Abstract
A multidrug efflux pump, SrpABC, plays a key role in Pseudomonas putida S12 tolerance to toxic organic solvents. SrpRS are putative regulators of the SrpABC efflux pump encoded upstream of the srpABC structural genes, and previous studies suggest that SrpS is a repressor of SrpABC expression. An S12 isolate able to withstand extreme solvent stress carries a novel insertion sequence, ISPpu21, interrupting srpS. This insertion preadapts S12 to extreme solvent conditions through constitutive SrpABC expression.
It has been noted that Pseudomonas species adapt to high concentrations of toxic solvents by using various mechanisms (22). An alteration of the cell envelope structure when the bacterium was exposed to organic solvents was observed previously (6). Weber and de Bont (23) detected an increase of trans-unsaturated fatty acid contents in cells grown with toluene. It was suggested that this isomerization of cis- into trans-unsaturated fatty acids plays an important role in solvent tolerance in bacteria (7, 17). A mutant lacking this cis-to-trans isomerization activity was sensitive to toluene (17). Pinkart et al. (16) observed a modification of lipopolysaccharide and an increase in total fatty acids in solvent-treated cells, in addition to an increase in the trans-unsaturated fatty acid content. Enhanced rigidity resulting from changes in cell membrane fatty acid and phospholipid composition was shown to exclude toluene from the cell membrane, and the removal of intracellular toluene by degradation contributes to the toluene tolerance phenotype of Pseudomonas putida DOT-T1 (18). Finally, Isken and de Bont were the first to demonstrate the presence of an active efflux system for toluene in P. putida S12 (8), and Kieboom et al. reported that this energy-dependent export mechanism is an efflux pump belonging to the resistance-nodulation-division efflux pump family (9). During the past decade, several other efflux systems have been discovered to be involved in bacterial tolerance to organic solvents (1, 13, 14, 19).
Bacterial efflux systems are energy consuming, and therefore, their expression must be tightly controlled under uninduced conditions. Because many multidrug efflux systems have a broad substrate range, it is suggested that they may even pump out useful metabolites from bacterial cells (12). For example, an Escherichia coli mutant strain lacking the repressor of the AcrAB-TolC efflux pump was shown to have a lower growth rate than the wild-type strain (15). Similarly, it was reported that the efflux pump-overproducing mutants of Pseudomonas aeruginosa and P. putida have reduced fitness compared with parental strains (20, 24). Thus, sustained overexpression of efflux pumps can be detrimental to cells.
The SrpABC efflux pump in P. putida S12, which is homologous to TtgGHI in P. putida DOT-T1E (19), was found to have two regulatory genes, srpR and srpS, situated upstream of and transcribed divergently from the structural genes srpABC. srpS encodes the putative pump repressor SrpS, while the function of srpR/SrpR is still undefined. In 2001, Wery et al. (24) discovered a 2.6-kb insertion sequence named ISS12 that can insert itself into the repressor gene srpS, thus blocking srpS expression and thereby enhancing the expression of the SrpABC efflux pump. The authors proposed that P. putida S12 employs ISS12 as a specific mutator element in order to generate mutations that allow the bacterium to swiftly adapt to extreme solvent conditions. Their results at least suggest that an insertion sequence is involved in the regulation of efflux pump expression. In this study, we identified and characterized a new P. putida S12 insertion sequence involved in the regulation of SrpABC expression.
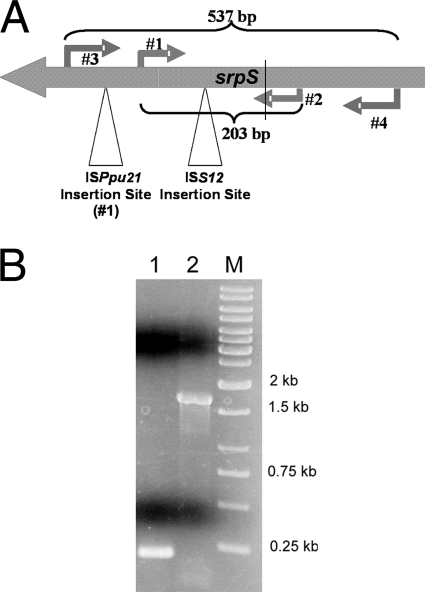
Wery et al. (24) reported that 93% of the P. putida S12 cells that survived a 1% (vol/vol) toluene shock carried an extra copy of the 2.6-kb insertion sequence ISS12 within the srpS gene. Our preliminary results showed much lower carriage of an insertion element in srpS. We therefore performed experiments in order to further understand the regulation of the SrpABC efflux pump. A single colony of P. putida S12 was used to inoculate 25 ml of Luria-Bertani (LB) broth, and the culture was grown overnight at 30°C. (Strains and plasmids used in this study are listed in Table 1.) One milliliter of the overnight culture was used to inoculate 50 ml of LB broth containing 1% (vol/vol) toluene. This shock dose of toluene killed most of the cells (approximately 99.99%) within 30 min. After shaking of the medium at 200 rpm and 30°C for 24 h, a new culture had grown up and total DNA was isolated from it. PCR was performed using a Mastercycler gradient thermocycler according to the recommendations of the manufacturer (Eppendorf, Hamburg, Germany). Total genomic DNA was isolated from P. putida strains by the hexadecyltrimethylammonium bromide procedure (2). Using primers 1 (TTGGAGGTGAATACTGG) and 2 (TCGGTCTGCCTGGCTTCT), the region between which includes the previously reported insertion site of ISS12, we amplified the DNA sample by PCR (Fig. 1A). Interestingly, the PCR result showed an srpS band of the original size (data not shown), indicating that ISS12 did not appear in this region. The more rigorous condition of a 20% (vol/vol) toluene shock was tested. Although the maximum aqueous solubility of toluene is exceeded at a 20% concentration, the toxic effect of toluene continues to increase with increasing toluene concentrations, due to increased frequency of cellular contact (25). However, even under these severe growth conditions, we did not find any insertion sequences within the region between primers 1 and 2 (Fig. 1B, lane 1). When a more inclusive primer pair, primers 3 (ACCACTCTGCCTCACTTCG) and 4 (ATCCAGGTCATCGCCAG), was used to amplify the DNA sample from cells that survived the 20% (vol/vol) toluene shock, the PCR result showed a band of 1.7 kb, much larger than the expected size of 537 bp (Fig. 1B, lane 2). DNA sequencing analysis identified a 1.2-kb insertion sequence, designated ISPpu21, located in the region between primers 1 and 3. Automated DNA sequencing was performed using a DYEnamic ET kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ) and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The newly isolated insertion sequence was determined to exhibit no homology to ISS12, and its insertion site in srpS is different from that of ISS12.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| P. putida S12 | Wild type; srpABC+srpR+srpS+ | 5 |
| P. putida JK1 | srpB::TnMod-KmO | 4 |
| P. putida S12TS | srpS::ISPpu21 | This study |
| P. putida S12DM | srpS::ISPpu21 srpB::TnMod-KmO | This study |
| Plasmids | ||
| pCR2.1-TOPO | TOPO TA cloning vector for direct insertion of PCR products and blue/white screening; Apr Kmr | Invitrogen |
| pJD101 | Derived from BamHI digestion of P. putida JK1 chromosome; contains srpR through partial ORF of TnMod-KmO plasposon-mutated srpB | 9 |
| pJD102 | Derived from PstI digestion of P. putida JK1 chromosome; contains partial ORF of TnMod-KmO plasposon-mutated srpB through srpC | 9 |
| pJD203 | Constructed by digestion (with EcoRV and BamHI) and ligation of pJD101 and pJD102; contains partial ORF of TnMod-KmO plasposon-mutated srpB through srpC | This study |
| pBBR1MCS-2 | Broad-host-range cloning vector; Kmr | 11 |
| pBBR1-2-srpR | Coding region of srpR cloned into pBBR1MCS-2 (using EcoRI and BamHI sites) | This study |
| pBBR1-2-srpS | Coding region of srpS cloned into pBBR1MCS-2 (using EcoRI and BamHI sites) | This study |
| pBBR1-2-srpRS | Coding regions of srpR and srpS cloned into pBBR1MCS-2 (using EcoRI and BamHI sites) | This study |
FIG. 1.
Discovery of the new insertion sequence ISPpu21 within the srpS gene in P. putida S12 by PCR. (A) Locations of the PCR primers (1 through 4), sizes of the amplified regions, and insertion sites of ISS12 and ISPpu21. (B) The products resulting from the PCR were separated on a 0.7% (wt/vol) agarose gel. Lanes: 1, region amplified with primers 1 and 2; 2, region amplified with primers 3 and 4; M, molecular size markers.
Subsequently, we tested whether individual cells surviving the 20% (vol/vol) toluene shock carry an insertion sequence within the repressor protein gene srpS. To ensure that the cells used for total DNA extraction were genetically homogenous, an aliquot of the culture incubated with 20% (vol/vol) toluene for 24 h was spread onto LB agar plates and the plates were incubated at 30°C in a sealed glass chamber with saturated toluene vapor. Thirty-three separate cultures grown from different single colonies were used for total DNA extraction, and subsequently, DNA samples were PCR amplified with primers 8 (GAGCACTGGCAATCTAAC) and 9 (AGAGTCTACCATTATGCGAT), which encompass the entire coding sequences of srpR and srpS. Among these samples, 22 (67%) carried no insertion sequence within the amplified region, 1 (3%) showed ISS12 located in the same site discovered previously (24), and the other 10 samples (30%) were found to contain ISPpu21. Further investigation by PCR analysis and DNA sequencing of the 10 samples that carried ISPpu21 showed two different insertion sites for ISPpu21 (Fig. 2): one (insertion site 1) located between primers 1 and 3 (utilized in 6 of 10 samples) and the other (insertion site 2) located downstream of primer 3 but still within srpS (utilized in 4 of 10 samples). These results indicate that only approximately one-third of the cells that survive toluene shock carry an insertion sequence within the efflux pump repressor gene srpS and that the other two-thirds of the cells do not require an srpS insertion mutation to survive the high-dose toluene shock.
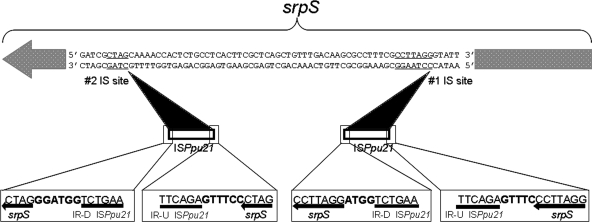
FIG. 2.
Schematic diagram of the insertion (IS) sites of ISPpu21 in the srpS gene in P. putida S12TS. The two black triangles indicate the two different insertion points of ISPpu21 in srpS. The sequences of the upstream and downstream terminal inverted repeats (IR-U and IR-D), as well as the sequences adjacent to these terminal repeats, have been marked with boxes. The nucleotides not belonging to either srpS or ISPpu21 are shown in boldface.
ISPpu21 is a 1,179-bp insertion sequence with a pair of 6-bp inverted repeats (TCTGAA) at its ends (Fig. 2). Interestingly, ISPpu21 elements introduced at insertion site 1 and insertion site 2 generate a pair of 7-bp and a pair of 4-bp target site duplication direct repeats, respectively. Only one open reading frame (ORF) was discovered in ISPpu21, putatively encoding a protein of 326 amino acids. The nucleotide sequence of ISPpu21 has 100% identity to a region of the P. aeruginosa serotype O15-ATCC putative O antigen biosynthesis gene cluster (GenBank accession no. AF498406), and the ORF in this ISPpu21 orthologue is transcribed in the opposite direction from the P. aeruginosa chromosomal gene cluster. The putative product of the ORF in ISPpu21 has homology (identities from 61 to 93%) to transposase proteins belonging to the IS5 family of transposases (Table 2).
TABLE 2.
Amino acid sequence identities between the putative protein encoded by ISPpu21 and transposase proteins from different organisms
| Protein | Identity (%) | Organism | Corresponding accession no. |
|---|---|---|---|
| IS5 family transposase | 93 | Pseudomonas resinovorans | NP_758545 |
| ISPssy transposase | 93 | Bordetella petrii | YP_001632795 |
| IS1384 transposase | 92 | P. putida | AAC98743 |
| ISPst5 transposase | 92 | Pseudomonas stutzeri | AAX14063 |
| IS5 family transposase | 91 | P. aeruginosa | ZP_00972095 |
| ISPsy2 transposase | 90 | Pseudomonas syringae | YP_275487 |
| IS4 family transposase | 90 | Azotobacter vinelandii | ZP_00415717 |
| IS5 family transposase | 89 | Yersinia enterocolitica | CAE46781 |
| IS1479 transposase | 65 | Xanthomonas campestris | YP_243093 |
| IS5 family transposase | 62 | Xanthomonas oryzae | YP_201728 |
| IS5 family transposase | 62 | Achromobacter xylosoxidans | YP_195864 |
| IS4 family transposase | 62 | Burkholderia vietnamiensis | YP_001109709 |
| ISBPH transposase | 61 | Achromobacter denitrificans | NP_990893 |
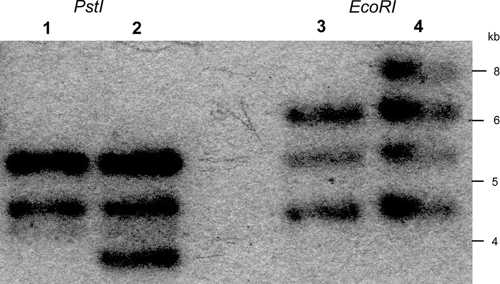
We examined the distribution of the new insertion sequence ISPpu21 in the genomes of wild-type P. putida S12 and the solvent-adapted strain carrying ISPpu21 within srpS, designated P. putida S12TS. For this purpose, total DNA was isolated from both P. putida S12 and P. putida S12TS. The DNA samples were digested with either PstI or EcoRI, which do not digest within the ISPpu21 DNA sequence, and Southern hybridization analysis was performed using an internal 352-bp DNA probe made from ISPpu21 with primers 6 (TGCCGTCCTTGTTCTTG) and 7 (ATCGAGCCGCACTACCC). From each parental sample, there were three hybridizing DNA bands, indicating three copies of ISPpu21 distributed over the P. putida S12 genome (Fig. 3). One extra hybridizing DNA fragment appears in each sample from P. putida S12TS, suggesting that transposition of ISPpu21 occurs by a replicative mechanism.
FIG. 3.
Southern hybridization analysis of total DNA from P. putida S12 and P. putida S12TS. The DNA samples were digested with PstI (lanes 1 and 2) or EcoRI (lanes 3 and 4) and analyzed by Southern hybridization with a 352-bp DNA probe made from ISPpu21. Lanes 1 and 3 show the results for wild-type P. putida S12; lanes 2 and 4 show the results for P. putida S12TS.
The authors of a previous study examining the survival frequencies of P. putida S12 and an srpS insertion mutant in response to toluene shock concluded that the increased survival rate of the mutant drops to wild-type levels only if both srpR and srpS are complemented (24). We obtained different results by performing a similar complementation experiment. DNA fragments containing the srpR, srpS, and srpRS genes were amplified using primers 12 (GGGAATTCTATGGCCAGAAAGAC) and 13 (CCGGATCCTCACTCGAAG), 14 (CCGAATTCTATGAACCAATCAG) and 15 (TTGGATCCTCTAGGGAGCTTTC), and 13 and 14, respectively. All three amplified fragments were ligated into pBBR1MCS-2 (11) by using incorporated BamHI and EcoRI sites. The corresponding constructs were designated pBBR1-2-srpR, pBBR1-2-srpS, and pBBR1-2-srpRS, respectively, and each was introduced into the S12TS mutant by electroporation using standard methods (3). Kanamycin was added to a final concentration of 250 μg/ml to maintain the plasmids when necessary. The survival frequencies were determined by measuring the numbers of CFU on LB plates after 20 h of incubation in LB medium with 20% (vol/vol) toluene or 20% (vol/vol) distilled water as a control. The survival frequency assay results are listed in Table 3. They show that the srpS::ISPpu21 mutant S12TS had a 17,000-fold-greater survival frequency in the presence of a 20% (vol/vol) toluene shock than wild-type S12, which was not influenced by the introduction of the empty vector pBBR1MCS-2. Interestingly, both S12TS(pBBR1-2-srpS) and S12TS(pBBR1-2-srpRS) showed survival frequencies close to parental levels, suggesting that the survival frequency of P. putida S12TS returns to the wild-type level when srpS is complemented. Complementation with srpR did not affect survival frequency, as the S12TS(pBBR1-2-srpR) value remained similar to that for S12TS (∼5%). Like SrpABC, the TtgGHI efflux pump of P. putida DOT-T1E has two regulatory genes, ttgV and ttgW, located upstream from the structural genes ttgGHI. These two regulatory genes are polycistronic, are transcribed divergently from the ttgGHI operon (18), and are 95% identical to srpRS at the DNA sequence level. A mutant deficient in ttgV, but not in ttgW, has much higher expression activity from the ttgGHI and ttgVW promoters than the wild type, suggesting that ttgV encodes a repressor for the expression of ttgGHI and itself. A similar result for a ttgVW double mutant has also been observed. However, the mutant lacking ttgW but carrying intact ttgV does not show any significant changes in the expression levels of ttgGHI and ttgVW. Thus, it was proposed that ttgW plays no major role in this regulatory system (18). Further investigation of the 0.0003% of wild-type cells surviving 20% (vol/vol) toluene for 30 min was performed. Total DNA was isolated from nine individual colonies and analyzed by PCR with primers 8 and 9. The results showed that four of nine samples had ISPpu21 within srpS, while the other five had intact srpS (data not shown). This finding again suggests that more than half of the surviving S12 cells do not need srpS inactivation in order to tolerate toluene shock.
TABLE 3.
Survival frequencies for different P. putida strains in response to a 20% (vol/vol) toluene shocka
| P. putida strainb | Survival frequency (%) |
|---|---|
| S12 | 0.0003 ± 0.0002 |
| S12TS | 5 ± 1.5 |
| S12TS(pBBR1MCS-2) | 5 ± 2.6 |
| S12TS(pBBR1-2-srpR) | 5 ± 2.3 |
| S12TS(pBBR1-2-srpS) | 0.0003 ± 0.0002 |
| S12TS(pBBR1-2-srpRS) | 0.0002 ± 0.0001 |
All assays were repeated three times with three separate cultures. The given values are the means ± standard deviations of triplicate measurements.
The names of the introduced plasmids are in parentheses.
Insertion sequences have been shown to be important for the genomic plasticity of certain bacteria. The literature contains many examples of insertion sequences causing the inactivation of genes by insertion, the activation of neighboring genes by the induction of outwardly directed promoters, or genomic rearrangements by homologous recombination. However, there are few published examples in which insertion sequence transposition is shown to be directly responsible for a change in cellular phenotype (21). The inactivation of srpS by ISPpu21 is a clear example of an insertion sequence inactivating a gene encoding a repressor directly responsible for controlling efflux pump expression, and therefore preadapting the bacterial cell for normally lethal organic solvent stress. Our detection of a second P. putida S12 insertion sequence element capable of inactivating srpS indicates that ISS12 is not a specific mutator element, as suggested previously by Wery et al. (24). It is also possible that other transposable elements besides ISS12 and ISPpu21 can insert themselves into srpS in order to generate constitutively expressed SrpABC efflux pump variants.
Based on our discovery of ISPpu21 in two different target sequences within the srpS ORF in different isolates of solvent-shocked S12, the target site recognition of the ISPpu21 transposase is not highly specific. At insertion site 1, the putative ISPpu21 target sequence in srpS is CCTTA; at insertion site 2, the putative target sequence is CTAG. In the P. aeruginosa putative O antigen biosynthesis cluster, the ISPpu21 target site appears to be TTT or TAG, although further experimental tests are required to confirm this. As shown in Fig. 2, determination of the ISPpu21 target sequence is complicated by the random addition of 4 to 6 bp at each end of the insertion that do not correspond to either target DNA (e.g., srpS) or ISPpu21. It is unknown whether other examples of IS5 family insertion elements behave similarly. Figure 3 illustrates the addition of one ISPpu21 copy into the genome after IS replication to give four copies, suggesting that ISPpu21 transposes by a replicative mechanism. It is expected that insertion sequence inactivation of srpS occurs prior to toluene shock. It has been demonstrated previously that toxic toluene accumulation in the P. putida S12 membranes occurs within 10 min of toluene introduction (8), whereas toluene induction of the srpABC promoter reaches half maximal transcription activation only after ∼60 min (10). The inability of 99.99% of cells to survive solvent shock relates to the inability of cells to express and assemble the SrpABC efflux pump in sufficient time to prevent membrane disruption and cell death. Interestingly, we found that only approximately 33% of toluene-tolerant P. putida S12 isolates contained IS insertions in the srpS gene, the majority of these insertions being ISPpu21 rather than ISS12. Sequence analysis of the other toluene-tolerant S12 isolates not containing an insertion sequence did not show mutations in srpS or in other, nearby DNA regions. This result is puzzling in light of the fact that immediate SrpABC expression appears to be a requirement for S12 solvent tolerance, and suggests that other mechanisms may be involved in controlling SrpABC expression. We are currently working to elucidate the role of SrpR in SrpABC expression in P. putida S12 under solvent stress conditions, especially with respect to S12 strains that have no insertion sequence interruption of srpS.
Nucleotide sequence accession number.
The DNA sequence of ISPpu21 has been deposited in GenBank under accession number FJ905320 and in the Insertion Sequence Database (http://www-is.biotoul.fr/).
Acknowledgments
We thank Zileena Zahir for initiating some of these experiments and Tracy Raivio for reviewing the manuscript and providing useful comments.
This work was funded by an operating grant from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. F. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Greene Publishing Associates, New York, NY.
- 3.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47:125-133. [DOI] [PubMed] [Google Scholar]
- 4.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmans, S., M. J. Van der Werf, and J. A. M. de Bont. 1990. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 56:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heipieper, H. J., and J. A. M. de Bont. 1994. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol. 60:4440-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heipieper, H. J., R. Diefenbach, and H. Keweloh. 1992. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isken, S., and J. A. de Bont. 1996. Active efflux of toluene in a solvent-resistant bacterium. J. Bacteriol. 178:6056-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieboom, J., J. J. Dennis, J. A. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85-91. [DOI] [PubMed] [Google Scholar]
- 10.Kieboom, J., J. J. Dennis, G. J. Zylstra, and J. A. de Bont. 1998. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180:6769-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. I. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 12.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 13.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X. Z., L. Zhang, and K. Poole. 1998. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 180:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 16.Pinkart, H. C., J. W. Wolfram, R. Rogers, and D. C. White. 1996. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to o-xylene. Appl. Environ. Microbiol. 62:1129-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos, J. L., E. Duque, J. J. Rodríguez-Herva, P. Godoy, A. Haïdour, F. Reyes, and A. Fernández-Barrero. 1997. Mechanisms for solvent tolerance in bacteria. J. Biol. Chem. 272:3887-3890. [DOI] [PubMed] [Google Scholar]
- 18.Rojas, A., A. Segura, M. E. Guazzaroni, W. Teran, A. Hurtado, M. T. Gallegos, and J. L. Ramos. 2003. In vivo and in vitro evidence that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J. Bacteriol. 185:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez, P., J. F. Linares, B. Ruiz-Díez, E. Campanario, A. Navas, F. Baquero, and J. L. Martínez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 21.Sokol, P. A., M. Z. Luan, D. G. Storey, and P. Thirukkumaran. 1994. Genetic rearrangement associated with in vivo mucoid conversion of Pseudomonas aeruginosa PAO is due to insertion elements. J. Bacteriol. 176:553-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber, F. J., and J. A. M. de Bont. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225-245. [DOI] [PubMed] [Google Scholar]
- 23.Weber, F. J., and J. A. M. de Bont. 1994. Cis/trans isomerization of fatty acids as a defence mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology 140:2013-2017. [DOI] [PubMed] [Google Scholar]
- 24.Wery, J., B. Hidayat, J. Kieboom, and J. A. de Bont. 2001. An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J. Biol. Chem. 276:5700-5706. [DOI] [PubMed] [Google Scholar]
- 25.Zahir, Z., K. D. Seed, and J. J. Dennis. 2006. Isolation and characterization of novel organic solvent-tolerant bacteria. Extremophiles 10:129-138. [DOI] [PubMed] [Google Scholar]