Abstract
Paenibacillus sp. (formerly Bacillus macerans) strain JJ-1b is able to grow on 4-hydroxybenzoate (4HB) as a sole source of carbon and energy and is known to degrade 4HB via the protocatechuate (PCA) 2,3-cleavage pathway. However, none of the genes involved in this pathway have been identified. In this study, we identified and characterized the JJ-1b genes for the 4HB catabolic pathway via the PCA 2,3-cleavage pathway, which consisted of praR and praABEGFDCHI. Based on the enzyme activities of cell extracts of Escherichia coli carrying praI, praA, praH, praB, praC, and praD, these genes were found to code for 4HB 3-hydroxylase, PCA 2,3-dioxygenase, 5-carboxy-2-hydroxymuconate-6-semialdehyde decarboxylase, 2-hydroxymuconate-6-semialdehyde dehydrogenase, 4-oxalocrotonate (OCA) tautomerase, and OCA decarboxylase, respectively, which are involved in the conversion of 4HB into 2-hydroxypenta-2,4-dienoate (HPD). The praE, praF, and praG gene products exhibited 45 to 61% amino acid sequence identity to the corresponding enzymes responsible for the catabolism of HPD to pyruvate and acetyl coenzyme A. The deduced amino acid sequence of praR showed similarity with those of IclR-type transcriptional regulators. Reverse transcription-PCR analysis revealed that praABEGFDCHI constitute an operon, and these genes were expressed during the growth of JJ-1b on 4HB and PCA. praR-praABEGFDCHI conferred the ability to grow on 4HB to E. coli, suggesting that praEGF were functional for the conversion of HPD to pyruvate and acetyl coenzyme A. A promoter analysis suggested that praR encodes a repressor of the pra operon.
Protocatechuate (PCA) is one of the key intermediate metabolites in the microbial catabolic pathways for various aromatic compounds, including phthalates, hydroxybenzoates, and lignin-derived aromatic compounds such as vanillate and ferulate. It is known that the aromatic ring fission of PCA is catalyzed by one of the three distinct dioxygenases PCA 3,4-dioxygenase (26), PCA 4,5-dioxygenase (36, 41), and PCA 2,3-dioxygenase (7). In the PCA 3,4-cleavage pathway, PCA is converted into 2-carboxy-cis,cis-muconate by the reaction catalyzed by PCA 3,4-dioxygenase, and the catabolic pathway for its product (β-ketoadipate pathway) has been reported in many bacteria (24, 26). In the case of the PCA 4,5-cleavage pathway, PCA is cleaved by PCA 4,5-dioxygenase to yield 4-carboxy-2-hydroxymuconate-6-semialdehyde, and then the product is degraded to 2-pyrone-4,6-dicarboxylate, 4-oxalomesaconate, and 4-carboxy-4-hydroxy-2-oxoadipate before entering the Krebs cycle (36). The genes and enzymes involved in this pathway have been recently characterized for several bacteria, such as Sphingobium (Sphingomonas) (36), Comamonas (47), Pseudomonas (35), and Arthrobacter (13) strains. On the other hand, no genetic information on the PCA 2,3-cleavage pathway has been reported since the finding of this pathway in some bacilli (7, 8).
In 1979, Crawford et al. reported the PCA 2,3-cleavage pathway of a 4-hydroxybenzoate (4HB) degrader, Paenibacillus sp. (formerly Bacillus macerans) strain JJ-1b, which was isolated from a 50°C hot spring (9). In this pathway, PCA is initially transformed to 5-carboxy-2-hydroxymuconate-6-semialdehyde (5CHMS) by PCA 2,3-dioxygenase (Fig. 1A). 5CHMS was proposed to be subject to decarboxylation, resulting in the formation of 2-hydroxymuconate-6-semialdehyde (HMS), which is finally degraded to pyruvate and acetyl coenzyme A (9). Among these pathway enzymes, only the PCA 2,3-dioxygenase was purified and characterized in 1993 (64); however, all the genes responsible for the PCA 2,3-cleavage pathway have not yet been identified.
FIG. 1.
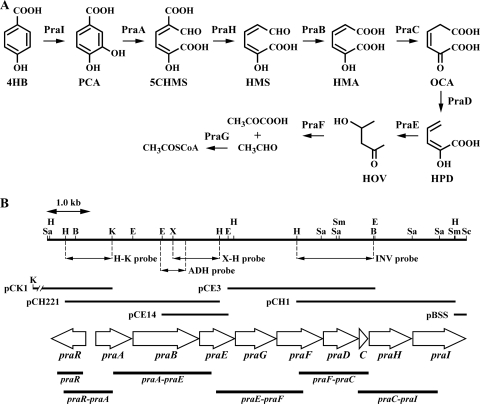
Catabolic pathway of 4HB in Paenibacillus strain sp. JJ-1b (A) and organization of the pra gene cluster (B). (A) PraI, 4HB 3-hydroxylase; PraA, PCA 2,3-dioxygenase; PraH, 5CHMS decarboxylase; PraB, HMS dehydrogenase; PraC, OCA tautomerase; PraD, OCA decarboxylase; PraE, HPD hydratase; PraF, HOV aldolase; and PraG, acetaldehyde dehydrogenase (acylating). (B) Open arrows indicate the sizes, locations, and transcriptional directions of ORFs. The locations of the cloned DNA fragments are indicated above the arrows representing the ORFs. Double-headed arrows indicate the positions of probes used in library screening. Boldface bars below the gene cluster diagram indicate the locations of the amplified RT-PCR products shown in Fig. 6. Abbreviations for restriction enzymes: B, BamHI; E, EcoRI; H, HindIII; K, KpnI; Sa, SacI; Sc, ScaI; Sm, SmaI; and X, XbaI.
In the present study, we identified and characterized the 4HB catabolic gene (pra gene) cluster, including the PCA 2,3-cleavage pathway genes, from Paenibacillus sp. strain JJ-1b. This is the first report on the identification and characterization of the PCA 2,3-cleavage pathway genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Paenibacillus sp. strain JJ-1b was grown in Luria-Bertani (LB) medium or in W minimal salt medium (44) containing 10 mM 4HB, 10 mM PCA, or 10 mM succinate at 37°C. Escherichia coli strains were grown in LB medium at 30°C or 37°C. For cultures of E. coli cells carrying the ampicillin (Ap) resistance marker, the media were supplemented with 100 mg of Ap/liter.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Paenibacillus sp. strain JJ-1b | Wild type (ATCC 35889) | 9 |
| E. coli | ||
| DH5 | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 21 |
| DH5α | Δ(lac)U169 φ80 Δ(lacZ)M15 hsdR17 endA1 gyrA96 recA1 relA1 supE44 thi-1 | 21 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | 59 |
| Plasmids | ||
| Charomid 9-36 | Apr cos | 50 |
| pBluescript II KS(+) | Cloning vector; Apr | 56 |
| pUC19 | Cloning vector; Apr | 65 |
| pT7Blue | Cloning vector; Apr T7 promoter | Novagen |
| pET21a(+) | Expression vector; Apr T7 promoter | Novagen |
| pColdIV | Expression vector; AprcspA promoter | Takara Bio |
| pQF50 | Broad-host-range transcriptional fusion vector containing a promoterless lacZ; Apr | 17 |
| pPR9TT | Broad-host-range vector containing lacZ without ATG; Apr Cmr | 53 |
| pPR9TZ | pPR9TT with a 3.6-kb SmaI-ScaI fragment containing lacZ from pQF50 replacing the 3.2-kb BamHI fragment | K. Takamura |
| pTE2G1 | pT7Blue with a 563-bp PCR-amplified fragment containing praB internal region | This study |
| pCH221 | Charomid 9-36 with a 3.4-kb HindIII fragment carrying praA and praB | This study |
| pCE14 | Charomid 9-36 with a 1.5-kb EcoRI fragment | This study |
| pCK1 | Charomid 9-36 with a 4.0-kb KpnI fragment | This study |
| pCE3 | Charomid 9-36 with a 3.3-kb EcoRI fragment | This study |
| pCH1 | Charomid 9-36 with a 3.5-kb HindIII fragment | This study |
| pBH3 | KS(+) with a 3.4-kb HindIII fragment of pCH221 | This study |
| pBE1F | KS(+) with a 1.5-kb EcoRI fragment of pCE14 | This study |
| pBSK | KS(+) with a 1.4-kb SacI-KpnI fragment of pCK1 | This study |
| pBE3F | KS(+) with a 3.3-kb EcoRI fragment of pCE3 | This study |
| pBH4F | KS(+) with a 3.5-kb HindIII fragment of pCH1 | This study |
| pTINVF | pT7Blue with a 3.3-kb PCR fragment generated by the INVF and INVR primer pair | This study |
| pBINV2H2 | KS(+) with a 2.0-kb HindIII fragment of PCR fragment generated by the INV2F and INV2R primer pair | This study |
| pBBH | KS(+) with a 1.8-kb BamHI-HindIII fragment of pBH4F | This study |
| pBSS | KS(+) with a 0.3-kb SmaI-ScaI fragment of pBINV2H2 | This study |
| pTN23 | pT7Blue with a 603-bp PCR fragment generated by the praA-F and praA-R primer pair | This study |
| pTNB | pT7Blue with a 706-bp PCR fragment generated by the praB-F and praB-R primer pair | This study |
| pTCR | pT7Blue with a 392-bp PCR fragment generated by the praC-F and praC-R primer pair | This study |
| pTDF | pT7Blue with a 817-bp PCR fragment generated by the praD-F and praD-R primer pair | This study |
| pTH2 | pT7Blue with a 155-bp PCR fragment generated by the praH-F and praH-R primer pair | This study |
| pTIR | pT7Blue with a 617-bp PCR fragment generated by the praI-F and praI-R primer pair | This study |
| pETA23 | pET21a(+) with a 0.8-kb NdeI-EcoRI fragment carrying praA | This study |
| pETB23 | pET21a(+) with a 1.9-kb NdeI-HindIII fragment carrying praB | This study |
| pETC23 | pET21a(+) with a 0.4-kb NdeI-EcoRI fragment carrying praC | This study |
| pC4D23 | pColdIV with a 0.8-kb NdeI-XhoI fragment carrying praD | This study |
| pETH23 | pET21a(+) with a 1.9-kb NdeI-HindIII fragment carrying praH | This study |
| pETI23 | pET21a(+) with a 1.4-kb NdeI-BamHI fragment carrying praI | This study |
| pETCH23 | pET21a(+) with a 2.2-kb NdeI-HindIII fragment carrying praC and praH | This study |
| pBRI23 | KS(+) with a 9.4-kb SacI-ScaI fragment carrying pra genes | This study |
| pPZAR | pPR9TZ with a 1.3-kb SacI-SacII fragment carrying praR and praA promoter region | This study |
| pPZA | pPR9TZ with a 0.6-kb PstI-SacII fragment carrying the praA promoter region | This study |
Apr and Cmr, resistance to ampicillin and chloramphenicol, respectively.
Cloning of the pra genes.
The E2GF and E2GR primer set (see Table S1 in the supplemental material) was used to amplify an aldehyde dehydrogenase gene sequence in JJ-1b. A 563-bp PCR-amplified fragment was used for colony hybridization as a probe to isolate the PCA 2,3-cleavage pathway genes from JJ-1b gene libraries, which were constructed using charomid 9-36 (50) with HindIII digests of the JJ-1b total DNA. The flanking sequences of the 3.4-kb HindIII fragment of pCH221 were obtained by using the 1.0-kb HindIII-KpnI fragment and the 1.0-kb XbaI-HindIII fragment as probes as shown in Fig. 1B. Colony and Southern hybridizations were performed using the digoxigenin system (Roche, Mannheim, Germany).
An inverse PCR was employed to amplify a DNA fragment containing the downstream region of pCE14 using the INVF and INVR primer set. Total DNA of JJ-1b was digested with BamHI and self-ligated, and then the ligation mixture was used as a template for the inverse PCR. The 1.7-kb HindIII-BamHI fragment of the amplified product was used for colony hybridization as a probe (INV probe shown in Fig. 1B) to isolate the 3.3-kb EcoRI fragment (pCE3) and the 3.5-kb HindIII fragment (pCH1) from gene libraries of JJ-1b as shown in Fig. 1B. The INV2F and INV2R primer set was designed for inverse PCR to amplify the region further downstream. Using BamHI-digested and self-ligated total DNA as a template, the DNA fragment was amplified. The 261-bp SmaI-ScaI fragment of the amplified region was cloned (pBSS) and sequenced. The nucleotide sequences of the primer sets are shown in Table S1 in the supplemental material.
DNA manipulations and nucleotide sequencing.
DNA manipulations, including total DNA isolation, 16S rRNA gene amplification, construction of deletion derivatives, and nucleotide sequencing, were performed as described in previous studies (30, 54). Analysis of nucleotide sequences was performed using the MacVector software (MacVector, Inc., Cary, NC). Homology searches, pairwise alignment, and ClustalW multiple-sequence alignment were performed as described previously (1, 29). Analysis of the 16S rRNA gene was performed using the Ribosomal Database Project release 10.10 online server (6). A distance matrix and phylogenetic trees were constructed by using the neighbor-joining method (51) and were visualized with the FigTree program (version 1.2; http://tree.bio.ed.ac.uk/software/figtree/).
Construction of expression plasmids of the pra genes.
praA, praB, praC, praD, praH, and praI were PCR amplified and subcloned into pT7Blue. In order to achieve ligation at the NdeI site of the expression vector, all of these genes were amplified with a forward primer designed to contain the NdeI site at its own ATG start codon (see Table S1 in the supplemental material). After the sequences were confirmed, each praA, praB, praC, praH, and praI fragment was ligated to an expression vector, pET21a(+), to form pETA23, pETB23, pETC23, pETH23, and pETI23, respectively. The praD fragment was ligated to pColdIV to construct pC4D23. To construct the praCH coexpression plasmid, pETCH23, the 1.8-kb EcoRI-HindIII fragment carrying praH was ligated into pETC23 digested with the same restriction enzymes.
Expression of the pra genes in E. coli.
The expression plasmids were introduced into E. coli BL21(DE3) cells. BL21(DE3) cells harboring pETA23 were grown in 10 ml of LB medium containing 100 mg of Ap/liter at 37°C, and the cells harboring pETB23, pETC23, pETH23, pETI23, and pETCH23 were grown in the same medium at 30°C. Expression of praA was induced for 4 h by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the absorbance of the culture at 600 nm (A600) reached 0.5. Expression of praB, praC, praH, and praI and coexpression of praC and praH were induced by adding 1 mM IPTG, and then growth was continued for 4 h. BL21(DE3) cells harboring pC4D23 were grown in 10 ml LB medium containing Ap at 37°C until the A600 reached 0.5, and the culture was placed at 15°C for 30 min and cultivated again at 15°C for 24 h after the addition of 0.1 mM IPTG. Cells were harvested, resuspended in 50 mM Tris-HCl buffer (pH 7.3), broken with an ultrasonic disintegrator (UD-201; Tomy Seiko Co., Tokyo, Japan), and centrifuged at 15,000 × g for 15 min. The resulting supernatants were used as crude enzymes.
Enzyme assays. (i) PCA 2,3-dioxygenase.
PCA 2,3-dioxygenase activity was assayed by measuring the substrate-dependent oxygen consumption rate. A 2-ml assay mixture contained 50 mM GTA buffer (pH 7.3) consisting of 50 mM 3,3-dimethylglutarate, 50 mM Tris, 50 mM 2-amino-2-methyl-1,3-propanediol, crude extract (2.5 μg of protein), and 100 μM PCA. The reaction mixture was incubated at 35°C, and the oxygen consumption rate was determined with an oxygen electrode (B-505; Iijima Electronics Manufacturing Co., Ltd., Aichi, Japan). One unit of enzyme activity was defined as the amount of enzyme that resulted in consumption of 1 μmol of O2 per min at 35°C. Specific activity was expressed in units per milligram of protein. PCA 2,3-dioxygenase activity was also monitored using a DU-7500 spectrophotometer (Beckman Coulter, Fullerton, CA). The reaction mixture (final volume, 1 ml) containing 50 mM Tris-HCl buffer (pH 7.3), 50 μM PCA, and crude extract (2.5 μg of protein) was preincubated without the substrate for 1 min at 35°C, and then the reaction was started by adding PCA.
For gas chromatography-mass spectrometry (GC-MS) analysis, the reaction mixture was acidified with 6 N hydrochloric acid to pH 2 and extracted with ethyl acetate. The extract was trimethylsilylated with the TMSI-H reagent (hexamethyldisilazane-trimethylchlorosilane-pyridine [2:1:10]; GL Science Inc., Tokyo, Japan) according to the procedure recommended by the manufacturer. The resulting trimethylsilyl (TMS) derivatives were analyzed by GC-MS.
(ii) 5CHMS decarboxylase.
5CHMS decarboxylase activity was assayed spectrophotometrically by monitoring the production of HMS from 5CHMS using a preassay mixture that consisted of 50 μM PCA and crude PraA (2.5 μg of protein) in 50 mM Tris-HCl buffer (pH 7.3) in a total volume of 900 μl. The mixture was incubated for 1 min at 35°C. After the reaction was completed, crude PraH (10 μg of protein) was added in a final volume of 1 ml, and then the decrease in the absorbance at 350 nm and the increase in the absorbance at 375 nm were monitored at 35°C.
(iii) HMS dehydrogenase.
HMS dehydrogenase activity was determined spectrophotometrically by monitoring the conversion of HMS to 2-hydroxymuconate (HMA) using a preassay mixture that consisted of 1 mM PCA and crude PraA (4 μg of protein) in 50 mM Tris-HCl buffer (pH 7.3) in a total volume of 50 μl. The reaction mixture was incubated for 30 min at 35°C, and the increase in the absorbance at 375 nm due to the formation of HMS from PCA was monitored. After the reaction was completed, crude PraB (25 μg of protein) and 50 μM NAD+ were added to the mixture. The reaction was carried out in a 1-ml reaction mixture at 35°C. For GC-MS analysis, the reaction mixture was acidified, extracted, and trimethylsilylated.
To determine the specific activity, 1 ml of reaction mixture containing 50 mM Tris-HCl buffer (pH 7.3), 50 μM HMS, crude PraB (25 μg of protein), 1.2 mM pyruvate, 1.0 U lactate dehydrogenase, and 500 μM NAD(P)+ was incubated at 35°C, and the decrease in the absorbance at 375 nm was monitored. The specific activity was calculated from the initial rates by using a molar extinction coefficient of 35,000 M−1 cm−1 for HMS. One unit of HMS dehydrogenase activity was defined as the amount of enzyme that converted 1 μmol of HMS per minute under the assay conditions used.
(iv) OCA tautomerase.
4-Oxalocrotonate (OCA) tautomerase activity was determined spectrophotometrically by measuring the formation of OCA at 236 nm (25). The reaction was carried out in a 1-ml reaction mixture containing 50 mM Tris-HCl buffer (pH 7.3), 50 μM NAD+, 50 μl of the PraA reaction mixture, crude PraB (25 μg of protein), and crude PraC (25 μg of protein) at 35°C.
(v) OCA decarboxylase.
OCA decarboxylase activity was determined by measuring the production of 2-hydroxypenta-2,4-dienoate (HPD) by high-performance liquid chromatography (HPLC) (Acquity Ultra Performance LC; Waters, Milford, MA). The assay was performed by using a preassay mixture that consisted of 50 μl of the PraA reaction mixture, 50 μM NAD+, crude PraB (25 μg of protein/ml), and crude PraC (25 μg of protein/ml) in 50 mM Tris-HCl buffer (pH 7.3) in a total volume of 990 μl. The reaction mixture was incubated for 30 min at 35°C; crude PraD (25 μg of protein) and 500 μM MgSO4 were then added to the mixture to obtain a final volume of 1 ml. The reaction mixture was further incubated at 35°C for 10 min, and the enzyme reaction was stopped by addition of methanol (final concentration, 25%). The stopped reaction mixture was then subjected to HPLC (Acquity Ultra Performance LC; Waters, Milford, MA) analysis.
(vi) 4HB 3-hydroxylase.
4HB 3-hydroxylase activity was measured spectrophotometrically by monitoring the decrease in the absorbance at 340 nm derived from the consumption of NADH or NADPH at 35°C. The assay mixture (final volume, 1 ml) contained 50 mM Tris-HCl buffer (pH 8.0), 200 μM NAD(P)H, 500 μM EDTA, 10 μM flavin adenine dinucleotide (FAD), 1 mM 4HB, and crude PraI (200 μg of protein). The assay mixture was preincubated without the substrate for 1 min at 35°C, and the reaction was started by adding 4HB. The specific activity was calculated from the initial rates by using molar extinction coefficients of 6,600 and 5,070 M−1 cm−1 for NADH and NADPH, respectively (37). One unit of 4HB 3-hydroxylase activity was defined as the amount of enzyme that consumed 1 μmol of NADH or NADPH per minute under the assay conditions used.
For GC-MS analysis, the assay was carried out in a 1-ml reaction mixture containing 50 mM Tris-HCl buffer (pH 8.0), 1 mM NADH, 500 μM EDTA, 10 μM FAD, 100 μM 4HB, and crude PraI (200 μg of protein). After the reaction, the reaction mixture was acidified, extracted, trimethylsilylated, and analyzed by GC-MS.
RNA preparation and reverse transcription-PCR (RT-PCR) analysis.
JJ-1b cells were grown in W minimal salt medium supplemented with 10 mM 4HB, PCA, or succinate at 37°C. When the A600 reached about 0.8, the cells were harvested by centrifugation at 5,000 × g at 4°C for 10 min. Total RNA was isolated with Isogen (Nippon Gene, Toyama, Japan) and then treated with DNase I (Takara Bio Inc.). Single-stranded cDNA was synthesized from 5.0 μg of total RNA by using PrimeScript reverse transcriptase (Takara Bio Inc.) with random primers in a 20-μl reaction mixture. PCR amplification was performed with a 1.0 μl of the cDNA mixture, specific primers (see Table S1 in the supplemental material), and Ex Taq DNA polymerase (Takara Bio Inc.). A control without reverse transcriptase was used for each reaction to verify the absence of genomic DNA contamination. PCR samples were electrophoresed on a 0.8% agarose gel and visualized with ethidium bromide.
Measurement of the promoter activity.
The 1.3-kb SacI-SacII fragment carrying praR and the potential pra operon promoter was cloned into the promoter-probe vector pPR9TZ to obtain pPZAR. The 0.6-kb PstI-SacII fragment was also cloned into pPR9TZ to create pPZA carrying only the potential pra operon promoter. Expression of the lacZ reporter gene was determined by β-galactosidase (β-Gal) assays performed as follows. E. coli DH5α cells harboring pPZAR or pPZA were grown in 10 ml of LB medium either with or without 10 mM 4HB or PCA at 37°C. After 12 h, cells were harvested and resuspended in Z buffer, consisting of 50 mM sodium phosphate buffer (pH 7.0), 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol. Toluene treatment and β-Gal assays using o-nitrophenyl-β-d-galactopyranoside were performed as described by Miller (38). The values presented are the averages and standard deviations (error bars) from at least three independent experiments.
Analytical methods.
The protein concentration was determined by the method of Bradford (5). The sizes of the proteins expressed in E. coli were examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE). The proteins in the gels were stained with Coomassie brilliant blue R-250. To determine the N-terminal amino acid sequence, the crude extract was subjected to SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). The enzyme band was cut out and analyzed on a Procise 494 HT protein sequencing system (Applied Biosystems, Foster City, CA). GC-MS analysis was performed with a model 5971A instrument equipped with an Ultra-2 capillary column (50 m by 0.2 mm; Agilent Technologies Co., Palo Alto, CA). The analytical conditions were the same as those described previously (29). HPLC analysis was performed with an Acquity Ultra Performance LC (Waters) equipped with a TSKgel ODS-140HTP column (2.1 by 100 mm; Tosoh, Tokyo, Japan). The mobile phase was 1% (vol/vol) acetonitrile in water containing 0.1% (vol/vol) phosphoric acid, and the flow rate was 0.5 ml/min. HMA, OCA, and HPD were detected at 303, 228, and 272 nm, respectively.
Nucleotide sequence accession numbers.
The nucleotide sequence reported in this paper has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB505863 and AB505864.
RESULTS
16S rRNA gene analysis of JJ-1b.
The nucleotide sequence of the 16S rRNA gene of JJ-1b showed significant similarity to those of Paenibacillus validus 197 (99.2% identity, 1,456-bp overlap; accession no. EU730934), Paenibacillus sp. strain Ao3 (97.0% identity, 1,515-bp overlap; EF208754), Paenibacillus sp. strain HM1 (96.9% identity, 1,515-bp overlap; AY283261), and Paenibacillus validus JCM 9077 (96.8% identity, 1,487-bp overlap; AB073203). Accordingly, we propose to rename Bacillus macerans JJ-1b as Paenibacillus sp. strain JJ-1b.
Cloning and nucleotide sequences of the PCA 2,3-cleavage pathway genes.
In order to isolate the PCA 2,3-cleavage pathway genes from JJ-1b, we first attempted to isolate the HMS dehydrogenase gene. Based on the sequence similarity among putative 5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenases from Bacillus cereus NVH 391-98 (ABS21296) and Geobacillus kaustophilus HTA426 (BAD77313) and a putative aldehyde dehydrogenase (AAU25432) from Bacillus licheniformis ATCC 14580, a primer set (E2GF and E2GR) was designed and used to amplify a 563-bp DNA fragment from total DNA of JJ-1b. The nucleotide sequence of this fragment showed 67% identity with that of AAU25432. The resulting PCR fragment was then used as a probe to screen a JJ-1b genomic library, and a positive clone, pCH221, carrying the 3.4-kb HindIII fragment was obtained. The nucleotide sequence of the 3.4-kb HindIII fragment was determined, and two open reading frames (ORFs), named praA and praB, were found. The homology search revealed that the praA and praB gene products showed similarity with homoprotocatechuate 2,3-dioxygenases and HMS dehydrogenases, respectively (Table 2). To acquire the flanking region of the 3.4-kb HindIII fragment, colony hybridization and inverse PCR were performed. As a result, five clones, pCK1, pCE14, pCE3, pCH1, and pBSS, were obtained (Fig. 1B). Using these clones, the nucleotide sequence of the 9,354-bp region was determined. In this region, 10 ORFs containing praA and praB were found. Nine of these ORFs are transcribed in the same direction; the exception is praR, which appeared to encode a putative transcriptional regulator. The gene products of praE, praG, praF, praD, praC, and praI showed similarity with HPD hydratase, acetaldehyde dehydrogenase (acylating), 4-hydroxy-2-oxovalerate (HOV) aldolase, OCA decarboxylase, OCA tautomerase, and 4HB 3-hydroxylase, respectively (Table 2). PraH showed similarity with 2-amino-3-carboxymuconate-6-semialdehyde (ACMS) decarboxylase (NbaD) from Pseudomonas fluorescens KU-7 (BAC65312), suggesting that praH encodes 5CHMS decarboxylase (Table 2).
TABLE 2.
Identification of pra gene functions
| Gene | Deduced molecular mass, Da (no. of amino acid residues) | Representative homolog | Identity (%)a | Accession no. |
|---|---|---|---|---|
| praR | 28,947 (258) | IclR-type transcriptional regulator from Bacillus licheniformis ATCC 14580 | 49 | AAU25433 |
| IclR-type transcriptional regulator (PcaU) from Acinetobacter baylyi ADP1 | 25 | AAC37157 | ||
| praA | 29,636 (266) | Hypothetical protein from B. licheniformis ATCC 14580 | 53 | AAU25429 |
| HPCA 2,3-dioxygenase (HpaD) from Pseudomonas sp. strain DJ-12 | 29 | AAL28115 | ||
| HPCA 2,3-dioxygenase (HpaD) from Burkholderia xenovorans LB400 | 28 | ABE33954 | ||
| HPCA 2,3-dioxygenase (HpcB) from Escherichia coli C | 26 | CAA38985 | ||
| praB | 53,188 (486) | Aldehyde dehydrogenase from B. licheniformis ATCC 14580 | 74 | AAU25432 |
| HMS dehydrogenase (NahI) from Pseudomonas putida G7 (NAH7) | 55 | BAE92168 | ||
| HMS dehydrogenase (XylG) from P. putida mt-2 (pWW0) | 54 | AAA26053 | ||
| praE | 27,687 (260) | HPD hydratase (BphH) from B. xenovorans LB400 | 52 | ABE37051 |
| HPD hydratase (NbaH) from Pseudomonas fluorescens KU-7 | 45 | BAC65306 | ||
| praG | 31,466 (294) | Acetaldehyde dehydrogenase (NahO) from Bacillus sp. strain JF8 | 61 | BAD08310 |
| Acetaldehyde dehydrogenase (NbaJ) from P. fluorescens KU-7 | 53 | BAC65307 | ||
| praF | 36,116 (337) | HOV aldolase (NahM) from Bacillus sp. strain JF8 | 58 | BAD08311 |
| HOV aldolase (XylK) from P. putida mt-2 (pWW0) | 51 | AAA25692 | ||
| praD | 28,125 (260) | Putative OCA decarboxylase from B. licheniformis ATCC 14580 | 64 | AAU25431 |
| OCA decarboxylase (XylI) from P. putida mt-2 (pWW0) | 46 | AAA25693 | ||
| praC | 7,012 (63) | Putative OCA tautomerase from B. licheniformis ATCC 14580 | 64 | AAU25430 |
| OCA tautomerase (DmpI) from Pseudomonas sp. strain CF600 | 49 | CAA43229 | ||
| praH | 35,570 (313) | Putative decarboxylase from B. licheniformis ATCC 14580 | 58 | AAU25434 |
| ACMS decarboxylase (NbaD) from P. fluorescens KU-7 | 24 | BAC65312 | ||
| praI | 44,262 (394) | Putative 4HB 3-hydroxylase from B. licheniformis ATCC 14580 | 66 | AAU25427 |
| 4HB 3-hydroxylase (PobA) from Arthrobacter sp. strain ATCC 51369 | 45 | BAE46572 |
Percent identity obtained by aligning the deduced amino acid sequences by use of the EMBOSS alignment tool.
Identification of PraA as PCA 2,3-dioxygenase.
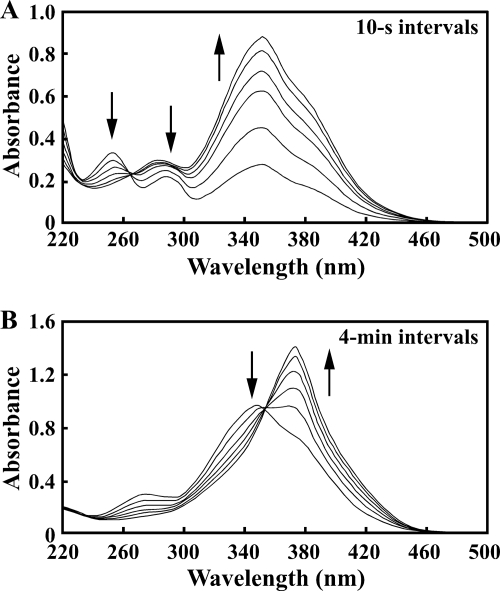
The praA gene was expressed in E. coli harboring pETA23, which carries praA in pET21a(+). Production of a 31-kDa protein in E. coli BL21(DE3) cells harboring pETA23 was observed by SDS-PAGE (see Fig. S1 in the supplemental material). Absorption spectral analysis revealed that PCA (λmax = 254 and 290 nm) was converted by crude PraA into a product having a spectrum with a maximum at 350 nm (Fig. 2A). This spectrum is characteristic of 5CHMS (9). Therefore, it was strongly suggested that praA encodes PCA 2,3-dioxygenase. The specific activity of crude PraA toward PCA was determined to be 32.9 ± 1.8 U/mg. When the reaction mixture was incubated for 20 min, a decrease in the absorbance at 350 nm and an increase in the absorbance at 375 nm were observed (Fig. 2B). According to previous studies (9, 52), this spectrum is characteristic of HMS.
FIG. 2.
Conversion of PCA to 5CHMS catalyzed by PraA. (A) The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.3), 50 μM PCA, and crude PraA (2.5 μg of protein), and it was incubated at 35°C in a final volume of 1 ml. UV-visible spectra were recorded at intervals of 10 s. (B) The reaction mixture was the same as described above, but UV-visible spectra were recorded at intervals of 4 min.
The PraA reaction mixture was analyzed by GC-MS. After 10 min of reaction, compound I, with a retention time of 18.5 min, was produced (see Fig. S2C in the supplemental material). The weight of the molecular ion of compound I (m/z 286) corresponded to that of TMS derivative of HMS. The major fragments at m/z 271, 257, 197, and 169 corresponded to M-CH3, M-CHO, M-OTMS, and M-COOTMS, respectively (see Fig. S2D in the supplemental material). These results indicated that HMS was formed from PCA by the consecutive reactions of 2,3-ring cleavage and spontaneous decarboxylation. The accumulation of 5CHMS was not observed under these conditions, suggesting that 5CHMS was converted to HMS during the extraction and/or analysis process.
Identification of PraH as 5CHMS decarboxylase.
The nucleotide sequence analysis predicted the presence of two consecutive ATG codons, which are possible praH start codons. The N-terminal amino acid sequence of PraH produced in E. coli harboring pETCH23, which carries praCH, was determined to be MYDVH; therefore, the second methionine was chosen as the start codon of praH to construct the praH expression plasmid, pETH23. SDS-PAGE analysis indicated the production of a 34-kDa protein in E. coli BL21(DE3) harboring pETH23 (see Fig. S1 in the supplemental material). 5CHMS produced from 50 μM PCA by the incubation with crude PraA was reacted with crude PraH. As shown in Fig. 3A, 5CHMS (λmax = 350 nm) was converted into a product having a spectrum with a maximum at 375 nm after 1 min. On the other hand, 5CHMS was never degraded under the same conditions without PraH (data not shown). These results strongly suggested that praH encodes 5CHMS decarboxylase.
FIG. 3.
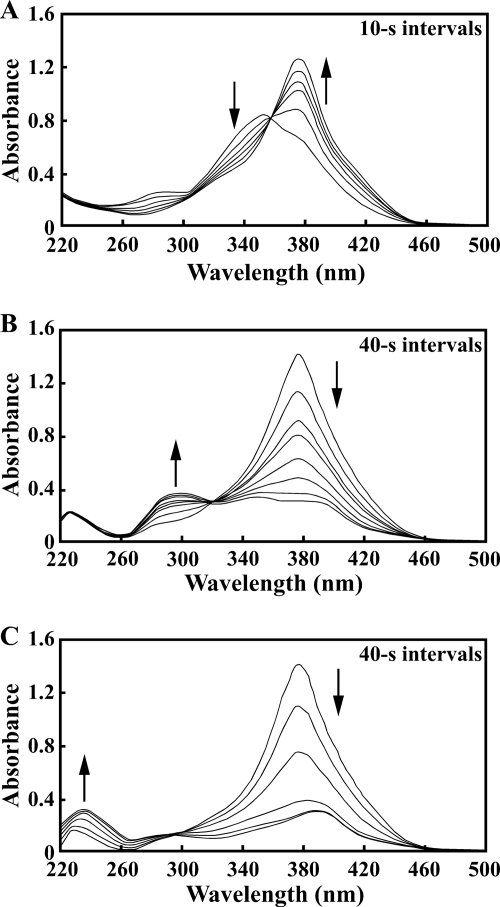
Spectral changes associated with the transformations of 5CHMS to HMS (A), HMS to HMA (B), and HMS to OCA (C). (A) The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.3), 50 μM PCA, crude PraA (2.5 μg of protein/ml), and crude PraH (10 μg of protein/ml). (B) The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.3), 50 μM HMS, 50 μM NAD+, and crude PraB (25 μg of protein/ml). (C) The reaction mixture contained 50 mM Tris-HCl buffer (pH 7.3), 50 μM HMS, 50 μM NAD+, crude PraB (25 μg of protein/ml), and crude PraC (25 μg of protein/ml). UV-visible spectra were recorded at intervals of 10 s (A) and 40 s (B and C).
Characterization of the praB, praC, and praD gene products.
The praB, praC, and praD genes were expressed in E. coli BL21(DE3) cells harboring pETB23, pETC23, and pC4D23, respectively. In SDS-PAGE analysis, the molecular masses of the products of praB, praC, and praD were estimated to be 58, 6, and 31 kDa, respectively (see Fig. S1 in the supplemental material).
The HMS dehydrogenase activity was assayed with crude PraB in the presence of 50 μM HMS, which was produced by the reaction catalyzed by PraA and spontaneous decarboxylation. The activity of crude PraB for HMS in the presence of NAD+ (1.19 ± 0.07 U/mg of protein) was about 11 times higher than that achieved with NADP+ (0.113 ± 0.009 U/mg of protein), indicating that PraB is highly specific for NAD+. Absorption spectral analysis revealed that HMS (λmax = 375 nm) was converted into a product having a spectrum with a maximum at 295 nm (Fig. 3B). This spectrum is characteristic of HMA (25, 52). When the reaction mixture was analyzed by GC-MS, the abundance of the TMS derivative of HMS decreased significantly at 5 min, and the accumulation of compound II with a retention time of 26.5 min was observed (see Fig. S3B and C in the supplemental material). The weight of the molecular ion of compound II (m/z 374) corresponded to that of the TMS derivative of HMA (see Fig. S3D in the supplemental material). These observations indicated that praB encodes HMS dehydrogenase.
During the transformation of HMS to HMA by the reaction catalyzed by PraB, crude PraC was added to the reaction mixture to allow the sequential reaction to occur. Absorption spectral analysis revealed that HMS was converted into a product having a spectrum with a maximum at 236 nm (Fig. 3C). This spectrum is characteristic of OCA (25); thus, HMA seemed to be converted to OCA by a tautomerization catalyzed by PraC.
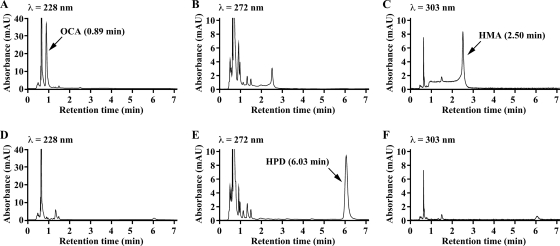
Crude PraD was added to the PraC reaction mixture. When the mixture was analyzed by HPLC immediately after the start of the reaction, peaks with retention times of 0.89 and 2.50 min were observed (Fig. 4A to C). These peaks showed a spectrum with maxima at 228 and 303 nm, respectively. Therefore, these peaks appeared to be OCA and HMA, respectively, that were formed by keto-enol tautomerization. After 10 min of incubation, a product having a spectrum with a maximum at 272 nm was formed in significant amounts with a retention time of 6.03 min (Fig. 4D to F). Since the absorption maximum at 272 nm at pH 2.0 is characteristic of HPD (52), it was strongly suggested that this compound was HPD. Harayama and coworkers reported that OCA but not HMA was the substrate for OCA decarboxylase (25). Therefore, PraD appeared to catalyze the decarboxylation of OCA to produce HPD.
FIG. 4.
HPLC chromatograms of the reaction product from OCA catalyzed by PraD. The reaction mixture contained the PraC reaction mixture, 500 μM MgSO4, and crude PraD (25 μg of protein/ml) in 50 mM Tris-HCl buffer (pH 7.3) in a total volume of 1 ml. The PraC reaction mixture was prepared as described in Materials and Methods. The reaction mixtures were analyzed by HPLC at the start (A to C) and after 10 min (D to F) of incubation. Compounds were monitored at 228 nm (A and D), 272 nm (B and E), and 303 nm (C and F).
Identification of PraI as 4HB 3-hydroxylase.
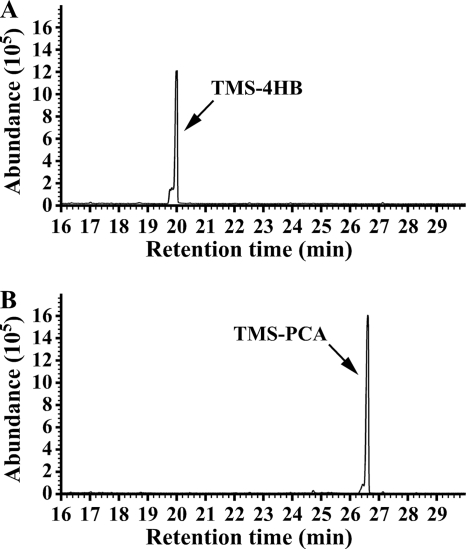
The praI gene was expressed in E. coli BL21(DE3) cells harboring pETI23. In SDS-PAGE analysis, the molecular mass of the praI product was estimated to be 41 kDa (see Fig. S1 in the supplemental material). The specific activities of crude PraI for 4HB in the presence of NADH and NADPH were determined to be 0.47 ± 0.01 and 0.36 ± 0.03 U/mg, respectively. These results indicated that PraI is able to utilize both NADH and NADPH as cofactors. GC-MS analysis of the PraI reaction mixture showed the disappearance of 4HB, and the formation of a compound with a retention time of 26.6 min was observed (Fig. 5). This compound was identified as PCA based on the comparison of retention times and mass fragmentation patterns with those of authentic PCA (data not shown). Thus, praI was concluded to encode 4HB 3-hydroxylase.
FIG. 5.
Conversion of 4HB to PCA catalyzed by PraI. The reaction mixture contained 50 mM Tris-HCl buffer (pH 8.0), 1 mM NADH, 500 μM EDTA, 10 μM FAD, 100 μM 4HB, and crude PraI (200 μg of protein/ml). Gas chromatograms of TMS derivatives of the reaction products at the start (A) and after 10 min (B) of incubation are shown.
E. coli acquires the ability to grow on 4HB by introduction of the pra gene cluster.
To test whether or not pra genes confer the ability to grow on 4HB to E. coli, DNA fragments derived from pBSK, pBH3, pBE1F, pBE3F, pBH4F, and pBSS were ligated into pBluescript II KS(+) as shown in Fig. S4 in the supplemental material. The resulting plasmid, pBRI23 carrying praR-praABEGFDCHI, was introduced into E. coli BL21(DE3) cells, and then the transformant was inoculated on M9 minimal salt medium containing 10 mM 4HB instead of glucose as the sole carbon source. The transformant obtained the ability to grow on 4HB within 10 days (see Fig. S4 in the supplemental material). This implies that praR-praABEGFDCHI is the complete gene set for the conversion of 4HB into pyruvate and acetyl coenzyme A.
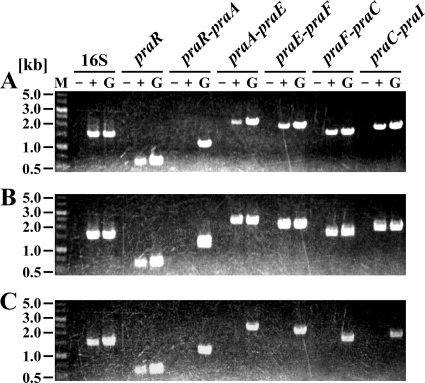
RT-PCR analysis of the pra genes.
To define the operon structure of the pra genes, RT-PCR analysis was carried out with total RNA harvested from JJ-1b cells grown on 4HB or PCA. RT-PCR amplification products of the expected sizes were detected for genes praA-praE, praE-praF, praF-praC, praC-praI, and praR (Fig. 6A and B). However, no transcript between praR and praA was observed. These results suggested that praABEGFDCHI are organized in the same transcriptional unit, whereas praR is transcribed separately. As shown in Fig. 6C, amplification products for the pra catabolic genes were not evident when RNA from succinate-grown cells was used. This indicates that the growth of JJ-1b on 4HB and PCA leads to an increase in the transcription of the pra catabolic operon.
FIG. 6.
RT-PCR analysis of the pra gene cluster in JJ-1b. Total RNA used for cDNA synthesis was isolated from JJ-1b cells grown on 4HB (A), PCA (B), and succinate (C). Agarose gel electrophoresis of RT-PCR assays with primers targeting praR (expected size, 572 bp), praR-praA (expected size, 1,092 bp), praA-praE (expected size, 2,190 bp), praE-praF (expected size, 1,940 bp), praF-praC (expected size, 1,553 bp), and praC-praI (expected size, 1,779 bp) are shown. Positions of primer pairs and primer sequences are indicated in Fig. 1B and in Table S1 in the supplemental material, respectively. Lane M, molecular weight markers; lanes + and −, RT-PCR with and without reverse transcriptase, respectively; lanes G, control PCR with the JJ-1b genomic DNA. RT-PCR of the 16S rRNA gene was used as a control to confirm equivalent quantities of template loading.
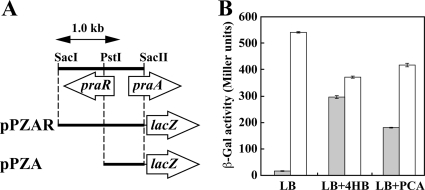
Transcriptional regulation of the pra operon.
PraR is similar in sequence to transcriptional regulators belonging to the IclR family from B. licheniformis ATCC 14580 (AAU25433) and Acinetobacter baylyi ADP1 (PcaU; AAC37157) (Table 2). To investigate whether PraR plays a role in the transcriptional regulation of pra genes, the level of expression of the praR-praA′-lacZ fusion in E. coli DH5α cells harboring pPZAR and exposed to 4HB or PCA was examined. β-Gal activity was increased 19-fold and 12-fold in the presence of 4HB and PCA, respectively (Fig. 7). These results suggested that both 4HB and PCA acted as inducers for the pra catabolic operon. E. coli cells harboring pPZA, which includes a praA upstream region but not praR, showed constitutive expression (Fig. 7), suggesting that PraR is a transcriptional repressor of the pra operon.
FIG. 7.
Regulation of the pra operon promoter activity by PraR. (A) Schematic representation of pPZAR and pPZA. (B) Promoter activities of the pra operon were measured in E. coli cells harboring pPZAR (shaded bars) or pPZA (open bars). These cells were grown in LB medium with or without 10 mM 4HB or PCA. Each value is the average ± standard deviation (error bars) based on at least three independent experiments.
DISCUSSION
In the present study, we identified the PCA 2,3-cleavage pathway genes involved in the 4HB catabolism of Paenibacillus sp. strain JJ-1b. The proposed 4HB catabolic pathway and the corresponding genes for the enzymes of JJ-1b are indicated in Fig. 1A.
4HB is transformed to PCA by the reaction catalyzed by PraI, which utilizes both NADH and NADPH as electron donors. 4HB 3-hydroxylases can be divided into two groups on the basis of the coenzyme specificity: (i) NADPH-specific and (ii) NAD(P)H-dependent 4HB 3-hydroxylases. A phylogenetic analysis of 4HB 3-hydroxylases indicated that NADPH-specific and NAD(P)H-dependent enzymes are located in the different branches (see Fig. S5A in the supplemental material). PraI forms a cluster with NAD(P)H-dependent enzymes. The deduced amino acid sequence alignment of PraI and NADPH-specific 4HB 3-hydroxylases indicated that Tyr38 and Arg42, which are essential for binding with NADPH (15, 16), are replaced by glutamate and threonine residues, respectively (see Fig. S5B in the supplemental material). These residues are highly conserved in the NAD(P)H-dependent 4HB 3-hydroxylases and are required for the recognition of NADH (15, 28).
PCA 2,3-dioxygenase had been previously purified and biochemically characterized (64), but the gene sequence has been unavailable. The praA gene product shares amino acid sequence similarity with extradiol dioxygenases. Extradiol dioxygenases have been divided into three families on the basis of amino acid sequence similarity (63). Type I extradiol dioxygenases belong to the vicinal oxygen superfamily, including a number of 2,3-dihydroxybiphenyl 1,2-dioxygenases (3, 27) and catechol 2,3-dioxygenases (40). Type II dioxygenases include several homoprotocatechuate 2,3-dioxygenases (49), the β subunit of PCA 4,5-dioxygenase (41, 47), gallate dioxygenase (30, 42), and 2-aminophenol 1,6-dioxygenase (61). Type III dioxygenases belong to the cupin superfamily, which includes enzymes such as the gentisate dioxygenase, homogentisate dioxygenase, and 1-hydroxy-2-naphthoate dioxygenase (12). These three types of enzymes have similar active sites and the same iron ligand, two histidines, and one glutamate (2-His-1-carboxylate structural motif), in spite of their different primary structures (31). As can be seen in the phylogenetic relationships of type II extradiol dioxygenases (see Fig. S6 in the supplemental material), PraA obviously belongs to the type II extradiol dioxygenase family and is closely related to a hypothetical protein (BL03909) of B. licheniformis ATCC 14580. However, the sequence of PraA formed a deep branch with those of the β subunit of PCA 4,5-dioxygenase, suggesting that they independently evolved from a common ancestor. Crystallographic studies revealed that the active site of the β subunit of PCA 4,5-dioxygenase (LigB) contains a nonheme iron coordinated by His12, His61, and Glu242, and His195 is thought to act as an active-site base to facilitate deprotonation of the hydroxyl group of the substrate (60). These amino acids are conserved among almost all the type II extradiol dioxygenases (14). The alignment of the deduced amino acid sequences of PraA and LigB revealed the presence of residues His11, His53, His180, and Glu227 of PraA, corresponding to His12, His61, His195, and Glu242, respectively, of LigB. These residues seemed to be involved in the roles described above.
When PCA was incubated with crude PraA, 5CHMS produced from PCA underwent spontaneous decarboxylation to HMS. However, no production of 5CHMS from PCA was observed in the reaction mixture containing the crude extract of JJ-1b (9). These facts suggested that 5CHMS decarboxylase encoded by praH plays a role in the decarboxylation of 5CHMS to HMS in JJ-1b cells. Based on the deduced amino acid sequence similarity, PraH seemed to be included in the metallo-dependent hydrolase (amidohydrolase_2) superfamily (33). This family includes ACMS decarboxylase from bacteria, animals, and humans (19, 39, 62); uracil-5-carboxylate decarboxylase of Neurospora crassa OR74A (57); the γ-resorcylate decarboxylase GraF from Rhizobium sp. strain MTP-10005 (66, 67); the 5-carboxyvanillate decarboxylase LigW (46); the biphenyl meta-cleavage compound hydrolase LigY (45); and the 4-oxalomesaconate hydratase LigJ (22) from Sphingobium sp. strain SYK-6. A phylogenetic analysis based on the amino acid sequences of the metallo-dependent hydrolase superfamily clearly indicated that PraH belongs to this superfamily but does not form a cluster with the other known members (see Fig. S7 in the supplemental material). It has been indicated for ACMS decarboxylase from P. fluorescens (NbaD) that the active-site Zn ion is directly coordinated by His9, His11, His177, Asp294, and the water molecule which is hydrogen bonded with His228 (32, 34). These residues are essential for the ACMS decarboxylase activity. In the case of PraH, His5, His7, His173, His222, and Asp288 seemed to be involved in the coordination of the Zn ion.
praE, praF, and praG are similar to the genes for HPD hydratase, HOV aldolase, and acetaldehyde dehydrogenase, respectively. The corresponding genes were also found in the clusters that encode the aromatic ring cleavage pathways for 2-aminophenol (61), 2-nitrobenzoate (39), and catechol (23, 58). According to the sequence similarity, these gene products most likely have the same functions. The facts that E. coli carrying praR-praABEGFDCHI on a plasmid was able to grow on 4HB as the sole carbon source (see Fig. S4 in the supplemental material) and that the transcription of these genes is induced during 4HB catabolism (Fig. 6) support this notion.
Based on the sequence similarity, PraR belongs to the IclR family of transcriptional regulators, which includes PcaU and PcaR, activators of β-ketoadipate pathway genes in Acinetobacter baylyi ADP1 (10, 20) and Pseudomonas putida PRS2000 (48), respectively, PobR, an activator of the 4HB 3-hydroxylase gene, pobA from A. baylyi ADP1 (11), as well as HmgR, a repressor of the homogentisate catabolic pathway genes of P. putida U (2). It has been reported that PCA, β-ketoadipate, 4HB, and homogentisate act as inducers for the PcaU, PcaR, PobR, and HmgR regulatory systems, respectively. RT-PCR analysis and β-Gal assay of the praA promoter (Fig. 6 and 7) indicated that both 4HB and PCA are inducers of the pra catabolic operon. The promoter activity was constitutively observed, in the absence of praR, suggesting that praR codes for a repressor of the pra catabolic operon.
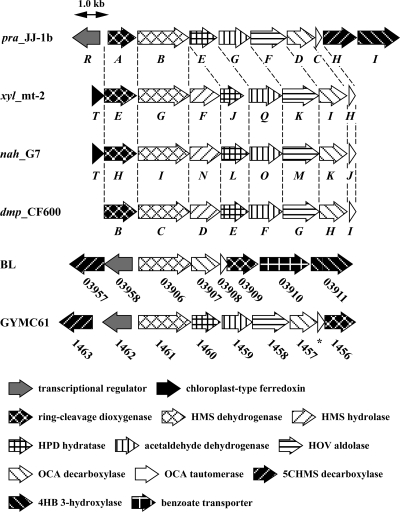
The gene organizations of the pra genes and the catechol meta-cleavage pathway genes on the TOL plasmid pWW0 of P. putida mt-2 (xyl) (18, 23), the naphthalene-catabolic plasmid NAH7 of P. putida G7 (nah) (58), and pVI150 of Pseudomonas sp. strain CF600 (dmp) (4, 43, 55) are almost identical, with the exception of the presence of praH and praI in the pra operon (Fig. 8). In the catechol meta-cleavage pathway gene clusters, the HMS hydrolase genes, xylF, nahN, and dmpD, are located between the HMS dehydrogenase genes (xylG, nahI, and dmpC) and the HPD hydratase genes (xylJ, nahL, and dmpE). However, the HMS hydrolase gene was missing in the pra gene cluster. Moreover, in the neighboring region of the pra gene cluster, the gene corresponding to xylT and nahT, which encode a chloroplast-type ferredoxin, was not observed.
FIG. 8.
Organizations of the gene clusters involved in the PCA 2,3-cleavage pathway and the catechol meta-cleavage pathway. pra_JJ-1b, the pra gene cluster in JJ-1b; xyl_mt-2, the xyl gene cluster in pWW0 of P. putida mt-2 (AJ344068); nah_G7, the nah gene cluster in NAH7 of P. putida G7 (AB237655); dmp_CF600, the dmp gene cluster in pVI150 of Pseudomonas sp. strain CF600 (M33263, X52805, and X60835); BL, the putative PCA 2,3-cleavage pathway gene cluster of B. licheniformis ATCC 14580 (CP000002); GYMC61, the putative PCA 2,3-cleavage pathway gene cluster of Geobacillus sp. strain Y412MC61 (ACED01000008). The ORF labeled with an asterisk does not appear in the Y412MC61 genome database.
In the B. licheniformis ATCC 14580 genome (CP000002), the genes related to praH, praR, praB, praD, praC, praA, and praI organize a gene cluster (Fig. 8). This cluster contains a gene encoding a putative benzoate transporter (BL03910), but the genes corresponding to praE, praF, and praG were absent. Recently, a draft genome sequence of Geobacillus sp. strain Y412MC61 has been reported (http://www.jgi.doe.gov/), and a gene cluster containing praH, praR, praB, praE, praG, praF, praD, praC, and praA orthologs was found (Fig. 8). This gene cluster has a complete set of the PCA 2,3-cleavage pathway genes except for the 4HB 3-hydroxylase gene. The presence of the pra gene cluster only in the genomes of bacilli suggested that the PCA 2,3-cleavage pathway genes might have evolved specifically in this particular group of bacteria.
Supplementary Material
Acknowledgments
We are grateful to S. Valla for the gift of pPR9TT.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abe, T., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernández, B. Galán, J. L. García, E. Díaz, and B. Miñambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186:5062-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asturias, J. A., L. D. Eltis, M. Prucha, and K. N. Timmis. 1994. Analysis of three 2,3-dihydroxybiphenyl 1,2-dioxygenases found in Rhodococcus globerulus P6. Identification of a new family of extradiol dioxygenases. J. Biol. Chem. 269:7807-7815. [PubMed] [Google Scholar]
- 4.Bartilson, M., and V. Shingler. 1989. Nucleotide sequence and expression of the catechol 2,3-dioxygenase-encoding gene of phenol-catabolizing Pseudomonas CF600. Gene 85:233-238. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, R. L. 1975. Novel pathway for degradation of protocatechuic acid in Bacillus species. J. Bacteriol. 121:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford, R. L. 1976. Pathways of 4-hydroxybenzoate degradation among species of Bacillus. J. Bacteriol. 127:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, R. L., J. W. Bromley, and P. E. Perkins-Olson. 1979. Catabolism of protocatechuate by Bacillus macerans. Appl. Environ. Microbiol. 37:614-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal, S., G. Trautwein, and U. Gerischer. 2005. Transcriptional organization of genes for protocatechuate and quinate degradation from Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 71:1025-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMarco, A. A., B. Averhoff, and L. N. Ornston. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sosa-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740-746. [DOI] [PubMed] [Google Scholar]
- 13.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppink, M. H., K. M. Overkamp, H. A. Schreuder, and W. J. Van Berkel. 1999. Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase. J. Mol. Biol. 292:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Eppink, M. H., H. A. Schreuder, and W. J. van Berkel. 1998. Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding. Eur. J. Biochem. 253:194-201. [DOI] [PubMed] [Google Scholar]
- 17.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuoka, S., K. Ishiguro, K. Yanagihara, A. Tanabe, Y. Egashira, H. Sanada, and K. Shibata. 2002. Identification and expression of a cDNA encoding human α-amino-β-carboxymuconate-ɛ-semialdehyde decarboxylase (ACMSD). A key enzyme for the tryptophan-niacine pathway and “quinolinate hypothesis”. J. Biol. Chem. 277:35162-35167. [DOI] [PubMed] [Google Scholar]
- 20.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harayama, S., P. R. Lehrbach, and K. N. Timmis. 1984. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J. Bacteriol. 160:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harayama, S., and M. Rekik. 1989. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J. Biol. Chem. 264:15328-15333. [PubMed] [Google Scholar]
- 25.Harayama, S., M. Rekik, K. L. Ngai, and L. N. Ornston. 1989. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J. Bacteriol. 171:6251-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 27.Hayase, N., K. Taira, and K. Furukawa. 1990. Pseudomonas putida KF715 bphABCD operon encoding biphenyl and polychlorinated biphenyl degradation: cloning, analysis, and expression in soil bacteria. J. Bacteriol. 172:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, Y., K. X. Zhao, X. H. Shen, C. Y. Jiang, and S. J. Liu. 2008. Genetic and biochemical characterization of a 4-hydroxybenzoate hydroxylase from Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 78:75-83. [DOI] [PubMed] [Google Scholar]
- 29.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 186:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koehntop, K. D., J. P. Emerson, and L. Que, Jr. 2005. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J. Biol. Inorg. Chem. 10:87-93. [DOI] [PubMed] [Google Scholar]
- 32.Li, T., H. Iwaki, R. Fu, Y. Hasegawa, H. Zhang, and A. Liu. 2006. α-Amino-β-carboxymuconic-ɛ-semialdehyde decarboxylase (ACMSD) is a new member of the amidohydrolase superfamily. Biochemistry 45:6628-6634. [DOI] [PubMed] [Google Scholar]
- 33.Liu, A., and H. Zhang. 2006. Transition metal-catalyzed nonoxidative decarboxylation reactions. Biochemistry 45:10407-10411. [DOI] [PubMed] [Google Scholar]
- 34.Martynowski, D., Y. Eyobo, T. Li, K. Yang, A. Liu, and H. Zhang. 2006. Crystal structure of α-amino-β-carboxymuconate-ɛ-semialdehyde decarboxylase: insight into the active site and catalytic mechanism of a novel decarboxylation reaction. Biochemistry 45:10412-10421. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama, K., T. Shibayama, A. Ichikawa, Y. Sakou, S. Yamada, and H. Sugisaki. 2004. Cloning and characterization of the genes encoding enzymes for the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 68:1434-1441. [DOI] [PubMed] [Google Scholar]
- 36.Masai, E., Y. Katayama, and M. Fukuda. 2007. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 37.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Muraki, T., M. Taki, Y. Hasegawa, H. Iwaki, and P. C. Lau. 2003. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakai, C., H. Kagamiyama, M. Nozaki, T. Nakazawa, S. Inouye, Y. Ebina, and A. Nakazawa. 1983. Complete nucleotide sequence of the metapyrocatechase gene on the TOL plasmid of Pseudomonas putida mt-2. J. Biol. Chem. 258:2923-2928. [PubMed] [Google Scholar]
- 41.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogales, J., Á. Canales, J. Jiménez-Barbero, J. L. García, and E. Díaz. 2005. Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J. Biol. Chem. 280:35382-35390. [DOI] [PubMed] [Google Scholar]
- 43.Nordlund, I., and V. Shingler. 1990. Nucleotide sequences of the meta-cleavage pathway enzymes 2-hydroxymuconic semialdehyde dehydrogenase and 2-hydroxymuconic semialdehyde hydrolase from Pseudomonas CF600. Biochim. Biophys. Acta 1049:227-230. [DOI] [PubMed] [Google Scholar]
- 44.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng, X., E. Masai, H. Kitayama, K. Harada, Y. Katayama, and M. Fukuda. 2002. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Providenti, M. A., J. Mampel, S. MacSween, A. M. Cook, and R. C. Wyndham. 2001. Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology 147:2157-2167. [DOI] [PubMed] [Google Scholar]
- 48.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roper, D. I., and R. A. Cooper. 1990. Subcloning and nucleotide sequence of the 3,4-dihydroxyphenylacetate (homoprotocatechuate) 2,3-dioxygenase gene from Escherichia coli C. FEBS Lett. 275:53-57. [DOI] [PubMed] [Google Scholar]
- 50.Saito, I., and G. R. Stark. 1986. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc. Natl. Acad. Sci. USA 83:8664-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Sala-Trepat, J. M., and W. C. Evans. 1971. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur. J. Biochem. 20:400-413. [DOI] [PubMed] [Google Scholar]
- 53.Santos, P. M., I. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 54.Sasoh, M., E. Masai, S. Ishibashi, H. Hara, N. Kamimura, K. Miyauchi, and M. Fukuda. 2006. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 72:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shingler, V., J. Powlowski, and U. Marklund. 1992. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J. Bacteriol. 174:711-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smiley, J. A., M. Kundracik, D. A. Landfried, V. R. Barnes, Sr., and A. A. Axhemi. 2005. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim. Biophys. Acta 1723:256-264. [DOI] [PubMed] [Google Scholar]
- 58.Sota, M., H. Yano, A. Ono, R. Miyazaki, H. Ishii, H. Genka, E. M. Top, and M. Tsuda. 2006. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188:4057-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 60.Sugimoto, K., T. Senda, H. Aoshima, E. Masai, M. Fukuda, and Y. Mitsui. 1999. Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions. Structure Fold. Des. 7:953-965. [DOI] [PubMed] [Google Scholar]
- 61.Takenaka, S., S. Murakami, R. Shinke, K. Hatakeyama, H. Yukawa, and K. Aoki. 1997. Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J. Biol. Chem. 272:14727-14732. [DOI] [PubMed] [Google Scholar]
- 62.Tanabe, A., Y. Egashira, S. Fukuoka, K. Shibata, and H. Sanada. 2002. Purification and molecular cloning of rat 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase. Biochem. J. 361:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaillancourt, F. H., J. T. Bolin, and L. D. Eltis. 2006. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 41:241-267. [DOI] [PubMed] [Google Scholar]
- 64.Wolgel, S. A., J. E. Dege, P. E. Perkins-Olson, C. H. Jaurez-Garcia, R. L. Crawford, E. Münck, and J. D. Lipscomb. 1993. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J. Bacteriol. 175:4414-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 66.Yoshida, M., N. Fukuhara, and T. Oikawa. 2004. Thermophilic, reversible γ-resorcylate decarboxylase from Rhizobium sp. strain MTP-10005: purification, molecular characterization, and expression. J. Bacteriol. 186:6855-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida, M., T. Oikawa, H. Obata, K. Abe, H. Mihara, and N. Esaki. 2007. Biochemical and genetic analysis of the γ-resorcylate (2,6-dihydroxybenzoate) catabolic pathway in Rhizobium sp. strain MTP-10005: identification and functional analysis of its gene cluster. J. Bacteriol. 189:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.