Abstract
We reported previously that a Salmonella enterica serovar Enteritidis dam mutant expressing a truncated Dam protein does not agglutinate in the presence of specific antibodies against O9 polysaccharide. Here we investigate the participation of Dam in lipopolysaccharide (LPS) synthesis in Salmonella. The LPS O-antigen profiles of a dam null mutant (SEΔdam) and the Salmonella serovar Enteritidis parental strain were examined by using electrophoresis and silver staining. Compared to the parental strain, SEΔdam produced LPS with shorter O-antigen polysaccharide chains. Since Wzz is responsible for the chain length distribution of the O antigen, we investigated whether Dam methylation is involved in regulating wzz expression. Densitometry analysis showed that the amount of Wzz produced by SEΔdam is threefold lower than the amount of Wzz produced by the parental strain. Concomitantly, the activity of the wzz promoter in SEΔdam was reduced nearly 50% in logarithmic phase and 25% in stationary phase. These results were further confirmed by reverse transcription-PCR showing that wzz gene expression was threefold lower in the dam mutant than in the parental strain. Our results demonstrate that wzz gene expression is downregulated in a dam mutant, indicating that Dam methylation activates expression of this gene. This work indicates that wzz is a new target regulated by Dam methylation and demonstrates that DNA methylation not only affects the production of bacterial surface proteins but also the production of surface polysaccharides.
Lipopolysaccharide (LPS) is a key component of the outer membrane of gram-negative bacteria that contributes to the stability and permeability barrier properties of this membrane. LPS is located on the outer leaflet of the outer membrane and consists of three regions: O-antigen polysaccharide, core oligosaccharide, and lipid A (47). The biogenesis of LPS is a complex process involving various steps that occur at the plasma membrane, followed by translocation of LPS molecules to the outer membrane (47, 48, 54, 58). The core oligosaccharide is assembled on preformed lipid A by sequential glycosyl transfer of monosaccharides, while the O antigen is independently assembled on undecaprenyl-phosphate (61). These pathways eventually converge by ligation of the O antigen to the outer core domain of the lipid A-core oligosaccharide acceptor (19, 20, 47, 48, 58, 61, 62). O-antigen assembly occurs by mechanisms referred to as Wzy (polymerase)-dependent and ATP-binding cassette-dependent pathways (for reviews, see references 47 and 58). The Salmonella O antigen is assembled by the Wzy-dependent pathway, in which the O-antigen repeating subunits are synthesized at the cytosolic side of the plasma membrane. Each subunit is subsequently translocated across the membrane by an ATP hydrolysis-independent mechanism mediated by the protein Wzx (27, 47, 49, 58). On the periplasmic side of the plasma membrane, translocated subunits polymerize to a certain length, unique to each O antigen, by the concerted actions of Wzy (O-antigen polymerase) and Wzz (O-antigen chain length regulator), and the polysaccharide is ultimately ligated to the lipid A-core oligosaccharide (31, 35, 38).
In Salmonella species, O-antigen length contributes to an effective barrier (39) and affects key virulence features, like serum resistance and entry into eukaryotic cells (17, 23, 40-42). Furthermore, O-antigen length can also modulate acquired immunity. Indeed, Phalipon and coworkers demonstrated that in Shigella flexneri induction of an O-antigen-specific antibody response depends on the length of the polysaccharide chain (45).
In gammaproteobacteria the DNA adenine methyltransferase (Dam) introduces a methyl group at the N6 position of the adenine of GATC sites in the newly synthesized DNA strand after DNA replication, generating methylated DNA (29, 30, 32, 63). At certain GATC sites, methylation of the newly synthesized strand is hindered by binding of proteins that protect the GATC sites from Dam methylase. The protection against methylation can either cause a temporary delay in methylation or generate GATC sites that are stably hemimethylated or unmethylated (30, 63). Thus, the DNA methylation status can affect the interactions between DNA and proteins such as RNA polymerase or transcription factors (63) that regulate (activate or repress) gene expression.
Dam is required for expression of virulence genes in certain bacteria (25), including Salmonella enterica (1, 4, 6, 13, 21, 24), but the virulence defects of dam mutants are pleiotropic and not completely known. It has been proposed that dam mutants could serve as live attenuated vaccines and that the Dam protein itself may provide a potential target for broad antimicrobial activity (21). Recently, dam mutants of S. enterica serovar Typhimurium have been analyzed as potential live vaccines to prevent salmonellosis in birds and cattle (10, 11). We have reported previously that an S. enterica serovar Enteritidis dam mutant expressing a truncated Dam protein is attenuated (15). This mutant has limited protective capacity as a live vaccine (55) and is unable to agglutinate in the presence of specific antibodies against O9 polysaccharide (14, 55), suggesting that there is a defective LPS. The aim of the present study was to investigate the participation of Dam methylation in LPS synthesis in Salmonella serovar Enteritidis. Compared to the LPS produced by the parental strain, the dam null mutant produced LPS with shorter O-antigen polysaccharide chains, indicating that Dam methylation regulates LPS gene expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Salmonella serovar Enteritidis #5694 was used to construct all of the mutant strains. Gene deletion was performed as described by Datsenko and Wanner (7). The mutagenic primers used are listed in Table 2. Bacteria were grown in Luria-Bertani (LB) broth (53) supplemented, as required, with antibiotics at the following final concentrations: ampicilin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 40 μg/ml; and tetracycline, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. enterica serovar Enteritidis strains | ||
| #5694 | Wild type | F. Collins collection |
| SEΔdam | Δdam | This study |
| SEΔwzz::lacZ | #5694 Δwzzst::lacZY, Kmr | This study |
| SEΔwzz::lacZΔdam | #5694 Δwzzst::lacZY Δdam, Kmr | This study |
| E. coli K-12 strain DH5α | F− φ80lacZM15 endA recA hsdR(rK− mK−) supE thi gyrA relA Δ(lacZYA-argF)U169 | Laboratory stock |
| Plasmids | ||
| pACYC184 | Cmr Tetr, cloning vector | 52 |
| pCE36 | ahp FRT lacZY+ this oriR6K | 12 |
| pCP20 | FLP+ λcI857+ λpR Repts Ampr Cmr | 7 |
| pIZ833 | E. coli dam gene, Ampr | 57 |
| pKD3 | Template plasmid for mutagenesis, Ampr Cmr | 7 |
| pKD4 | Template plasmid for mutagenesis, Ampr Kmr | 7 |
| pKD46 | γ, β, and exo from λ phage, araC-ParaB, Ampr | 7 |
| pwzz | wzz, Cmr (pACYC184 backbone) | This study |
TABLE 2.
Oligonucleotide primers used in this studya
| Gene targeted | Primerb | Sequence (5′→3′)c |
|---|---|---|
| Gene deletion | ||
| dam | dam::Cm (F) | TTCTCCACAGCCGGAGAAGGTGTAATTAGTTAGTCAGCATGTGTGTAGGCTGGAGCTGCTTC |
| dam::Cm (R) | GGCAATCAAATACTGTTTCATCCGCTTCTCCTTGAGAATTACATATGAATATCCTCCTTAG | |
| Chromosomal lacZY fusion | ||
| wzz | wzz::Km (F) | TACACTGTCTCCAGCTTCATCCTTTTTTTAGTTAGGGTATCTAGTGTAGGCTGGAGCTGCTTCG |
| wzz::Km (R) | TACCTTTCGAAGCCGACCACCATCCGGCAAAGAAGCTTACATATGAATATCCTCCTTAG | |
| Gene cloning | ||
| wzz | wzz-F | GCTTACAAGGCTTTTGGC |
| wzz-R | TAGGGTATCTATGACAGTGGAT | |
| Verification of predicted construction | ||
| dam | rpe | TACGACAACCTGAACGGTTG |
| damX | GCAGCGTGCGGTCAACATG | |
| wzz | His | GCGGCCACCGTCAATGATCG |
| Ugd | CATTATTTCCAACAGGATGGCGGC | |
| Kmr | CCATGTTGGAATTTAATCGCGGCC | |
| LacZrevcheck | ACCAGGCAAAGCGCCATTCG | |
| Real-time PCR | ||
| 16S rRNA gene | q-16S-F | GCCGCAAGGTTAAAACTCAA |
| q-16S-R | AAGGCACCAATCCATCTCTG | |
| wzz | q-wzz-F | CGTCGCTTCGTTCTGTATCA |
| q-wzz-R | AGGATGTTACCCAGGACACG |
Primers were purchased from Invitrogen Inc. and were designed using the DNA sequence information available for the Salmonella serovar Enteritidis strain (Salmonella sp. comparative sequencing blast server BLAST Server Database at www.sanger.ac.uk).
F, forward primer; R, reverse primer.
Underlining indicates a sequence homologous to pKD3, pKD4, or pCE36.
Bacterial transformation.
Salmonella serovar Enteritidis was transformed by electroporation as previously described (9). Briefly, bacteria grown for 24 h were subcultured in 5 ml LB broth for 2 h, harvested into 10% glycerol, and washed twice. Forty-five microliters of bacteria was mixed with 5 to 10 μl plasmid or PCR product and subjected to electroporation in a 0.2-cm electroporation cuvette (Bio-Rad) at a voltage of 2.5 kV, a resistance of 200 Ω, and a capacitance of 25 μF. After this, for recovery, 800 μl of SOC broth was added, and bacteria transformed with a plasmid and bacteria transformed with a PCR product were incubated for 2 and 18 h, respectively. Then 70 μl of bacteria was transferred onto selective plates containing the appropriate antibiotic.
Determination of the DNA methylation status.
Genomic DNA was isolated using a Puregene DNA isolation kit (Gentra Systems). Two micrograms of DNA was incubated with MboI, Sau3AI, and DpnI for 4 h at 37°C. The buffer mixtures were the mixtures recommended by the manufacturer (Promega). Restricted DNA was analyzed by electrophoresis on 0.7% (wt/vol) agarose gels along with DNA subjected to the same conditions in the absence of enzyme. After electrophoresis the gels were visualized with UV illumination.
Sensitivity to 2-aminopurine.
The sensitivity to the base analogue 2-aminopurine was monitored using LB medium plates containing 100 mg/ml 2-aminopurine (16, 44).
O-antigen agglutination.
Individual bacterial colonies were emulsified in 50 μl of saline and checked for autoagglutination. When no agglutination was observed, an equal volume of specific antiserum (Difco) was added. Slides were incubated at room temperature with gentle agitation for 5 min, and agglutination was recorded based on the flocculation of bacteria. Saline and Salmonella serovar Enteritidis #5694 were used as negative and positive controls for O9 agglutination, respectively.
Introduction of a lacZY transcriptional fusion into the chromosomal wzz locus.
The wzz gene of Salmonella serovar Enteritidis strain #5694 was disrupted using the method described by Ellermeier et al. (12). FRT sites generated by excision of kanamycin resistance cassettes were used to integrate plasmid pCE36 (12), generating transcriptional lacZY fusions to the wzz gene promoter. The resulting strain was designated SEΔwzz::lacZ. We also constructed an SEΔwzz::lacZ Δdam derivative by using the method of Datsenko and Wanner (7). For complementation experiments, pIZ833 carrying a dam gene was introduced into the mutant strains by electroporation.
Molecular cloning of Salmonella wzz gene.
PCR amplification was performed using Platinum Pfx DNA polymerase (Invitrogen). The blunt-ended PCR product was purified using a gel extraction kit (Qiagen) and ligated with T4 DNA ligase (New England BioLabs) into pACYC184, which was digested with EcoRV, blunt ended, and dephosphorylated with shrimp alkaline phosphatase (Roche Diagnostics). The ligation mixture was used to transform competent DH5α cells. Plasmids were isolated from Cmr transformants and screened with restriction endonucleases for inserts that were the appropriate size and correct orientation. The integrity of the insert was confirmed by sequencing (Macrogen Inc.), and the insert was analyzed with Sequencher (Gene Codes Corporation) and Vector NTI software.
LPS analysis.
LPS was extracted as described by Marolda et al. (33). Briefly, the optical densities at 600 nm (OD600) of samples (final volume, 100 μl) from an overnight plate culture were adjusted to 2.0. Then the samples were suspended in lysis buffer containing proteinase K as described by Hitchcock and Brown (22), which was followed by hot phenol extraction and subsequent extraction of the aqueous phase with ether. LPS was resolved by electrophoresis in 14% polyacrylamide gels using a Tricine-sodium dodecyl sulfate (SDS) system (26, 56) and was visualized by silver staining. Each well was loaded with the same LPS concentration, as determined by the keto-deoxyoctulosonic (KDO) assay (43). A densitometry analysis was performed using ImageJ software. The ratio of the relative intensity of the lipid A-core band to the average intensity of the bands corresponding to total O antigen and core + n was calculated by quantifying the pixels in a narrow window across the center of each lane. The densitometric analysis was calibrated by determining the ratio of the relative intensity of the lipid A-core region to the average intensity of the O-antigen bands.
β-Galactosidase assays.
The expression of the lacZY transcriptional fusion was quantified spectrophotometrically as described elsewhere (36, 46). Enzymatic activity, which was expressed in Miller units and normalized for bacterial density (OD600), was calculated using the following equation: [(A420 − 1.75A550) × 1,000]/(reaction time × culture volume × OD600), where the reaction time was expressed in minutes and the culture volume was expressed in milliliters. Each sample was analyzed in triplicate for three independent experiments.
Protein analysis.
Total membrane fractions were prepared from cells grown in LB medium and harvested at an OD600 of 1 as previously described (34). Samples were mixed with 3× protein tracking dye, incubated for 30 min at 45°C, separated on a 14% SDS-polyacrylamide gel electrophoresis (PAGE) gel, and transferred to nitrocellulose membranes. The same membrane was incubated with anti-Wzz affinity-purified polyclonal rabbit antibodies (34) and anti-Flag monoclonal antibodies (Sigma). The reacting bands were detected by fluorescence with an Odyssey infrared imaging system (Li-cor Biosciences) using IRDye800CW affinity-purified anti-rabbit antibodies (Rockland, Pennsylvania) and Alexa Fluor 680 anti-mouse antibodies (Molecular Probes). Densitometry analysis was performed using the Odyssey software and digital images of the membranes, which were incubated simultaneously with both anti-Wzz and anti-Flag antibodies. This resulted in protein bands with two fluorescence colors, and since the anti-Flag antibody cross-reacted with an unknown constitutively expressed membrane protein, we used the density of pixels of this protein band as an internal loading standard for normalization.
RT and real-time PCR.
Bacteria were grown at 37°C with agitation to an OD600 of 0.6. Cells were lysed, and total RNA was isolated using Trizol reagent (Invitrogen). Contaminating DNA was digested with RNase-free DNase I (Epicentre Biotechnologies), and the purity of all RNA preparations was confirmed by subjecting them to reverse transcription-PCR (RT-PCR) analysis using primers specific for the gene encoding the 16S rRNA (Table 2). After inactivation of DNase, RNA was used as a template for RT. Complementary cDNA was synthesized using random hexamer primers (Invitrogen), deoxynucleoside triphosphates, and Moloney murine leukemia virus reverse transcriptase (Invitrogen). Relative quantitative real-time PCR was performed with an appropriate primer set, cDNAs, and Mezcla Real (Biodynamics) that contained nucleotides, polymerase, reaction buffer, and Green dye, using a Rotor-Gene 6000 real-time PCR machine (Corbett Research). The amplification program consisted of an initial incubation for 3 min at 95°C, followed by 40 cycles of 95°C for 20 s, 60°C for 30 s, and 72°C 20 s. We used primers q-wzz-F and q-wzz-R for wzz and primers q-16S-F and q-16S-R for the 16S rRNA gene (Table 2). For the relative gene expression analysis, a comparative cycle threshold method (ΔΔCT) was used (28). The number of copies of each sample transcript was determined with the aid of the software. Briefly, the amplification efficiencies of the genes of interest and the 16S rRNA gene used for normalization were tested. Then each sample was first normalized for the amount of template added by comparison to the 16S rRNA gene (endogenous control). The normalized values were further normalized using the wild-type sample (calibrator treatment). Hence, the results were expressed relative to the value for the calibrator sample, which was 1.
RESULTS AND DISCUSSION
Abnormal O-antigen chain length distribution in a Salmonella serovar Enteritidis dam deletion mutant.
We observed previously that Salmonella serovar Enteritidis mutant SD1 expressing a truncated Dam protein (15) does not agglutinate with anti-O9 serum (14). To better understand the role of Dam in LPS expression, we constructed a dam deletion mutant of Salmonella serovar Enteritidis strain #5694 (SEΔdam) (5). The dam null phenotype of SEΔdam was verified by digesting its chromosomal DNA with appropriate restriction enzymes. SEΔdam was susceptible to 2-aminopurine and formed filaments when it was cultured in liquid media, both of which are characteristic phenotypes of dam mutants (data not shown). Like the SD1 mutant (14, 55), SEΔdam also showed a reduced level of agglutination with anti-O9 serum, as demonstrated by a 1- to 2-min delay in agglutination compared to the parental strain, which agglutinated almost immediately.
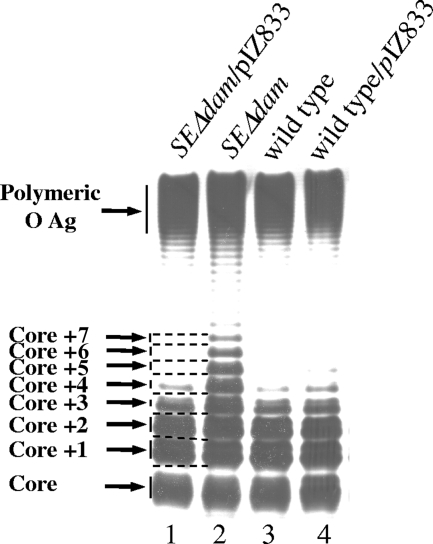
The LPS O-antigen profiles of the SEΔdam and parental strains were examined by electrophoresis and silver staining. A different banding pattern was observed for the LPS O antigen from SEΔdam, which had many more visible bands in the intermediate region of the gel (Fig. 1, lane 2) compared to the banding pattern of the wild-type LPS (Fig. 1, lane 1). This region contains O polymers with lower molecular masses, usually consisting of one to five O-antigen units. On the other hand, the amount of molecules in the polymeric O-antigen region was not substantially different (Fig. 1). Therefore, the observed difference in the SEΔdam LPS pattern is consistent with an increased amount of short polysaccharide chains, suggesting that this mutant has an altered O-antigen polysaccharide chain length distribution. To determine if this defect is associated with the absence of Dam function, we also examined the O-antigen banding pattern of the SEΔdam strain containing pIZ833, which contains a functional dam gene. The banding pattern of this strain was similar to the parental banding pattern (Fig. 1, lane 4). Densitometric quantification of LPS gels revealed no significant differences between SEΔdam and the wild-type strain in the amount of total O antigen relative to the lipid A-core region (3.365 ± 0.105 and 3.115 ± 0.165, respectively). This result suggests that the shorter polysaccharide chains observed in the mutant are not synthesized at the expense of longer chains. We concluded that Dam methylation has an effect on the O-antigen polysaccharide chain length in Salmonella serovar Enteritidis. This phenomenon was not unique to Salmonella, as we also found that a lack of Dam affects the LPS pattern in Escherichia coli, increasing the amount of shorter polysaccharides (as seen in SEΔdam) and modifying the size of the banding pattern of the O-antigen region (data not shown). Also, Fälker et al. showed that overproduction of Dam in Yersinia enterocolitica results in an increased amount of rough LPS molecules (13). Moreover, these authors suggested that Dam methylation affects the stability of shorter LPS species or influences the addition of the first O-antigen units to a growing chain. Taken together, these findings indicate that Dam methylation plays a role in O-antigen LPS expression in enterobacteria.
FIG. 1.
LPS analysis of Salmonella serovar Enteritidis strains. LPS from wild-type strains (wild type and wild type/pIZ833) and dam deletion mutants (SEΔdam and SEΔdam/pIZ833) of Salmonella serovar Enteritidis were studied. Equal amounts of LPS were loaded in each lane and analyzed by Tricine-SDS-PAGE on a 14% (wt/vol) acrylamide gel, followed by silver staining. The concentration of LPS was determined by measuring the KDO using the purpald assay. Plasmid pIZ833 bears the dam gene. The gel shown is representative of nine independent experiments. O Ag, O antigen.
Dam participates in the regulation of Wzz synthesis.
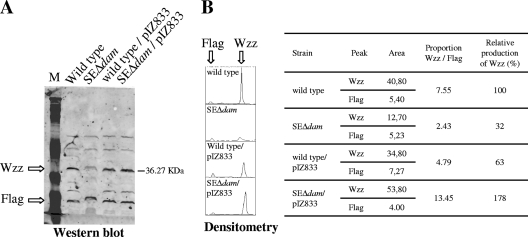
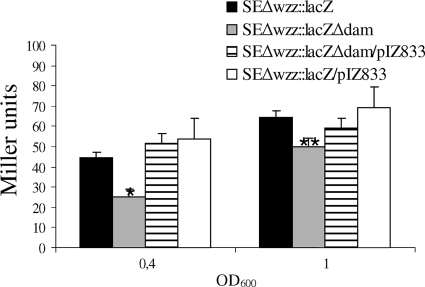
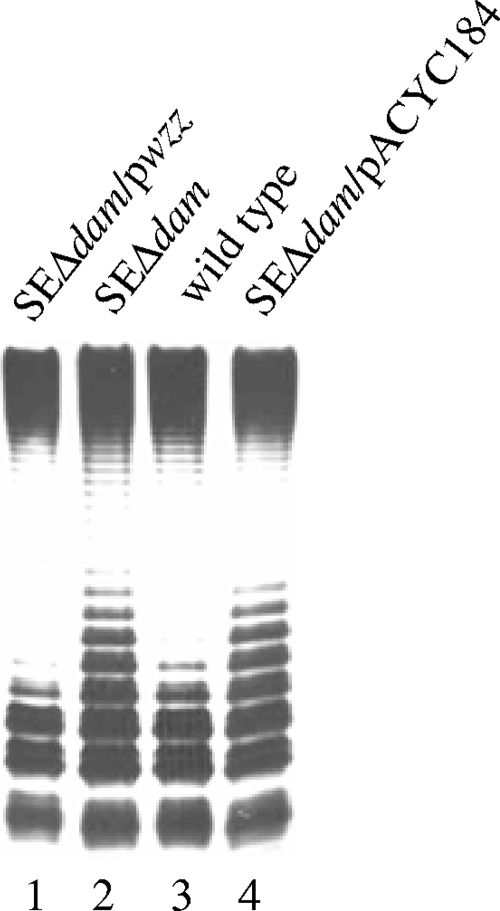
Since Wzz is responsible for the chain length distribution of the O antigen (37, 47), we investigated whether Dam methylation is involved in regulating wzz gene expression. Membrane protein preparations from the parental strain, SEΔdam, and a strain complemented with pIZ833 were examined by Western blotting using anti-Wzz serum, and the results of a representative experiment are shown in Fig. 2A. We also treated the blots simultaneously with the anti-Flag antibody, since this antibody cross-reacts with an unknown constitutively expressed membrane protein. Therefore, the density of the pixels for this protein band serves as an internal loading standard. Normalized densitometry analysis showed that the relative level of the Wzz protein produced by the dam mutant was threefold lower (32%) than the level of the Wzz protein produced by the parental strain (Fig. 2B). To investigate wzz gene transcription, we determined the galactosidase activities produced by the parental strain and the SEΔdam mutant harboring a wzz::lacZY transcriptional fusion in the chromosome. β-Galactosidase activity was measured using cells harvested from exponential- and stationary-phase cultures (OD600, 0.4 and 1.0, respectively). As shown in Fig. 3, the transcription of the wzz gene in SEΔwzz::lacZΔdam was nearly 50% lower in the logarithmic phase and 25% lower in the stationary phase than the transcription in parental strain SEΔwzz::lacZ (P < 0.01 and P < 0.05, respectively). These results were confirmed by RT-PCR, which revealed that wzz gene expression was threefold lower in the dam mutant (relative amount of mRNA, 0.313 ± 0.025) than in the parental strain. Next, we investigated whether increased expression of wzz in SEΔdam could restore the wild-type O-antigen banding pattern. To do this, strain SEΔdam was transformed with plasmid pwzz bearing the wzz gene under regulation of the tetracycline resistance gene promoter. As shown in Fig. 4, overexpression of wzz in SEΔdam reduced the amount of short polysaccharide chains observed in the dam mutant (lanes 1 and 2), resulting in an LPS pattern comparable to that observed for the parental strain (lane 3). Transformation with a plasmid vector (pACYC184) resulted in no changes in the O-antigen LPS pattern of strain SEΔdam (Fig. 4, lane 4).
FIG. 2.
Wzz protein in Salmonella serovar Enteritidis dam mutant. Bacterial strains were cultured in LB medium and harvested at an OD600 of 1. Bacterial proteins were extracted, and the Wzz protein was analyzed by Western blotting (A). The intensities of Wzz and FLAG bands were determined by densitometry (B). Plasmid pIZ833 bears the dam gene. Prestained SDS-PAGE standards (Bio-Rad) were used as molecular weight markers. The data are representative data for three independent experiments. Lane M contained broad-range prestained SDS-PAGE standards (Bio-Rad).
FIG. 3.
Activity of wzz gene promoter of Salmonella serovar Enteritidis strains. The β-galactosidase activities (in Miller units) expressed by strains harboring chromosomal lacZY transcriptional fusions to the wzz gene in the exponential (OD600, 0.4) and stationary (OD600, 1) phases were determined. The transcriptional activities in SEΔwzz::lacZ and in the dam deletion mutant SEΔwzz::lacZΔdam of Salmonella serovar Enteritidis were investigated. Strains harboring a plasmid bearing the dam gene (SEΔwzz::lacZ/pIZ833 and SEΔwzz::lacZΔdam/pIZ833) were included as controls. The data are means ± standard deviations of three independent experiments performed in triplicate. *, P < 0.01 for a comparison with the wild type; **, P < 0.05 for a comparison with the wild type.
FIG. 4.
Production of LPS in Salmonella serovar Enteritidis dam mutant transformed with the wzz gene. Equal amounts of LPS were loaded in the lanes and analyzed by Tricine-SDS-PAGE on a 14% (wt/vol) acrylamide gel, followed by silver staining. The concentration of LPS was determined by measuring KDO using the purpald assay. The data are representative data for seven independent experiments.
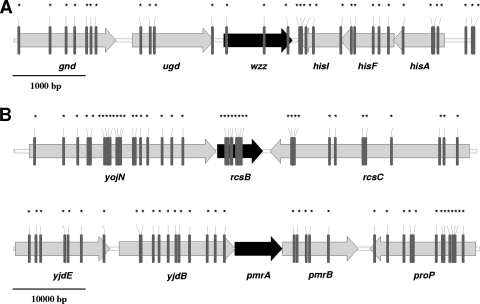
Collectively, the experiments described above demonstrated that Dam activity modulates wzz gene expression. Therefore, it is expected that the wzz gene locus contains GATC sequences with the potential to be methylated by Dam. Certainly, an in silico analysis (Fig. 5A) revealed the presence of three GATC motifs in the coding sequence of wzz and four GATC sequences upstream of this gene. Dam methylation of any of these GATC clusters could regulate wzz expression, although through different mechanisms. On the one hand, it has been proposed that a GATC cluster located in the coding sequence of a given gene affects DNA stability, depending on the methylated state (50). On the other hand, Dam methylation in three GATC motifs (18, 59), two GATC motifs (2), or even a single GATC motif (3, 51) located upstream of a given gene can affect the interaction between regulatory proteins and the DNA binding site. Moreover, Wallecha et al. demonstrated that transcription of the agn43 gene in E. coli is regulated by a GATC motif located downstream of the promoter (60). It remains to be determined whether Dam regulation of the wzz gene is also mediated in an indirect manner. For instance, Dam methylation could regulate the expression of other genes whose products are involved in wzz transcription. It is known that the PmrA/PmrB and RcsC/YojN/RcsB two-component systems independently promote transcription of the wzz gene since a pmrA rcsB double mutant of Salmonella serovar Typhimurium does not express Wzz (8). In theory, the expression of pmrA and rcsB could be regulated by Dam methylation since GATC motifs are present upstream of both genes (Fig. 5B). Interestingly, no GATC motifs are present in the pmrA sequence, whereas rcsB contains eight GATC sequences downstream of the +1 codon in a 224-bp interval, a density threefold higher than the density expected from a random distribution. The regulation of wzz in Salmonella serovar Enteritidis is highly complex and not completely understood; therefore, further experiments are necessary to investigate whether the expression of pmrA, rcsB, or a different unidentified factor(s) is regulated by Dam methylation.
FIG. 5.
Distribution of GATC sequences. (A) wzz gene region. (B) pmrA and rcsB gene region. The vertical lines and asterisks indicate the locations of GATC motifs. The diagram is based on the Refseq NC_011294 sequence of S. enterica serovar Enteritidis.
In summary, our results unequivocally show that wzz gene expression is downregulated in the dam mutant of Salmonella serovar Enteritidis. The presence of GATC motifs in wzz, as well as in the pmrA and rcsB gene clusters, strongly suggests that Dam methylation is involved in the regulation of LPS synthesis.
Acknowledgments
We are very grateful to M. I. Bernal for technical assistance and to C. Quiroga and P. Lahiry for useful discussions.
This work was supported in part by grants from Universidad de Buenos Aires, Buenos Aires, Argentina (grant UBACyT M009), and Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina (grants PIP 5534 and 0992) to M.C.C. and from the Canadian Institutes of Health Research to M.A.V.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Balbontín, R., G. Rowley, M. G. Pucciarelli, J. López-Garrido, Y. Wormstone, S. Lucchini, F. García-Del Portillo, J. C. Hinton, and J. Casadesús. 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:8160-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braaten, B. A., J. V. Platko, M. van der Woude, B. H. Simons, F. K. de Graaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camacho, E. M., and J. Casadesús. 2005. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 57:1700-1718. [DOI] [PubMed] [Google Scholar]
- 4.Campellone, K. G., A. J. Roe, A. Løbner-Olesen, K. C. Murphy, L. Magoun, M. J. Brady, A. Donohue-Rolfe, S. Tzipori, D. L. Gally, J. M. Leong, and M. G. Marinus. 2007. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 63:1468-1481. [DOI] [PubMed] [Google Scholar]
- 5.Cerquetti, M. C., E. Hovsepian, S. H. Sarnacki, and N. B. Goren. 2008. Salmonella enterica serovar Enteritidis dam mutant induces low NOS-2 and COX-2 expression in macrophages via attenuation of MAPK and NF-κB pathways. Microbes Infect. 10:1431-1439. [DOI] [PubMed] [Google Scholar]
- 6.Chessa, D., M. G. Winter, S. P. Nuccio, C. Tükel, and A. J. Bäumler. 2008. RosE represses Std fimbrial expression in Salmonella enterica serotype Typhimurium. Mol. Microbiol. 68:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 90:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado, M. A., C. Mouslim, and E. A. Groisman. 2006. The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol. Microbiol. 60:39-50. [DOI] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine 21:3249-3258. [DOI] [PubMed] [Google Scholar]
- 11.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants prevent colonization of newly hatched chickens by homologous and heterologous serovars. Int. J. Food Microbiol. 80:153-159. [DOI] [PubMed] [Google Scholar]
- 12.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 13.Fälker, S., J. Schilling, M. A. Schmidt, and G. Heusipp. 2007. Overproduction of DNA adenine methyltransferase alters motility, invasion, and the lipopolysaccharide O-antigen composition of Yersinia enterocolitica. Infect. Immun. 75:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giacomodonato, M. N., S. H. Sarnacki, F. Sisti, R. L. Caccuri, and M. C. Cerquetti. 2003. Salmonella enteritidis dam mutant of leaky phenotype as a potential vaccine strain, abstr. 106A. Abstr. ASM Conf. Salmonella: pathogenesis, epidemiology, and vaccine development. American Society for Microbiology, Washington, DC.
- 15.Giacomodonato, M. N., S. H. Sarnacki, R. L. Caccuri, D. O. Sordelli, and M. C. Cerquetti. 2004. Host response to a dam mutant of Salmonella enterica serovar Enteritidis with a temperature-sensitive phenotype. Infect. Immun. 72:5498-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glickman, B., P. van den Elsen, and M. Radman. 1978. Induced mutagenesis in dam− mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol. Gen. Genet. 163:307-312. [DOI] [PubMed] [Google Scholar]
- 17.Grossman, N., M. A. Schmetz, J. Foulds, E. N. Klima, V. Jiminez, L. L. Leive, and K. A. Joiner. 1987. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 169:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 19.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 20.Heinrichs, D. E., J. A. Yethon, P. A. Amor, and C. Whitfield. 1998. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharides of Escherichia coli. The R1 core-specific beta-glucosyltransferase provides a novel attachment site for O-polysaccharides. J. Biol. Chem. 6:29497-29505. [DOI] [PubMed] [Google Scholar]
- 21.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoare, A., M. Bittner, J. Carter, S. Alvarez, M. Zaldivar, D. Bravo, M. A. Valvano, and I. Contreras. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect. Immun. 74:1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakomin, M., D. Chessa, A. J. Bäumler, and J. Casadesús. 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190:7406-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 27.Liu, D., R. A. Cole, and P. R. Reeves. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 178:2102-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
28.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the
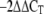 method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar] - 29.Løbner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154-160. [DOI] [PubMed] [Google Scholar]
- 30.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marino, P. A., B. C. McGrath, and M. J. Osborn. 1991. Energy dependence of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:3128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 33.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 172:3590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marolda, C. L., E. R. Haggerty, M. Lung, and M. A. Valvano. 2008. Functional analysis of predicted coiled-coil regions in the Escherichia coli K-12 O-antigen polysaccharide chain length determinant Wzz. J. Bacteriol. 190:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath, B. C., and M. J. Osborn. 1991. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics, p.352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Morona, R., L. Van Den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 177:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulford, C. A., and M. J. Osborn. 1983. An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 80:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata, T., W. Tseng, T. Guina, S. I. Miller, and H. Nikaido. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:7213-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella Typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 41.Murray, G. L., S. R. Attridge, and R. Morona. 2005. Inducible serum resistance in Salmonella Typhimurium is dependent on wzz(fepE)-regulated very long O antigen chains. Microbes Infect. 7:1296-1304. [DOI] [PubMed] [Google Scholar]
- 42.Murray, G. L., S. R. Attridge, and R. Morona. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osborn, M. J. 1963. Studies in the Gram-negative cell wall. I. Evidence for the role of 2-keto-3-deoxyoctonoate in the lipopolysaccharide of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 50:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, B. R., and M. N. Marinus. 1994. The dam and dcm strains of Escherichia coli. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 45.Phalipon, A., C. Costachel, C. Grandjean, A. Thuizat, C. Guerreiro, M. Tanguy, F. Nato, B. Vulliez-Le Normand, F. Belot, K. Wright, V. Marcel-Peyre, P. J. Sansonetti, and L. A. Mulard. 2006. Characterization of functional oligosaccharide mimics of the Shigella flexneri serotype 2a O-antigen: implications for the development of a chemically defined glycoconjugate vaccine. J. Immunol. 176:1686-1694. [DOI] [PubMed] [Google Scholar]
- 46.Putnam, S. L., and A. L. Koch. 1975. Complication in the simplest cellular enzyme assay: lysis of Escherichia coli for the assay of β-galactosidase. Anal. Biochem. 63:350-360. [DOI] [PubMed] [Google Scholar]
- 47.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rick, P. D., K. Barr, K. Sankaran, J. Kajimura, J. S. Rush, and C. J. Waechter. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278:16534-16542. [DOI] [PubMed] [Google Scholar]
- 50.Riva, A., M. O. Delorme, T. Chevalier, N. Guilhot, C. Hénaut, and A. Hénaut. 2004. The difficult interpretation of transcriptome data: the case of the GATC regulatory network. Comput. Biol. Chem. 28:109-118. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43:117-130. [DOI] [PubMed] [Google Scholar]
- 52.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Samuel, J. E., K. Kiss, and S. Varghees. 2003. Molecular pathogenesis of Coxiella burnetii in a genomics era. Ann. N. Y. Acad. Sci. 990:653-663. [DOI] [PubMed] [Google Scholar]
- 55.Sarnacki, S. H., M. N. Giacomodonato, M. Noto Llana, M. A. Valvano, and M. C. Cerquetti. 2006. Diminished innate and acquired immunity response to dam mutants of Salmonella enterica could be related to a defective lipopolysaccharide (LPS) synthesis, abstr. B052. Abstr. 106th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 56.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 57.Torreblanca, J., S. Marques, and J. Casadesus. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452-471. [DOI] [PubMed] [Google Scholar]
- 59.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 60.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitfield, C., and M. A. Valvano. 1993. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv. Microb. Physiol. 35:135-246. [DOI] [PubMed] [Google Scholar]
- 62.Whitfield, C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3:178-185. [DOI] [PubMed] [Google Scholar]
- 63.Wion, D., and J. Casadesús. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]