Abstract
We report on the identification of a glycosyltransferase (GT) from Mycobacterium tuberculosis H37Rv, Rv3779, of the membranous GT-C superfamily responsible for the direct synthesis of polyprenyl-phospho-mannopyranose and thus indirectly for lipoarabinomannan, lipomannan, and the higher-order phosphatidyl-myo-inositol mannosides.
The mycobacterial cell envelope consists of a multilayered structure of covalently linked peptidoglycan, arabinogalactan, and mycolic acids (the mAGP complex) and, among other important constituents, various noncovalently bound glycosylated lipids, notably the phosphatidyl-myo-inositol mannosides (PIMs) and their more glycosylated end products lipomannan (LM) and lipoarabinomannan (LAM) (6, 8). These glycolipids and lipoglycans exhibit a broad range of immunomodulatory activities implicated in the pathogenesis of tuberculosis and leprosy (for recent reviews, see references 5, 8, and 10).
Many steps in the biosynthesis of these phosphoinositides have been defined (for recent reviews, see references 3 and 12). Mannosyltransferases (ManTs) responsible for the polymerization aspects of the synthesis of the higher-order extracytoplasmic PIMs (PIM4 to PIM6), LM and LAM, are of the glycosyltransferase C (GT-C) multi-transmembrane domain superfamily, whose members are dependent on polyprenyl-phospho-mannopyranose (polyprenyl-P-Manp), specifically C35-P-Man or C50-P-Man, as the Manp donor, which is in contrast with what occurs in the early steps of the synthesis of the simpler PIMs, which directly utilize GDP-Man (3, 4, 12). However, the biosynthetic origins of C35/C50-P-Man have not been fully explored. In this report, we identify Rv3779, an unassigned GT-C, as a ManT directly responsible for the origins of polyprenyl-P-Man and indirectly for the synthesis of the more polar PIMs, LM and LAM.
A survey of the Mycobacterium tuberculosis H37Rv genome for genes with predicted (poly)saccharide-associated functions led to the identification of a cluster of 31 “cell wall biosynthetic genes” (2, 3), including not only arabinogalactan synthetic genes (embCA and embCB, glf, glfT1, and glfT2) but the genes for the putative ABC transporter proteins and mycolyl transferases and Rv3779, among five open reading frames, apparently encoding polyprenyl-P sugar-dependent GT-Cs (3). In particular, the integral membrane protein Rv3779 (666 amino acids) was identified as a putative GT-C, due to a conserved hallmark DXD motif and 12 to 14 predicted membrane-spanning domains (3). However, unlike with other GT-C enzymes, which typically carry the signature DXD motif on extracytoplasmic loops, the DLD motif of Rv3779 (at amino acid position 82) is predicted to map to the second loop and to be on the cytosolic side of the plasma membrane. This observation suggests that unlike other GT-C superfamily enzymes, Rv3779 utilizes a cytosolic sugar donor, presumably a nucleotide sugar. Rv3779 has orthologs in all slow-growing mycobacteria whose genome sequences are available but is not found in Mycobacterium smegmatis or in any fast-growing mycobacteria, with the notable exception of Mycobacterium abscessus. Orthologs of this gene are also not found in any other members of the suborder Corynebacterineae.
Since Rv3779 is not naturally present in M. smegmatis, M. smegmatis strain mc2155 was transformed with a multicopy plasmid (pVV16-Rv3779), allowing the expression of a recombinant C-terminal His6-tagged Rv3779 protein under the control of the phsp60 promoter (13). A cell-free ManT assay using membranes or whole lysate, similar to the one described by Korduláková et al. (13), was then conducted to determine if Rv3779 is involved in some aspect of PIM/LM/LAM biosynthesis. Thin-layer chromatography (TLC) autoradiography demonstrated an approximately threefold increase in a time-dependent manner in the incorporation of [14C]Man from GDP-[14C]Man into C35-P-Man and C50-P-Man by mc2155/pVV16-Rv3779 extracts compared with that of the control (Fig. 1A). The two accumulated Man-containing glycolipids were mildly alkali stable and mildly acid labile (data not shown), with TLC mobility properties confirming their identities as precursor glycolipids of the mycobacterial polyisoprenyl-P class (4). An important and substrate concentration-dependent increase in mannosyl transfer following Rv3779 overexpression was also observed when C50-P, the only form of polyprenylphosphate apparently produced by M. tuberculosis (7), was added as an acceptor substrate to the reaction mixture (Fig. 1B and C). This increase in activity over the control was about 27-fold during the first 30 min of the reaction when 0.05 mM C50-P was used in the assay (Fig. 1C). The background polyprenyl-P-Manp syntase activity detected in the control strain most likely resulted from MSMEG_3859/MSMEG_3860 (Ppm1/Ppm2), an M. smegmatis ortholog of the Ppm1 synthase from M. tuberculosis (68% identity, as determined with a 781-amino-acid overlap) (1, 9, 11). Unfortunately but not unexpectedly in light of the reported difficulty of expressing polytopic membrane GTs in Escherichia coli (14), attempts to produce Rv3779 in an active form in E. coli using different expression systems, host strains, growth conditions, and induction protocols were unsuccessful.
FIG. 1.
Effect of Rv3779 overexpression on the incorporation of [14C]Man from GDP-[14C]Man into mannolipids by cell extracts from M. smegmatis. (A) TLC analysis of an in vitro cell-free assay using GDP-[14C]Man and crude cell lysates from mc2155/pVV16 and mc2155/pVV16-Rv3779. Lysates were incubated with GDP-[14C]Man at 37°C for the indicated periods of time. The synthesized mannolipids were extracted with CHCl3-CH3OH (2:1, vol/vol), and a 10% aliquot of each sample was analyzed by TLC followed by autoradiography. The TLC plate was developed in CHCl3-CH3OH-H2O-NH4OH (65:25:4:0.5). Lanes: C, mc2155/pVV16; O, mc2155/pVV16-Rv3779. (B and C) Incorporation of [14C]Man from GDP-[14C]Man into exogenous decaprenyl-phosphate (C50)-P using membrane extracts from mc2155/pVV16 and mc2155/pVV16-Rv3779. (B) TLC analysis of the first 15 min of the in vitro cell-free assays using 0.5 mM of (C50)-P. (C) Quantification of the Man incorporated by the control (open symbols) and overexpressor (filled symbols) into C50-P-Man over time. The concentrations of (C50)-P used in the assays were 0 (circles), 0.005 (rectangles), and 0.05 (diamonds) mM. The inset shows a closeup of results of the assays using 0 and 0.005 mM (C50)-P.
Incubation of whole-cell lysate with GDP-[14C]Man for more-extended periods (4 to 24 h) and extraction of products with more-polar solvents (e.g., hot phenol) provides a ready estimate of the degree of synthesis of at least the LM metabolic end product. A comparison of the products synthesized by mc2155/pVV16 and mc2155/pVV16-Rv3779 (Fig. 2) showed little synthesis of [14C]LM at 4 h but the production of some low-molecular-weight [14C]LM at 24 h by the control lysate. In contrast, there was a distinct production of [14C]LM populations by the lysate from mc2155/pVV16-Rv3779 at both times, indicative of a secondary stimulatory effect of Rv3779 overexpression on LM, and presumably LAM, synthesis.
FIG. 2.
In vitro LM biosynthesis in M. smegmatis overexpressing Rv3779. Crude cell extracts (4 mg protein) of mc2155/pVV16 and mc2155/pVV16-Rv3779 were incubated with 1.0 μCi of GDP-[14C]Man (specific activity, 305 mCi/mmol) for 4 and 24 h. The reaction was stopped by adding CHCl3-CH3OH (2:1, vol/vol), and the LM/LAM contained in the cell pellet was extracted with hot phenol. LM/LAM separated on 10 to 20% Tricine gel and subsequently blotted to a nitrocellulose membrane was revealed by autoradiography. The leftmost column shows molecular mass markers (in kilodaltons).
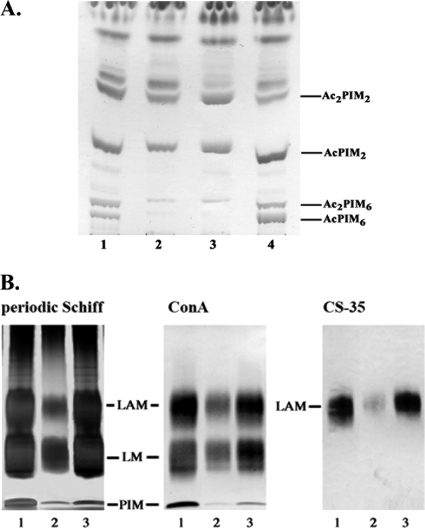
To confirm these effects of Rv3779, an M. tuberculosis H37Rv Rv3779 knockout mutant (H37RvΔRv3779) was generated by homologous recombination using standard procedures (15) (see Fig. S1 in the supplemental material). H37RvΔRv3779 grew at a much lower rate than WT H37Rv (see Fig. S2 in the supplemental material). Moreover, when examined by scanning electron microscopy, the mutant cells were found to be significantly shorter than their wild-type (WT) parent (Fig. 3). Normal growth rate and cell length were restored in the mutant upon complementation with a WT copy of Rv3779 (Fig. 3; see Fig. S2 in the supplemental material). TLC profiles of lipid extracts and matrix-assisted laser desorption ionization-time of flight mass spectrometry analyses showed a profound decrease in the amounts of acyl-PIM6 (AcPIM6) and Ac2PIM6 in H37RvΔRv3779 compared to those of the WT strain (Fig. 4A; see Fig. S3 in the supplemental material). Importantly, the synthesis of the simpler PIMs, those arising directly from GDP-Man, was not altered in the mutant (Fig. 4A; see Fig. S3 in the supplemental material). The examination by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with the monoclonal antibody CS-35 and concanavalin A of the phenol-extracted LM/LAM fraction from WT H37Rv and H37RvΔRv3779 also revealed considerably smaller amounts of both LM and LAM in the mutant (Fig. 4B). Complementation of H37RvΔRv3779 with a WT copy of Rv3779 restored both a normal polar PIM and a normal LM/LAM profile to the mutant.
FIG. 3.
Scanning electron micrographs of M. tuberculosis H37Rv (WT), the mutant H37RvΔRv3779, and the complemented mutant H37RvΔRv3779/pVV16-Rv3779 cultured in 7H9-oleic acid-albumin-dextrose-catalase-Tween 80 broth at 37°C. The length of the H37RvΔRv3779 mutant cells was on average 1.2 ± 0.1 μm, compared to 2.3 ± 0.1 μm for the WT strain and 2.1 ± 0.3 μm for the complemented mutant.
FIG. 4.
Analysis of polar PIMs LM and LAM from WT M. tuberculosis H37Rv, the H37RvΔRv3779 mutant, and H37RvΔRv3779/pVV16-Rv3779. (A) Equal amounts of total cellular lipids from WT H37Rv (lane 1), the Rv3779 mutant H37RvΔRv3779 (lane 2), the mutant carrying an empty plasmid, H37RvΔRv3779/pVV16 (lane 3), and the complemented mutant H37RvΔRv3779/pVV16-Rv3779 (lane 4) were analyzed by TLC developed in CHCl3-CH3OH-H2O-NH4OH (65:25:4:0.5). (B) LM and LAM extracted from equal-weight cells of WT H37Rv (lane 1), the Rv3779 mutant H37RvΔRv3779 (lane 2), and the complemented mutant H37RvΔRv3779/pVV16-Rv3779 (lane 3) were separated on a 10 to 20% Tricine gel and revealed by periodic acid-Schiff staining. The Western blot analyses were performed on the same samples using concanavalin A (ConA) reacting with the t-Manp residues of LM/LAM, as well as the CS-35 monoclonal antibody known to react with the arabinan segment of LAM.
From this evidence, we conclude that Rv3779 is involved in the synthesis of the polar PIMs LM and LAM through its primary role as a polyprenyl-P-Man synthase. The disruption of this gene, which results in important alterations in PIM, LM, and LAM profiles, also has profound effects on cell growth and shape. Rv3779 is the second polyprenyl-P-Man synthase involved in PIM/LM/LAM biosynthesis identified in M. tuberculosis (11).
Supplementary Material
Acknowledgments
Our research was supported by grants AI018357 and AI064798 from the NIH/NIAID.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baulard, A. R., S. S. Gurcha, J. Engohang-Ndong, K. Gouffi, C. Locht, and G. S. Besra. 2003. In vivo interaction between the polyprenol phosphate mannose synthase Ppm1 and the integral membrane protein Ppm2 from Mycobacterium smegmatis revealed by a bacterial two-hybrid system. J. Biol. Chem. 278:2242-2248. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis, p. 191-202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.
- 3.Berg, S., D. Kaur, M. Jackson, and P. J. Brennan. 2007. The glycosyltransferases of Mycobacterium tuberculosis—roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17:35R-56R. [DOI] [PubMed] [Google Scholar]
- 4.Besra, G. S., C. B. Morehouse, C. M. Rittner, C. J. Waechter, and P. J. Brennan. 1997. Biosynthesis of mycobacterial lipoarabinomannan. J. Biol. Chem. 272:18460-18466. [DOI] [PubMed] [Google Scholar]
- 5.Briken, V., S. A. Porcelli, G. S. Besra, and L. Kremer. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53:391-403. [DOI] [PubMed] [Google Scholar]
- 6.Crick, D. C., L. E. Quadri, and P. J. Brennan. 2008. Biochemistry of the cell envelope of Mycobacterium tuberculosis, p. 1-39. In S. H. E. Kaufmann and E. Rubin (ed.), Handbook of tuberculosis. Wiley-VCH, Weinheim, Germany.
- 7.Crick, D. C., M. C. Schulbach, E. E. Zink, M. Macchia, S. Barontini, G. S. Besra, and P. J. Brennan. 2000. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 182:5771-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffé, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 9.Gibson, K. J. C., L. Eggeling, W. N. Maughan, K. Krumbach, S. S. Gurcha, J. Nigou, G. Puzo, H. Sahm, and G. S. Besra. 2003. Disruption of Cg-Ppm1, a polyprenyl monophosphomannose synthase, and the generation of lipoglycan-less mutants in Corynebacterium glutamicum. J. Biol. Chem. 278:40842-40850. [DOI] [PubMed] [Google Scholar]
- 10.Gilleron, M., M. Jackson, J. Nigou, and G. Puzo. 2008. Structure, biosynthesis, and activities of the phosphatidyl-myo-inositol-based lipoglycans, p. 75-105. In M. Daffé and J.-M. Reyrat (ed.), The mycobacterial cell envelope. ASM Press, Washington, DC.
- 11.Gurcha, S. S., A. R. Baulard, L. Kremer, C. Locht, D. B. Moody, W. Muhlecker, C. E. Costello, D. C. Crick, P. J. Brennan, and G. S. Besra. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem. J. 365:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur, D., M. Guerin, H. Škovierová, P. J. Brennan, and M. Jackson. 2009. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv. Appl. Microbiol. 69:23-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korduláková, J., M. Gilleron, K. Mikušová, G. Puzo, P. J. Brennan, B. Gicquel, and M. Jackson. 2002. Definition of the first mannosylation step in phosphatidylinositol synthesis: PimA is essential for growth of mycobacteria. J. Biol. Chem. 277:31335-31344. [DOI] [PubMed] [Google Scholar]
- 14.Morita, Y. S., C. C. B. Sena, R. F. Waller, K. Kurokawa, M. F. Sernee, F. Nakatani, R. E. Haites, H. Billman-Jacobe, M. J. McConville, Y. Maeda, and T. Kinoshita. 2006. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281:25143-25155. [DOI] [PubMed] [Google Scholar]
- 15.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.