that proper flagellar biosynthesis is more sensitive to decreases in GTP hydrolysis by FlhF than σ54-dependent flagellar gene expression.
FIG. 6.
Model for role of FlhF in activation of expression of σ54-dependent flagellar genes and the GTPase activity of FlhF in proper flagellar biosynthesis. Construction of the FEA is hypothesized to be required for the formation of a signal sensed by the FlgS sensor kinase for phosphorelay to the cognate FlgR response regulator to activate σ54 for expression of a subset of flagellar genes (20, 23, 24). An activity of FlhF independent of GTP hydrolysis is hypothesized to be required for a late step in regulating the expression of σ54-dependent flagellar genes which may include coactivation mechanisms with FlgR to stimulate σ54 in RNA polymerase, activation of transcription initiation by σ54-RNA polymerase holoenzyme, or stability of σ54-dependent mRNA transcripts. The GTP hydrolysis activity of FlhF is hypothesized to be required at an early step in flagellar biosynthesis by possibly influencing the FEA so that a flagellum is constructed and only a single flagellum forms at each pole.
In our in vitro biochemical assays, we identified a requirement of residues K295, D321, and R324 of FlhF for full GTPase activity. K295 is located in the P loop (G1 region) of the GTPase domain, whereas D321 and R324 are in a conserved DXXR motif of the G2 region that has been postulated to assist in GTP hydrolysis (2). However, these residues in any FlhF protein had not been analyzed to determine if they are required for GTPase activity until this study. Further characterization of these mutant FlhF proteins will be necessary to determine if these proteins are blocked only at the hydrolysis step or have defects in GTP binding as well. Because evidence from structural studies with FlhF of B. subtilis suggests that FlhF may have an unusual mechanism for GTP hydrolysis compared to other SIMIBI family members, these mutant proteins we have created may have the potential to provide insights into the biochemical mechanism of GTP hydrolysis by FlhF (34). In eukaryotic systems, GTPases are often regulated by GTPase-activating proteins (GAPs) and GTPase exchange factors (GEFs) (reviewed in reference 43). Bacterial versions of these GAPs or GEFs have not been identified for the other SIMIBI members, FtsY and Ffh. Rather, heterodimer formation by FtsY and Ffh has been found to directly stimulate the GTPase activity of each individual protein (12, 39). More-extensive biochemical analysis will be required to determine if FlhF has an associated GAP- or GEF-like protein or if the GTPase activity of FlhF is influenced by possible homodimer formation or heterodimer formation with another GTPase.
Because the GTPase domain of FlhF proteins is most similar to those of the bacterial FtsY and Ffh SRP GTPases, some speculation has been made that FlhF may function as a SRP protein specific for flagellar proteins that compose the FEA (5, 28, 30, 37). The bacterial SRP system is required for targeting many proteins to the general secretory (Sec) system (25, 46). These proteins predominantly include those to be inserted in the inner membrane or some proteins that are to be secreted into the periplasm. Considering our analysis of the ΔflhF mutant, we do not favor that FlhF functions exclusively in the targeting to the inner membrane of flagellar proteins, such as components of the FEA, that are essential for flagellar biosynthesis and flagellar gene expression. In the ΔflhF mutant, we detected at least two FEA proteins localized to the membrane fraction. Furthermore, in a small minority of individual ΔflhF mutant bacteria, a flagellum was detected, suggesting that C. jejuni does not have an absolute dependence on FlhF for FEA formation and FEA-mediated flagellar protein secretion. Further indirect evidence that FlhF is likely not required to form the FEA is the observation that the ΔflhF mutant shows 12- to 15-fold-reduced expression of σ54-dependent flagellar genes but a mutant lacking a component of the FEA (such as a ΔflhB mutant) generally demonstrates at least a 25- to 60-fold reduction in expression of these genes (20). Thus, if a ΔflhF mutant is lacking one of the FEA components in the inner membrane, we would have expected to see a deficiency in expression of σ54-dependent flagellar genes equivalent to that typically seen in an FEA mutant.
Monitoring formation of the FEA by measuring FEA-dependent secretion of flagellar proteins in the ΔflhF mutant would be one method to study if FlhF affects formation of the FEA, but this approach would be difficult since many of the genes that encode the rod, hook, and flagellin proteins that are secreted by the FEA are part of the σ54-dependent regulon and by consequence are dependent on FlhF and the FEA for expression. Future studies will focus on characterizing FEA-mediated secretion in flhF mutants ectopically expressing genes for the rod, hook, and flagellin proteins from nonnative, FEA- and FlhF-independent promoters to determine if there is a functional relationship between FlhF and FEA formation.
It is possible that FlhF, and more specifically its GTPase activity, may be required for modulating the activity of the FEA. In support of this hypothesis, the flhF(D321A) and flhF(R324A) mutants were impaired for proper flagellar biosynthesis, since approximately 60 to 65% of these bacteria (compared to ∼7% of wild-type bacteria) were unable to produce flagella, produced flagella at incorrect positions on the bacterial surface, or produced multiple flagella at a single pole. Since these mutants have a greater propensity for these phenotypes than wild-type bacteria, the GTPase activity of FlhF may be required at early initiation steps with respect to the FEA in flagellar biosynthesis (Fig. 6). These steps may include one or more of the following: the proper positioning of the FEA at a pole, ensuring that only one FEA is formed at a pole, monitoring the FEA so that it properly secretes flagellar proteins in the correct order to build a flagellum, or assisting the FEA in more efficiently secreting flagellar proteins. Some evidence for the first hypothesis exists in V. cholerae, since FlhF appears to assist in properly localizing at least one FEA component, the FliF MS ring, to the old pole in a bacterium (14). If FlhF of C. jejuni functions in an analogous process, it will be interesting to study this localization process, since C. jejuni usually constructs a flagellum at both the old and new poles. Furthermore, it would be interesting to study the localization of the FEA components in the flhF(D321A) and flhF(R324A) mutants, which presumably mislocalize the FEA to produce nonpolar flagella or allow for multiple polar FEA formation to contribute to more than one flagellum at a single pole. All of these studies depend on visualization of proteins in C. jejuni using fluorescence microscopy with labeled proteins, which has been difficult with some strains of C. jejuni, including the strain used in this study, C. jejuni strain 81-176 (33; our unpublished observations). To study the other hypotheses, development of new and better reagents to characterize FEA formation and to monitor flagellar protein secretion in flhF mutants is required to provide any insights into how FlhF may influence the activity of the FEA.
We found a more severe defect in flagellar gene regulation and biosynthesis with a mutant lacking flhF than with the GTP hydrolysis-hindered flhF(D321A) and flhF(R324A) mutants. The severity of the biosynthesis defect in the ΔflhF mutant is most likely related to the fact that expression of at least three σ54-dependent flagellar genes (and probably all other σ54-dependent flagellar genes) was reduced at least 5- to 15-fold. The decreased expression of multiple genes would greatly reduce the levels of the encoded proteins essential for construction of flagella. Since flhF(D321A) and flhF(R324A) were not significantly defective, if at all, for expression of σ54-dependent flagellar genes, these results indicate that other domains or activities of FlhF independent of GTP hydrolysis are required for processes leading to a full level of expression of σ54-dependent flagellar genes. These activities may be related to the B or N domain. To determine if the B and N domains of FlhF are specifically required for expression of σ54-dependent flagellar genes, we attempted to create an flhF mutant of C. jejuni lacking these domains, but this mutant protein was unstable in the bacterium. We continued our analysis of FlhF and revealed that the protein is not required for formation of the FEA or the FlgSR two-component system. Furthermore, we provided data that suggest that the FlgSR system does not function downstream of FlhF for activation of σ54 and that the lack of flhF and the FEA contributes to greater defects in expression of σ54-dependent flagellar genes than either mutation alone. Thus, our results indicate that FlhF may function in a separate pathway that converges with or acts downstream of the FEA-FlgSR pathway to stimulate σ54 (Fig. 6). Future studies will determine if FlhF is required at the step of FlgR-dependent activation of σ54 or at a more downstream step, such as σ54-RNA polymerase initiation or stability of σ54-dependent mRNAs.
With continued investigation into the FlhF proteins of polarly flagellated bacteria, it is evident that these proteins are an essential component of regulatory systems for expression of flagellar genes, flagellar biosynthesis, or both. For instance, flhF mutants of P. aeruginosa are only slightly affected in expression of the major flagellin in P. aeruginosa, but this defect is not detrimental to flagellar biosynthesis (34). Instead, FlhF is more specifically required for the polar placement of flagella. Furthermore, expression of flhF is dependent on σ54, rather than being required for expression of the σ54 flagellar regulon, in P. aeruginosa (11). flhF is also dependent on σ54 for expression in V. cholerae, but the FlhF protein is required for expression of other σ54-dependent flagellar genes and consequently flagellar biosynthesis (10). In C. jejuni and H. pylori, no evidence exists to indicate that flhF expression is σ54 dependent, and therefore, expression may be constitutive or regulated by a yet-unknown factor. However, FlhF is required for wild-type-level expression of the σ54-dependent flagellar rod and hook genes in H. pylori and C. jejuni and flagellar biosynthesis (36). Therefore, polarly flagellated bacteria have acquired flhF and adapted the encoded protein to be involved at different specific steps in flagellar gene regulation and biosynthesis that behoove the individual species. Thus, exploration of the role of FlhF in diverse bacterial species will be required to fully understand the biochemical properties of FlhF and how these activities function to ensure proper flagellar biosynthesis. Our study has provided a foundation for the molecular characterization of FlhF and how biological properties of FlhF are linked to distinct steps in flagellar gene regulation and biosynthesis.
Acknowledgments
We thank Deborah Ribardo for assistance in protein purification.
This work was supported by NIH grant R01 AI065539 and by National Research Initiative grants 2006-35201-17382 and 2009-35201-05039 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bange, G., G. Petzold, K. Wild, R. O. Parlitz, and I. Sinning. 2007. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc. Natl. Acad. Sci. USA 104:13621-13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingham-Ramos, L. K., and D. R. Hendrixson. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, P. B., D. W. Hanlon, and G. W. Ordal. 1992. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol. Microbiol. 6:2705-2713. [DOI] [PubMed] [Google Scholar]
- 6.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, United States, 2005. MMWR Morb. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 8.Chevance, F. F., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 10.Correa, N. E., F. Peng, and K. E. Klose. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 12.Focia, P. J., I. V. Shepotinovskaya, J. A. Seidler, and D. M. Freymann. 2004. Heterodimeric GTPase core of the SRP targeting complex. Science 303:373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, C. R., R. M. Hoekstra, M. Samuel, R. Marcus, J. Bender, B. Shiferaw, S. Reddy, S. D. Ahuja, D. L. Helfrick, F. Hardnett, M. Carter, B. Anderson, and R. V. Tauxe. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3):S285-S296. [DOI] [PubMed] [Google Scholar]
- 14.Green, J. C. D., C. Kahramanoglou, A. Rahman, A. M. C. Pender, N. Charbonnel, and G. M. Fraser. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391:679-690. [DOI] [PubMed] [Google Scholar]
- 15.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 139:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 17.Hendrixson, D. R. 2008. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70:519-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 19.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 20.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 21.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 23.Joslin, S. N., and D. R. Hendrixson. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191:2656-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joslin, S. N., and D. R. Hendrixson. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol. 190:2422-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755-775. [DOI] [PubMed] [Google Scholar]
- 26.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 27.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 28.Kusumoto, A., K. Kamisaka, T. Yakushi, H. Terashima, A. Shinohara, and M. Homma. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J. Biochem. 139:113-121. [DOI] [PubMed] [Google Scholar]
- 29.Kusumoto, A., A. Shinohara, H. Terashima, S. Kojima, T. Yakushi, and M. Homma. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390-1399. [DOI] [PubMed] [Google Scholar]
- 30.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 31.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29:970-972. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. D., H. D. Bernstein, and P. Walter. 1994. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature 367:657-659. [DOI] [PubMed] [Google Scholar]
- 33.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nachamkin, I., X.-H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 37.Pandza, S., M. Baetens, C. H. Park, T. Au, M. Keyhan, and A. Matin. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414-423. [DOI] [PubMed] [Google Scholar]
- 38.Peluso, P., S. O. Shan, S. Nock, D. Herschlag, and P. Walter. 2001. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40:15224-15233. [DOI] [PubMed] [Google Scholar]
- 39.Powers, T., and P. Walter. 1995. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science 269:1422-1424. [DOI] [PubMed] [Google Scholar]
- 40.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 41.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 42.Shan, S. O., R. M. Stroud, and P. Walter. 2004. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2:e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siderovski, D. P., and F. S. Willard. 2005. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 1:51-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommerlad, S. M., and D. R. Hendrixson. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459-469. [DOI] [PubMed] [Google Scholar]
- 46.Stroud, R. M., and P. Walter. 1999. Signal sequence recognition and protein targeting. Curr. Opin. Struct. Biol. 9:754-759. [DOI] [PubMed] [Google Scholar]
- 47.Wassenaar, T. M., B. A. M. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 48.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 49.Wosten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 50.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]