Abstract
In all domains of life Oxa1p-like proteins are involved in membrane protein biogenesis. Bacillus subtilis, a model organism for gram-positive bacteria, contains two Oxa1p homologs: SpoIIIJ and YqjG. These molecules appear to be mutually exchangeable, although SpoIIIJ is specifically required for spore formation. SpoIIIJ and YqjG have been implicated in a posttranslocational stage of protein secretion. Here we show that the expression of either spoIIIJ or yqjG functionally compensates for the defects in membrane insertion due to YidC depletion in Escherichia coli. Both SpoIIIJ and YqjG complement the function of YidC in SecYEG-dependent and -independent membrane insertion of subunits of the cytochrome o oxidase and F1Fo ATP synthase complexes. Furthermore, SpoIIIJ and YqjG facilitate membrane insertion of F1Fo ATP synthase subunit c from both E. coli and B. subtilis into inner membrane vesicles of E. coli. When isolated from B. subtilis cells, SpoIIIJ and YqjG were found to be associated with the entire F1Fo ATP synthase complex, suggesting that they have a role late in the membrane assembly process. These data demonstrate that the Bacillus Oxa1p homologs have a role in membrane protein biogenesis rather than in protein secretion.
The YidC/OxaI/Alb3 protein family plays a crucial role in membrane protein biogenesis by facilitating the insertion of a specific subset of membrane proteins (for reviews, see references 20 and 24). In mitochondria, the OxaI protein is essential for insertion of both nucleus- and mitochondrion-encoded proteins into the inner membrane (39). The OxaI homolog of Escherichia coli, designated YidC, is known to play a role in two different membrane protein insertion pathways. Some proteins, such as subunit c of the rotary domain of the F1Fo ATP synthase (Foc) (47), MscL (10), M13 (34), and Pf3 (5), insert via the YidC-only pathway. YidC also functions in concert with the protein-conducting channel SecYEG in membrane insertion of subunit a of cytochrome o oxidase (CyoA) (8, 44) and subunit a of the F1Fo ATP synthase (23, 53, 54). In addition, YidC has been implicated in the folding of a membrane-inserted lactose permease (30) and the binding protein-dependent maltose ABC transporter (50).
Members of the YidC/OxaI/Alb3 protein family are found in all three domains of life, and the number of paralogs per cell or organelle ranges from one (most gram-negative bacteria) to six (Arabidopsis thaliana). The length of Oxa1p-like proteins varies considerably, from just over 200 amino acids (in most gram-positive bacteria) to 795 amino acids (Chlamydophila pneumoniae) (52). However, in all Oxa1p proteins, a conserved region consisting of about 200 amino acids can be recognized, which comprises five putative transmembrane segments, as experimentally demonstrated for E. coli YidC (33). Overall, the amino acid sequence conservation among Oxa1p homologs is low (17). Bacillus subtilis contains two membrane proteins, SpoIIIJ and YqjG, with significant similarity to proteins belonging to the YidC/OxaI/Alb3 family. Previous gene inactivation analysis showed that a single paralog is sufficient for cell viability during vegetative growth of B. subtilis, while a double knockout led to a lethal phenotype (29, 41). SpoIIIJ is essential for activation of a prespore-specific sigma factor (9, 36), and cells with spoIIIJ deleted are incapable of spore formation. Sporulation is blocked at stage III, directly after completion of prespore engulfment (9). YqjG cannot complement SpoIIIJ in this process, but the exact reason for the specific requirement for SpoIIIJ is unknown. Previous studies indicated that the stability of various secretory proteins (e.g., LipA and PhoA) was strongly affected under YqjG- and SpoIIIJ-limiting conditions, while the insertion or stability of a number of membrane proteins tested appeared to be unaffected (41). These data suggested that YqjG and SpoIIIJ, unlike the other Oxa1p-like proteins, play a role in protein secretion. Here we show that both YidC homologs in B. subtilis complement the E. coli growth defect due to a YidC depletion and functionally replace YidC in Sec-dependent and -independent membrane protein insertion. In vitro insertion assays demonstrated that membrane insertion of Foc of both E. coli and B. subtilis is mediated by SpoIIIJ and YqjG. In addition, the entire F1Fo ATP synthase of B. subtilis was found to copurify with both SpoIIIJ and YqjG, suggesting that these proteins have a role in a late stage of the assembly of this membrane protein complex.
MATERIALS AND METHODS
Antisera.
Antisera against PspA, Foc, LepB, and YidC were kind gifts from J. Tommassen (University of Utrecht, Utrecht, The Netherlands), G. Deckers-Hebestreit (University of Osnabrück, Osnabrück, Germany), W. Wickner (Dartmouth Medical School, Hanover, NH), and J. W. L. de Gier (Stockholm University, Stockholm, Sweden), respectively.
Strains and plasmids.
In order to (over)express spoIIIJ and yqjG in E. coli, these genes were amplified from genomic DNA (B. subtilis 168) using primer pairs FoSpoIIIJ/ReSpoIIIJ and FoYqjG/ReYqjG (Table 1), respectively. A sequence for a six-histidine tag at the 3′ end of the genes was inserted by PCR amplification using primer pairs FoSpoIIIJ/ReSpoIIIJHis and FoYqjG/ReYqjGHis (Table 1). PCR products were cleaved with NcoI and XbaI and ligated into the corresponding sites of the pTrc99A vector, yielding pTrcspoIIIJ, pTrcspoIIIJHis, pTrcyqjG, and pTrcyqjGHis. For construction of pTrcyidC, the yidC gene was amplified from genomic DNA derived from E. coli SF100 using primers FoYidC and ReYidC. The amplified product was cleaved with XbaI and HindIII and ligated into the corresponding sites of pTrc99A.
TABLE 1.
Primers
| Primer | Sequence (5′→3′) |
|---|---|
| FoSpo | GCTGCCATGGTGTTGAAAAGGAGAATAGGG |
| ReSpo | GCGTCTAGATCACTTTTTCTTTCCTCCGGCTTTTTGCGGC |
| ReSpoHis | GCGTCTAGATCAGTGATGGTGATGGTGATGCTTTTTCTTTCCTCCGGCTTTTTGCGGC |
| FoYqjG | GCTGCCATGGTAAAAACATATCAAAAACTTTTGGC |
| ReYqjG | GCGTCTAGATTATTTCACCGACTCAGTAAGAGCGGCTGTTTTTTTAC |
| ReYqjGHis | GCGTCTAGATTAGTGATGGTGATGGTGATGTTTCACCGACTCAGTAAGAGCGGCTGTTTTTTTAC |
| FoYidC | GCTCTAGATGGATTCGCAACGC |
| ReYidC | GGGAAGCTTCAGGATTTTTTCTTCTCGCGGC |
| FoBatpE | GGGAATTCCATATGAATTTAATAGCAGC |
| ReBatpE | CGGAATTCTTAGCCAAAGAACGC |
For overexpression of SpoIIIJ and YqjG in B. subtilis the SURE (subtilin-regulated) expression system was used. The same PCR products that were used for cloning into pTrc99A were digested with NcoI and XbaI and ligated into the corresponding sites of the pNZ8901 vector. The resulting constructs were designated pNZspoIIIJ, pNZspoIIIJHis, pNZyqjG, and pNZyqjGHis (Table 1). The lantibiotic subtilin, the inducer for expression in the SURE system, was prepared from the culture supernatant of B. subtilis ATCC 6633 as described previously (3). B. subtilis pNZ8900 was used as a host strain for expression of SpoIIIJ and YqjG. The DNA sequence of spoIIIJ amplified by PCR using genomic DNA of B. subtilis 168 as the template exhibited a silent A-G point mutation at position 144. In the amplified yqjG gene a C-T point mutation at position 557 introduces an alanine-to-valine substitution at amino acid position 186 of the overexpressed YgjG protein.
For studies with E. coli, strain FTL10 was used, in which expression of the yidC gene is under control of the araBAD operator/promoter (13). Since YidC is an essential protein, E. coli FTL10 is viable only when the growth medium is supplemented with arabinose (0.2%, wt/vol). Strains and plasmids used in this study are listed in Table 2.
TABLE 2.
Strains and plasmids
| Plasmid or strain | Relevant properties | Reference |
|---|---|---|
| Plasmids | ||
| pTrc99A | Expression vector for E. coli based on pKK233-2, carries a hybrid trp/lac promoter and the multiple cloning site of pUC18; Apr | 1 |
| pTrcspoIIIJ | Like pTrc99A, contains B. subtilis spoIIIJ | This study |
| pTrcspoIIIJHis | Like pTrc99A, contains B. subtilis spoIIIJ with His6 tag at 3′ end | This study |
| pTrcyqjG | Like pTrc99A, contains B. subtilis yqjG | This study |
| pTrcyqjGHis | Like pTrc99A, contains B. subtilis yqjG with His6 tag at 3′ end | This study |
| pTrcyidC | Like pTrc99A, contains E. coli yidC | This study |
| pNZ8901 | Expression vector for B. subtilis, mutated Pspa promoter; Cmr | 3 |
| pNZspoIIIJ | Like pNZ8901, contains B. subtilis spoIIIJ | This study |
| pNZspoIIIJHis | Like pNZ8901, contains B. subtilis spoIIIJ with His6 tag at 3′ end | This study |
| pNZyqjG | Like pNZ8901, contains B. subtilis yqjG | This study |
| pNZyqjGHis | Like pNZ8901, contains B. subtilis yqjG with His6 tag at 3′ end | This study |
| pET20batpE | pET20b (Novagen) containing E. coli atpE | 47 |
| pET20bBatpE | pET20b containing B. subtilis atpE | This study |
| E. coli strains | ||
| DH5α | supE44 hsdR14 recA1 endA1 gyrA96 thi-1 relA1 ΔlacU169 (φ80lacZΔM15); K-12 derivative | 12 |
| SF100 | F− ΔlacX74 galK thi rpsL (strA) ΔphoA(pvuII) ΔompT | 2 |
| FTL10 | ΔyidC attB::(araC+ PBADyidC+); Kanr | 13 |
| B. subtilis strains | ||
| 168 | trpC2 | 25 |
| ATCC 6633 | Subtilin producer | 7 |
| pNZ8900 | 168 amyE::spaRK; Kmr | 3 |
For in vitro expression of B. subtilis Foc, the atpE gene was amplified by PCR using primer pair FoBatpE/ReBatpE (Table 1) and genomic DNA from B. subtilis 168 as the template. The PCR product was cleaved by NdeI and EcoRI and subsequently cloned into the corresponding sites of pET20b (Novagen). The resulting construct was designated pET20bBatpE. For expression of E. coli Foc plasmid pET20batpE (47) was used.
Cell growth and isolation of IMVs.
E. coli strain FTL10 (13) was transformed with plasmid pTrc99A (wild-type Ara and YidC depleted), pTrcyidC (wild-type Glu), pTrcspoIIIJ (SpoIII), or pTrcyqjG (YqjG). The cultures were grown overnight at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) and 0.2% (wt/vol) arabinose (wild-type Ara and ΔYidC) or 0.2% (wt/vol) glucose (wild-type Glu, SpoIII, and YqjG). Cultures were diluted 1:100 in fresh medium and then incubated at 37°C. When the cultures reached an optical density at 660 nm (OD660) of 0.8, cells were harvested by centrifugation (10 min, 7,500 × g). To obtain YidC-depleted cells, the pellet was washed once in prewarmed LB medium and the cells were resuspended to an OD660 of 0.4 in LB medium containing 0.2% (wt/vol) glucose. When an OD660 of 0.8 was reached, cells were diluted twofold with LB medium containing 0.2% (wt/vol) glucose. This procedure was repeated until the cells stopped growing (mostly after the fifth generation). Inverted inner membrane vesicles (IMVs) were isolated as described previously (19).
Protein expression and purification.
In order to express His-tagged SpoIIIJ and YqjG in E. coli, the pTrcspoIIIJHis and pTrcyqjGHis constructs were transformed into E. coli SF100. Overnight cultures grown on LB medium containing 100 μg/ml ampicillin were diluted 40-fold, and cell growth was continued until the OD660 was 0.7. Expression was induced by addition of 0.5 mM IPTG (isopropyl-β-thiogalactopyranoside), and cells were grown for an additional 2.5 h. Cells were harvested by centrifugation (8,000 × g, 15 min) and resuspended in 50 mM Tris-HCl (pH 8.0) containing 20% (wt/vol) sucrose, and IMVs were isolated as described previously (19).
For expression experiments with B. subtilis, the empty vector pNZ8901 or either the pNZspoIIIJ, pNZspoIIIJHis, pNZyqjG, or pNZyqjGHis construct was transformed into B. subtilis pNZ8900. Cells were grown in TY medium (19) supplemented with kanamycin (10 μg/ml) and chloramphenicol (5 μg/ml). Overnight cultures were diluted 40-fold in fresh medium and grown to an OD660 of 1.0. Protein expression was induced by addition of subtilin (0.1%, vol/vol). Cell growth was continued for 2 h, after which cells were harvested by centrifugation (8,000 × g, 15 min), resuspended in protoplasting buffer (50 mM Tris-HCl [pH 8.0], 20% [wt/vol] sucrose, 0.1 mM phenylmethanesulfonyl fluoride [PMSF], 1 mg/ml lysozyme), and incubated for 30 min at 37°C. Protoplasts were harvested by centrifugation, and the resulting pellet was resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], DNase, 0.1 mM PMSF). Subsequently, the cells were disrupted by sonication (Soniprep; MSE), and after the cell debris was removed (8,000 × g 10 min) the membranes were collected by centrifugation (180,000 × g, 30 min), resuspended in 50 mM Tris-HCl (pH 8.0) containing 20% (vol/vol) glycerol, and stored at −80°C.
For protein purification, E. coli or B. subtilis membranes were diluted to obtain a protein concentration of 1 mg/ml with solubilization buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, 5 mM imidazole, 20% [vol/vol] glycerol, 2% [wt/vol] dodecyl maltoside [DDM]) and incubated for 1 h at 4°C. After unsolubilized material was removed by centrifugation (100,000 × g, 30 min), the supernatant was mixed with Ni2+-nitrilotriacetic acid (NTA) agarose beads (Qiagen) and incubated overnight at 4°C. The mixture was poured into a column, and unbound material was removed by draining. Beads were subsequently washed with 5 column volumes of 10 mM Tris-HCl (pH 8.0), 100 mM KCl, 30 mM imidazole, 20% (vol/vol) glycerol, 0.1% (wt/vol) DDM. Finally, bound proteins were eluted with 10 mM Tris-HCl (pH 7.0), 100 mM KCl, 400 mM imidazole, 20% (vol/vol) glycerol, 0.1% (wt/vol) DDM and stored at −80°C.
ΔpH measurement.
The generation of a transmembrane pH gradient (ΔpH) by IMVs was monitored by use of the fluorescent indicator 9-amino-6-chloro-2-methoxyacridine (ACMA). Measurements were obtained with a Perkin Elmer LS50B luminescence spectrophotometer at 30°C using excitation and emission wavelengths of 409 and 474 nm, respectively, with slit width of 3 nm. After IMVs (50 μg/ml) were added to the assay buffer (100 mM HEPES/KOH [pH 8.0], 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT]), the indicator ACMA was added to a final concentration of 1 μM, and the reaction was started by adding either ATP (1 mM) or NADH (1.25 mM). To convert the transmembrane electrical potential into a ΔpH, valinomycin (1 μM) was added, while nigericin (1 μM) was used to dissipate the ΔpH.
ATPase activity assay.
The ATPase activity of IMVs was determined as described previously (26), using the phosphate-sensitive dye malachite green. IMVs were diluted in assay buffer (50 mM HEPES/KOH [pH 7.5], 50 mM KCl, 2.5 mM MgCl2, 2 mM DTT) to obtain a concentration of 15 μg/ml and incubated at 37°C. The reaction was started by adding ATP (final concentration, 1 mM) and was stopped after 15 min by addition of EDTA (final concentration, 10 mM).
Oxygen consumption measurement.
Cytochrome o oxidase activity was determined by measuring the oxygen consumption of IMVs as described previously (27), using a Clarke electrode (Yellow Springs Instrument Co., Inc., Yellow Springs, OH). The reaction mixtures contained 100 mM potassium phosphate buffer (pH 6.6), 5 mM MgCl2, 2.5 mM potassium ascorbate (electron acceptor), and 35 μg/ml IMVs. Reactions were started by addition of the artificial electron donor phenomethosulfate (final concentration, 10 μM). All values determined were normalized using the endogenous oxygen consumption.
Cytochrome absorption spectra.
Cytochrome absorption spectra of air-oxidized and dithionate-reduced IMVs (0.5 mg/ml in 10 mM HEPES/KOH [pH 7.5]) were measured at 25°C using a Carry-Varian spectrophotometer. The spectra were recorded in the wavelength range from 400 to 500 nm.
In vitro transcription, translation, and membrane insertion.
For in vitro synthesis of E. coli and B. subtilis Foc, a cell lysate was prepared by inoculating 50 ml double-strength YPTG medium (21) with a single colony of strain BL21(DE3) Rosetta (Novagen). After overnight incubation at 37°C and 250 rpm, this preculture was used to inoculate a 4-liter Erlenmeyer containing 1 liter of 2× YPTG medium (21). When the OD600 was 0.6, the culture was chilled in an ice-water bath. Cells were harvested by centrifugation (15 min, 7,500 rpm, JLA 10-500 rotor, 4°C), washed with buffer A (10 mM Tris-acetate [pH 8.0], 14 mM magnesium acetate, 60 mM KCl, 50 μg/ml PMSF), and finally suspended in 2 ml buffer A per g of cells. Cells were lysed by passage through a cell disruptor (10,000 lb/in2; Constant Cell Disruption Systems, United Kingdom). Cell debris was removed by centrifugation (10,000 rpm, SS34 rotor, 4°C). The supernatant was transferred to an MLA-80 tube and centrifuged for 30 min at 25,000 rpm in an MLA-80 rotor (4°C). The supernatant was transferred to a 5-ml Falcon tube and supplemented with 55 mM sodium pyruvate, 45 μM coenzyme A, and 110 μM NAD. After 90 min of incubation in a water bath (37°C) in the dark, membranes were removed by ultracentrifugation (30 min, 52,000 rpm, MLA-80 rotor, 4°C). The cell lysate was dialyzed for 24 h against 1 liter of buffer A without PMSF (4°C; molecular mass cutoff, 6 to 8 kDa) with three replacements of the buffer. After dialysis, the lysate was aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C.
In vitro transcription-translation reactions were performed as described previously (15) with DTT present at a final concentration of 4 mM. Each reaction was started by addition of T7 polymerase from Fermentas and the Easytag express protein labeling mixture (Perkin Elmer). Coupled in vitro synthesis and insertion were carried out for 30 min at 37°C in the presence of 10 μg of IMVs per reaction mixture. Subsequently, a 10% synthesis control was collected, and the remainder of the mixture was subjected to proteinase K treatment (2 mg/ml, 30 min, 37°C) in order to remove noninserted proteins. All consecutive steps were performed as described previously (46).
BN-PAGE.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed using Criterion gradient gels (4 to 20%) from Bio-Rad under conditions described previously (35). Elution fractions of His-tagged purified SpoIIIJ and YqjG containing 30 μg (total amount) of protein each were incubated in 1% (wt/vol) sodium dodecyl sulfate (SDS) or 1% (wt/vol) DDM for 30 min prior to electrophoresis.
In-gel tryptic digestion.
To identify the proteins that copurify with His-tagged SpoIIIJ and YqjG, proteins eluted from the Ni2+-NTA agarose beads were separated by SDS-PAGE and stained with Coomassie brilliant blue. Protein bands were excised from the gel and washed in 50% acetonitrile in 50 mM NH4HCO3 at room temperature, until destaining was complete. The gel slices were dehydrated by incubation for 5 min in 100% acetonitrile, which was followed by rehydration in a trypsin solution (Promega) (10 ng/μl in 40 mM NH4HCO3, 10% acetonitrile). The slices were incubated for 2 h at 37°C. Subsequently, 10 μl of 25 mM NH4HCO3 was added to prevent drying, and the preparation was incubated overnight at 37°C. The tryptic peptides were recovered by three extractions with 30 μl of 20%, 50%, and 70% acetonitrile in 1% trifluoroacetic acid (TFA). The extracted peptides were pooled and concentrated under a vacuum.
Liquid chromatography-mass spectrometry.
Mass spectrometric analysis was performed using a model 4800 matrix-assisted laser desorption ionization-time of flight-time of flight proteomics analyzer (Applied Biosystems, Foster City, CA). Dried peptide mixtures from in-gel trypsin digestions were resuspended in 20 μl 0.1% TFA and separated on a C18 capillary column (75 μm by 150 mm; particle size, 3 μm; LC-Packing, Amsterdam, The Netherlands) mounted on an Ultimate 3000 nanoflow liquid chromatography system (LC-Packing, Amsterdam, The Netherlands). Buffer A (0.05% TFA) and buffer B (80% acetonitrile, 0.05% TFA) were used for elution. A gradient from 4 to 60% buffer B in 45 min was used at a flow rate of 300 nl·min−1. The column effluent was mixed 1:4 (vol/vol) with a matrix solution of 2.3 mg/ml α-cyano-4-hydroxycinnamic acid (LaserBio Labs, Sophia-Antipolis, France) containing 2 nM angiotensin II and 4 nM ACTH 18-36 (Sigma, Zwijndrecht, The Netherlands) as internal standards for mass calibration. Fractions that were 12 s wide were spotted on a blank matrix-assisted laser desorption ionization target with a Probot system (LC Packings, Amsterdam, The Netherlands). Mass spectrometric data acquisition was performed in positive-ion mode. During acquisition, peptides with a signal-to-noise ratio greater than 100 were selected for tandem mass spectrometry.
Database search.
Protein identification was carried out with the ProteinPilot 2.0 software using the Paragon algorithm (Applied Biosystems/MDS Sciex, Foster City, CA), searching against the UniProtKB/Swiss-Prot protein sequence database (40). Trypsin specificity and all default options were included in the search. ProteinPilot does not use fixed mass tolerances but employs feature probabilities and a “sequence temperature value” (38). The results were manually inspected, and protein identifications based on at least two peptides identified independently with probability greater than 99% were accepted.
RESULTS
B. subtilis spoIIIJ and yqjG complement the E. coli YidC depletion growth defect.
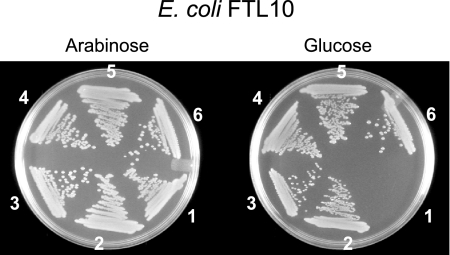
To determine whether the B. subtilis Oxa1p homologs SpoIIIJ and YqjG can complement the E. coli growth defect due to YidC depletion, plasmids pTrcspoIIIJ and pTrcyqjG (and their His-tagged versions) were transformed into the YidC depletion strain E. coli FTL10. In addition, cells were transformed with the empty vector pTrc99A and with the wild-type YidC-expressing vector pTrcyidC (Table 2), which were used as negative and positive controls, respectively. In strain FTL10, the yidC gene on the chromosome was replaced by a functional yidC gene under control of the araBAD promoter. Therefore, FTL10 has a lethal phenotype when it is grown on LB medium containing glucose, while normal growth occurs on medium supplemented with arabinose. As expected, the negative control (empty plasmid) grew only on LB medium supplemented with arabinose, and the pTrcyidC-transformed cells were viable on both glucose- and arabinose-containing media. pTrcspoIIIJ, pTrcyqjG, and the plasmids coding for the His-tagged proteins restored the growth of E. coli FTL10 on LB medium containing glucose (Fig. 1). These data demonstrate that SpoIIIJ and YqjG are able to functionally substitute for YidC in E. coli.
FIG. 1.
Complementation of the growth defect of the YidC-depleted strain E. coli FTL10 by B. subtilis SpoIIIJ and YqjG. Cells were grown on LB plates containing 100 μg/ml ampicillin and either 0.2% (wt/vol) glucose or 0.2% (wt/vol) arabinose. Cells containing pTrc99A (negative control) (section 1), pTrcyidC (positive control) (section 2), pTrcspoIIIJ (section 3), pTrcspoIIIJHis (section 4), pTrcyqjG (section 5), and pTrcyqjGHis (section 6) were grown under repression (glucose) and induction (arabinose) conditions.
Repression of PspA stress response.
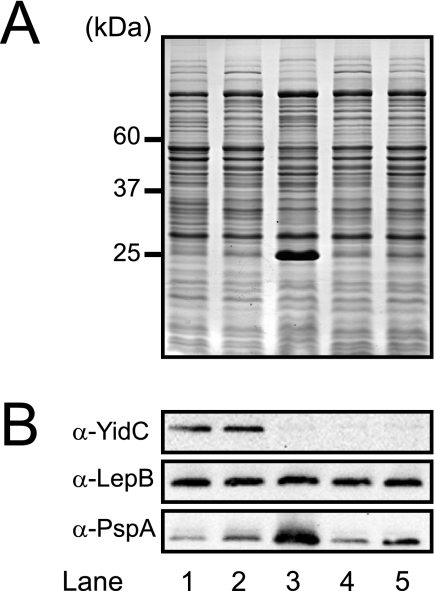
Depletion of YidC in E. coli results in increased expression of PspA, a phage shock protein that is overexpressed in cells under membrane stress conditions, including a reduction in the proton motive force (PMF) (28, 48). In order to determine if SpoIIIJ and YqjG relieve this stress response, IMVs were isolated from E. coli FTL10 cells transformed with pTrc99A, pTrcyidC, pTrcspoIIIJ, and pTrcyqjG after growth on LB medium in the presence of glucose. The overall and global protein patterns of the IMVs derived from the different cultures on SDS-PAGE gels appeared to be very similar, except that a ∼25-kDa prominent protein was present in the YidC-depleted membrane (Fig. 2, lane 3). Immunoblotting verified that this protein was PspA. The level of PspA in the YidC-depleted membranes containing SpoIIIJ or YqjG (Fig. 2, lanes 4 and 5, respectively) was comparable to wild-type levels (lanes 1 and 2), indicating that there was effective suppression of the PspA stress response. However, the PspA level in YidC-depleted IMVs containing YqjG (lane 5) was slightly altered compared with the SpoIIIJ level (lane 4), which could have been due to either a lower level of expression or partial complementation of the YidC function by YqjG in E. coli. Additionally, the levels of YidC and the signal peptidase LepB were monitored by immunoblotting. As expected, YidC could be detected only in wild-type IMVs (Fig. 2, lanes 1 and 2). The levels of YidC expressed from the chromosomal copy and from pTrcyidC were identical (Fig. 2, lanes 1 and 2). The levels of LepB were similar in all membrane preparations. These data indicate that SpoIIIJ and YqjG relieve the membrane stress response that occurs upon YidC depletion. Since LepB is not dependent on YidC for membrane insertion, the levels determined by immunoblotting were used to normalize the membrane concentrations (32) in the functional assays described below.
FIG. 2.
SpoIIIJ or YqjG represses the PspA stress response when YidC is depleted. (A) SDS-PAGE of IMVs (10 μg) isolated from E. coli FTL10 cells grown on LB medium with arabinose (lane 1) or LB medium with glucose (lanes 2 to 5). Lane 1, wild-type Ara; lane 2, wild-type Glu; lane 3, YidC-depleted IMVs; lane 4, YidC-depleted IMVs plus SpoIIIJ; lane 5, YidC-depleted IMVs plus YqjG. The approximate molecular masses of YidC (60 kDa), LepB (37 kDa), and PspA (25 kDa) are indicated on the left. (B) Immunoblots of SDS-PAGE gel obtained using antisera against YidC (α-YidC), LepB (α-LepB), and PspA (α-PspA).
SpoIIIJ and YqjG restore the PMF generation defect caused by YidC depletion.
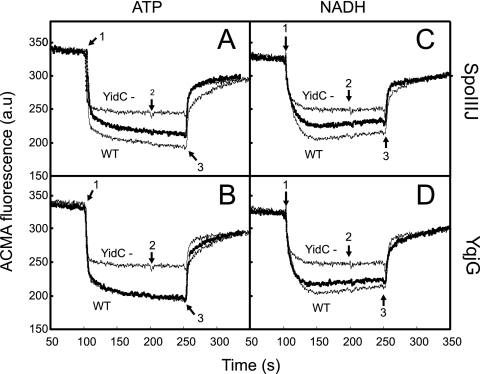
The absence of the PspA stress response in the YidC-depleted IMVs containing either SpoIIIJ or YqjG suggests that the B. subtilis homologs complement the role of YidC in assembly of energy-transducing complexes responsible for the generation of a PMF. To validate restoration of a PMF, the generation of a transmembrane ΔpH was determined in vitro by use of IMVs and the fluorescent dye ACMA. In the presence of valinomycin, a K+ ionophore, the PMF consists of only a ΔpH. To activate the F1Fo ATPase or the respiratory chain, IMVs were energized by addition of either ATP or the electron donor NADH. With both substrates, YidC-depleted IMVs showed a clear defect in the ability to generate a PMF (Fig. 3, compare the wild-type IMVs with the YidC-depleted IMVs). However, in IMVs derived from YidC-depleted cells expressing SpoIIIJ (Fig. 3A and C) or YqjG (Fig. 3B and D), the PMF was largely restored. In general, YqjG appeared to be somewhat more efficient for complementation than the paralog SpoIIIJ, regardless of the nature of the energy source. These experiments demonstrated that both SpoIIIJ and YqjG are able to restore the PMF generation defect caused by YidC depletion.
FIG. 3.
SpoIIIJ and YqjG restore the defect in PMF generation due to YidC depletion. The ΔpH of IMVs was measured by using ACMA fluorescence in the presence of 1 mM ATP (A and B) or 1.25 mM NADH (C and D) added at the time indicated by arrow 1. Valinomycin (arrow 2) and nigericin (arrow 3) were each added at a final concentration of 1 μM. The ΔpH traces generated by wild-type IMVs (WT) and YidC-depleted IMVs (YidC-) are indicated by thin lines, and the ΔpH generated by YidC-depleted IMVs containing SpoIIIJ (A and C) and YqjG (B and D) are indicated by thick lines. a.u, arbitrary units.
SpoIIIJ and YqjG restore ATPase and cytochrome o oxidase activity of YidC-depleted membranes.
In E. coli, the impaired PMF in YidC-depleted cells and membranes derived from these cells originates from defective insertion of the large energy-transducing complexes into the membrane, particularly F1Fo ATPase and cytochrome o oxidase (48). To quantify the recovery when SpoIIIJ and were YqjG expressed, the ATPase and cytochrome o oxidase activities in IMVs were determined.
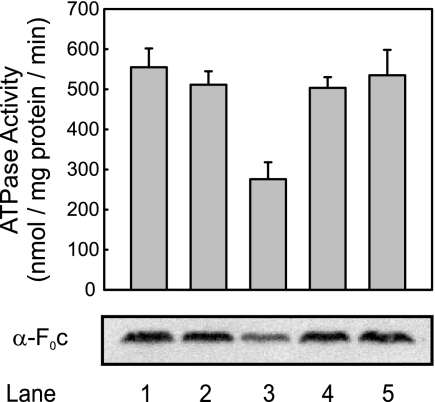
The reduction in the ATPase activity under YidC-limiting conditions is due to a defect in membrane insertion of the ring-forming subunit Foc, a substrate of the YidC-only pathway (48). In fact, the ATPase activity in vesicles lacking YidC (Fig. 4, lane 3) was about 50% of the activity in wild-type IMVs; likewise, the level of Foc was reduced, as assessed by immunoblotting (Fig. 4, lanes 1 and 2). In contrast, in IMVs derived from YidC-depleted cells expressing SpoIIIJ or YqjG, both the ATPase activity and the insertion of Foc were restored to wild-type levels (Fig. 4, lanes 4 and 5). Therefore, we concluded that SpoIIIJ and YqjG can functionally replace YidC in the YidC-only pathway.
FIG. 4.
SpoIIIJ and YqjG restore reduction of ATPase activity due to YidC depletion. The activity of the F1Fo ATPase was determined by measuring the release of inorganic phosphate upon addition of ATP to IMVs. Lane 1, wild-type Ara; lane 2, wild-type Glu; lane 3, YidC-depleted IMVs; lane 4, YidC-depleted IMVs plus SpoIIIJ; lane 5, YidC-depleted IMVs plus YqjG. The levels of Foc were determined by immunoblotting. α-F0c, anti-Foc.
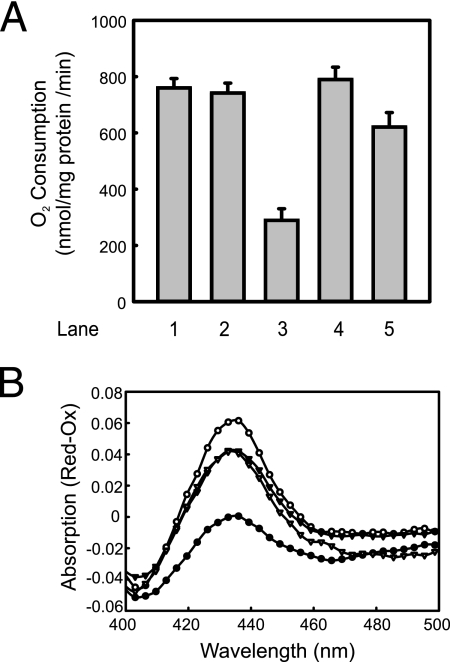
Membrane insertion of subunit a of the cytochrome o oxidase complex (CyoA) is both YidC and SecYEG dependent (8). To determine if SpoIIIJ and YqjG also complement the function of YidC in Sec-dependent insertion, the content of functional cytochrome o oxidase and the oxygen consumption of the IMVs were determined. When NADH was used as the electron donor, the oxygen consumption in YidC-depleted vesicles (Fig. 5A, bar 3) was only about 30% of that in the wild-type vesicles (bars 1 and 2). Respiration was restored fully in YidC-depleted IMVs containing SpoIIIJ (bar 4) and partially (80%) in IMVs containing YqjG (bar 5). The content of functional cytochrome o oxidase complexes in IMVs was determined by measuring difference absorption spectra of dithionate-reduced minus air-oxidized IMVs (Fig. 5B). This assay measured the amounts of functionally incorporated heme, which were found to be similar for IMVs derived from YidC-depleted cells expressing SpoIIIJ and IMVs derived from YidC-depleted cells expressing YqjG (Fig. 5B) and slightly lower than in YidC-containing IMVs. On the other hand, YidC depletion resulted in a major reduction in the cytochrome levels. These data suggest that the B. subtilis SpoIIIJ and YqjG proteins also functionally complement the Sec-dependent function of YidC.
FIG. 5.
SpoIIIJ and YqjG restore the reductions in the cytochrome o oxidase activity and CyoA levels caused by YidC depletion. (A) Cytochrome o oxidase activity of IMVs was determined by measuring the oxygen consumption of IMVs in the presence of the phenomethosulfate-ascorbate artificial electron donor system. Bar 1, wild-type Ara; bar 2, wild-type Glu; bar 3, YidC-depleted IMVs; bar 4, YidC-depleted IMVs plus SpoIIIJ; bar 5, YidC-depleted IMVs plus YqjG. (B) Heme levels of cytochrome o oxidase in membranes as determined by spectrometry. Difference spectra (dithionite reduced minus air oxidized) of wild-type IMVs (open circles), YidC-depleted IMVs (filled circles), SpoIIIJ-containing IMVs (open triangles), and YqjG-containing IMVs (filled triangles) were recorded and plotted as a function of the wavelength.
In vitro membrane insertion of B. subtilis and E. coli Foc is mediated by SpoIIIJ and YqjG.
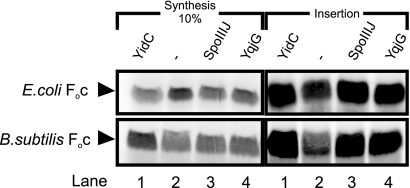
In E. coli, the membrane insertion of Foc is dependent solely on YidC, as shown previously by in vitro insertion assays (47). In order to investigate whether SpoIIIJ and YqjG can replace YidC in Foc insertion, we tested the IMVs mentioned above in a coupled in vitro transcription, translation, and membrane insertion assay. In addition, membrane insertion of B. subtilis Foc was tested. In these assays, noninserted Foc was removed by proteinase K digestion, while membrane-inserted Foc was analyzed by SDS-PAGE and detected by autoradiography (Fig. 6, right panels). As expected, insertion of E. coli Foc into YidC-depleted IMVs (Fig. 6, upper right panel, lane 2) was strongly reduced compared to insertion of E. coli Foc into YidC-containing IMVs (Fig. 6, upper right panel, lane 1). Insertion of E. coli Foc into YidC-depleted IMVs was restored to nearly wild-type levels by expression of SpoIIIJ or YqjG (Fig. 6, upper right panel, lanes 3 and 4). Essentially similar results were obtained when B. subtilis Foc was used as a substrate. Compared to YidC-containing IMVs (Fig. 6, lower right panel, lane 1), only low levels of insertion of B. subtilis Foc were observed with YidC-depleted IMVs (lane 2), while expression of SpoIIIJ and expression of YqjG (Fig. 6, lower right panel, lanes 3 and 4, respectively) restored the membrane insertion of Foc, albeit seemingly at levels lower than those observed with YidC. The latter finding is most likely due to the high level of Foc synthesis in the presence of wild-type IMVs (Fig. 6, lower left panel, lane 1) compared to the levels in the other reactions (lane 2 to 4). Normalization of total Foc synthesis with membrane-inserted Foc results in almost equal levels of SpoIIIJ-, YqjG-, and YidC-inserted B. subtilis Foc. These findings provide strong evidence for the mutual exchangeability of YidC, SpoIIIJ, and YqjG and demonstrate a function of these proteins in membrane protein insertion.
FIG. 6.
SpoIIIJ and YqjG promote membrane insertion of in vitro-synthesized Foc from E. coli and B. subtilis into IMVs derived from E. coli. Lane 1, wild-type Glu; lane 2, YidC-depleted IMVs; lane 3, YidC-depleted IMVs plus SpoIIIJ; lane 4, YidC-depleted IMVs plus YqjG. The panels on the left show 10% of the total protein synthesis, and the panels on the right show the amount of membrane-inserted and radiolabeled Foc. The upper panels show data for Foc from E. coli, and the lower panels show data for Foc from B. subtilis.
B. subtilis F1Fo ATP synthase copurifies with SpoIIIJ and YqjG.
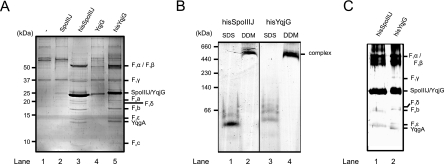
In order to investigate which proteins physically interact with SpoIIIJ and YqjG, we overexpressed and affinity purified both of these proteins under relatively mild detergent solubilization conditions from inner membranes of B. subtilis. Vectors bearing the C-terminally His-tagged SpoIIIJ and YqjG proteins were transformed into B. subtilis pNZ8900, and IMVs containing the overexpressed proteins were isolated. As controls, vectors expressing the untagged SpoIIIJ and YqjG proteins and the empty overexpression vector pNZ8901 were used in parallel experiments. The His-tagged SpoIIIJ and YqjG proteins are functional as they complemented the E. coli growth defect due to the YidC depletion (Fig. 1). IMVs were solubilized with the detergent DDM and passed over an Ni-NTA affinity resin. Several proteins were found to coelute specifically with His-tagged SpoIIIJ and YqjG, while only nonspecific association was observed with nontagged protein or with empty vector controls (Fig. 7A). The specifically bound and coeluted proteins were identified with high confidence by mass spectrometry. Remarkably, almost all proteins identified were subunits of the B. subtilis F1Fo ATP synthase. All subunits of the F1 domain (F1α, F1β, F1γ, F1δ, and F1ɛ) and all known Fo subunits (Foa, Fob, and Foc) could be identified (Fig. 7A and Table 3). Thus, all structural components of the B. subtilis F1Fo ATP synthase complex copurify with His-tagged SpoIIIJ and YqjG. Furthermore, the B. subtilis protein YqgA, which has no known function or similarity to other proteins, specifically coelutes with both SpoIIIJ and YqjG. It should be noted that SpoIIIJ and YqjG also copurify with F1α and F1β of the E. coli ATP synthase when these proteins are overexpressed and purified in E. coli cells (data not shown). This is consistent with recent copurification data showing that E. coli YidC also copurifies with F1α and F1β of the E. coli ATP synthase (43).
FIG. 7.
SpoIIIJ and YqjG form a stable complex with B. subtilis F1Fo ATP synthase. (A) Specific copurification of B. subtilis F1Fo ATP synthase with SpoIIIJ and YqjG. Membrane vesicles of B. subtilis pNZ8900 transformed with different plasmids were isolated and subjected to His tag purification. Proteins were identified by mass spectrometry, as indicated on the right. The F1Fo ATP synthase elutes with His-tagged SpoIIIJ or YqjG but not with the non-His-tagged versions, showing specific binding of B. subtilis ATP synthase to SpoIIIJ and YqjG. Lane 1, pNZ8901 (empty vector); lane 2, pNZspoIIIJ; lane 3, pNZspoIIIJHis; lane 4, pNZyqjG; lane 5, pNZyqjGHis. (B) The native conformation of SpoIIIJ/YqjG in complex with the F1Fo ATP synthase was monitored by BN-PAGE. Affinity-purified SpoIIIJ (lanes 1 and 2) or YqjG (lanes 3 and 4) was treated with either SDS (lanes 1 and 3) or DDM (lanes 2 and 4) prior to BN-PAGE. (C) SpoIIIJ and YqjG are part of the high-molecular-weight complexes. The high-molecular-weight bands indicated in panel B were excised from the BN-PAGE gel and subjected to nonnative SDS-PAGE and silver staining. Identification of the proteins indicated was based on alignment with the protein pattern in panel A.
TABLE 3.
Summary of mass spectrometry identification of SpoIIIJ- and YqjG-associated proteinsa
| Accession no. | Protein | No. of unique peptides | Coverage (%) |
|---|---|---|---|
| P37808|ATPA_BACSU | ATP synthase F1 subunit α | 23 | 52.8 |
| P37809|ATPB_BACSU | ATP synthase F1 subunit β | 21 | 64.7 |
| P37810|ATPG_BACSU | ATP synthase F1 subunit γ | 18 | 57.2 |
| P37811|ATPD_BACSU | ATP synthase F1 subunit δ | 8 | 42 |
| P37812|ATPE_BACSU | ATP synthase F1 subunit ɛ | 7 | 59.8 |
| P37813|ATP6_BACSU | ATP synthase Fo subunit a | 4 | 16.4 |
| P37814|ATPF_BACSU | ATP synthase Fo subunit b | 6 | 81.2 |
| P37815|ATPL_BACSU | ATP synthase Fo subunit c | 3 | 22.9 |
| P54484|YQGA_BACSU | Hypothetical protein YqgA | 14 | 73.9 |
Protein identification was carried out using the Paragon algorithm, which employs feature probabilities and a “sequence temperature value,” for a sequence database search. All proteins identified originated from B. subtilis.
To further investigate the complex consisting of SpoIIIJ/YqjG and the F1Fo ATP synthase, BN-PAGE experiments were performed. Elution fractions obtained by purification of His-tagged SpoIIIJ (Fig. 7B, lanes 1 and 2) and YqjG (lanes 3 and 4) were incubated with either SDS (lanes 1 and 3) or DDM (lanes 2 and 4) prior to electrophoresis. The DDM-treated SpoIIIJ (lane 2) and YqjG (lane 4) migrated as a single high-molecular-mass complex with the F1Fo ATP synthase at an apparent molecular mass of 550 kDa. No unbound, monomeric SpoIIIJ or YqjG could be detected in the native samples after Coomassie brilliant blue (Fig. 7B) or silver staining (data not shown). On the other hand, the SDS treatment resulted in denaturation of the complex, and proteins migrated at low molecular weights (lanes 1 and 3). The protein bands corresponding to the native complex were excised from the BN-PAGE gel and subjected to nonnative SDS-PAGE. This resulted in protein patterns (Fig. 7C) similar to that observed in the affinity purification experiment, including the bands for SpoIIIJ and YqjG (Fig. 7A). Based on the protein staining density in the gels, it appears that superstoichiometric amounts of SpoIIIJ and YqjG participate in the formation of a stable complex with the F1Fo ATP synthase.
DISCUSSION
Members of the YidC/OxaI/Alb3 protein family exhibit rather low primary sequence similarity. Nevertheless, to a large extent they are functionally conserved and even appear to be generally exchangeable (18, 31, 45). Our results confirm this general trend, since the two YidC homologs from the gram-positive bacterium B. subtilis completely restore growth of YidC-depleted E. coli cells both on agar plates and in liquid medium. YqjG and SpoIIIJ complement YidC to a high degree, as control and complemented cells grew equally well and with similar PspA expression levels. In contrast, a recent study showed that the YidC homologs of Streptococcus mutans barely complemented the E. coli growth defect on plates, whereas the growth defect could be only partially restored in liquid culture (6). In contrast, the SpoIIIJ- or YqjG-complemented YidC-depleted strain exhibited wild-type growth rates (data not shown). This shows that YidC homologs from different gram-positive bacteria complement the essential function of YidC, even though SpoIIIJ and YqjG appear to be more proficient than the S. mutans YidC paralogs. The results of various in vivo assays further support the high level of complementation; in these assays the activities of the YidC-depleted vesicles containing either SpoIIIJ or YqjG were almost the same as those of YidC-containing IMVs.
Another conclusion derived from the in vivo studies is that both SpoIIIJ and YqjG compensate for YidC in the YidC-only and Sec-dependent pathway. This is evident from the restoration of the ATPase activity, the correct insertion of Foc, a known substrate of the YidC-only pathway (47). The restoration of the cytochrome o oxidase activity and levels provide evidence for a functional CyoA, a protein that requires both SecYEG and YidC for insertion (8). Like Oxa1p, the mitochondrial YidC homolog, SpoIIIJ and YqjG lack the large periplasmic loop at the N terminus. Previous studies showed that a region in this periplasmic loop is needed for SecF binding, while only a few amino acids in the extreme N-terminal part of the loop are essential for the insertase function of YidC (51). Therefore, we concluded that SecF binding is not an essential feature of the YidC function.
Previously, SpoIIIJ and YqjG have been implicated in protein secretion rather than in membrane protein insertion (41). It was observed that several secreted proteins of B. subtilis were affected by SpoIIIJ depletion in cells lacking YqjG. Interestingly, the depletion also affected two membrane proteins, one of which is CtaC, the CyoA homolog in B. subtilis, a known YidC substrate. Recently, a role for SpoIIIJ in membrane protein insertion was suggested based solely on its sequence similarity to E. coli YidC, although no direct experimental evidence for such a function was provided (4). Another report suggested that SpoIIIJ could be involved in the maturation of SpoIIIAE (37). Both proteins are essential for activation of a forespore-specific transcription factor and could be cross-linked in vivo. However, the level of membrane-inserted SpoIIIAE was not affected in a SpoIIIJ deletion strain (37), possibly suggesting a function in a postinsertional stage of SpoIIIAE biogenesis. Our report now demonstrates that both SpoIIIJ and YqjG mediate membrane insertion of in vitro-synthesized E. coli and B. subtilis Foc and supports the universal hypothesis that Oxa1p-like proteins are required for membrane insertion and biogenesis of subunits of respiratory chain complexes and other energy-transducing complexes.
Remarkably, when the His-tagged SpoIIIJ and YqjG proteins are expressed in B. subtilis, a number of proteins are found to specifically copurify during Ni2+-NTA affinity chromatography. All subunits of the F1Fo ATP synthase are present in the complex with SpoIIIJ and YqjG. A specific role of YidC in membrane insertion of the ring-forming subunit Foc is well established, whereas both YidC and SecYEG are essential for membrane insertion of Foa (23, 53). However, it is still not clear whether YidC also has a role downstream in membrane assembly of F1Fo ATP synthase. Our observation that SpoIIIJ and YqjG form a complex with the entire F1Fo ATP synthase suggests that these proteins have a role in the final assembly of this energy-transducing complex. In yeast (Saccharomyces cerevisiae), OxaI has recently been shown to interact with an Atp9(Foc)-F1 subcomplex, and it has been proposed that OxaI acts as a chaperone to ensure correct assembly of the Atp9p (Foc) ring and coassembly with Atp10-chaperoned Atp6p (Foa) (14, 42). Thus, in yeast, the incorporation of Atp6p (Foa) into the complex seems to be one of the last steps in the assembly process. Remarkably, the complex consisting of F1Fo ATPase with SpoIIIJ and YqjG is not active in ATP hydrolysis (M. J. Saller, unpublished results). We hypothesized that this complex stalls at a late stage of biogenesis, possibly at the stage of functional incorporation of Foa (49), which is also assumed to be the last step in assembly in bacteria.
OxaI and several bacterial YidC homologs have an elongated C-terminal tail that has been implicated in direct ribosome binding (22) and/or biogenesis of the ATP synthase (11). Interestingly, both in mitochondria and in some bacteria, there are two paralogs, one having an elongated C-terminal tail (OxaI and YidC2) and one lacking this tail (Cox18 and YidC1). In yeast, OxaI, but not Cox18, was found to specifically copurify with the mitochondrial F1Fo ATP synthase (16). Furthermore, in S. mutans, a defect in the ATPase activity due to the absence of YidC2 could be complemented by OxaI, but not by Cox18. This led Funes et al. to conclude that the elongated C-terminal tail of YidC2 is required for biogenesis of the F1Fo ATPase complex (11). Our results contradict this conclusion as both B. subtilis YidC homologs lack the elongated C-terminal tail but still copurify with the F1Fo ATPase complex and complement its biogenesis defect in YidC-depleted E. coli cells. The other protein that copurifies with SpoIIIJ and YqjG is YqgA. This is a small membrane protein with no known function, and homologs are found only in a number of other Bacillus species. The yqgA gene is not essential in B. subtilis. Future experiments should address whether YqgA plays a role in the F1Fo ATP synthase assembly process, but because of its limited distribution among bacterial species, the role of this protein may be specific to bacilli.
Taken together, our complementation, in vivo, and in vitro assays demonstrate a function for SpoIIIJ and YqjG in membrane protein insertion, which extends the general conserved function of the YidC/OxaI/Alb3 protein family to gram-positive bacteria.
Acknowledgments
We thank Wim H. C. Huibers for technical assistance and David J. F. du Plessis for providing protocols.
This work was supported by the European Science Foundation within the framework of the BACELL EuroSCOPE program, by The Netherlands Proteomics Centre, and by European Community grant LSHG-CT-2004-504601 (E-Mep).
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx, F., and G. Georgiou. 1990. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 172:491-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bongers, R. S., J. W. Veening, M. Van Wieringen, O. P. Kuipers, and M. Kleerebezem. 2005. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71:8818-8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camp, A. H., and R. Losick. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, M., J. C. Samuelson, F. Jiang, M. Muller, A. Kuhn, and R. E. Dalbey. 2002. Direct interaction of YidC with the Sec-independent Pf3 coat protein during its membrane protein insertion. J. Biol. Chem. 277:7670-7675. [DOI] [PubMed] [Google Scholar]
- 6.Dong, Y., S. R. Palmer, A. Hasona, S. Nagamori, H. R. Kaback, R. E. Dalbey, and L. J. Brady. 2008. Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J. Bacteriol. 190:2458-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Plessis, D. J., N. Nouwen, and A. J. M. Driessen. 2006. Subunit A of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281:12248-12252. [DOI] [PubMed] [Google Scholar]
- 9.Errington, J., L. Appleby, R. A. Daniel, H. Goodfellow, S. R. Partridge, and M. D. Yudkin. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138:2609-2618. [DOI] [PubMed] [Google Scholar]
- 10.Facey, S. J., S. A. Neugebauer, S. Krauss, and A. Kuhn. 2007. The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365:995-1004. [DOI] [PubMed] [Google Scholar]
- 11.Funes, S., A. Hasona, H. Bauerschmitt, C. Grubbauer, F. Kauff, R. Collins, P. J. Crowley, S. R. Palmer, L. J. Brady, and J. M. Herrmann. 2009. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. USA 106:6656-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hatzixanthis, K., T. Palmer, and F. Sargent. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49:1377-1390. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann, J. M., R. A. Stuart, E. A. Craig, and W. Neupert. 1994. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 127:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewett, M. C., and J. R. Swartz. 2004. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 86:19-26. [DOI] [PubMed] [Google Scholar]
- 16.Jia, L., M. K. Dienhart, and R. A. Stuart. 2007. OxaI directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol. Biol. Cell 18:1897-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, F., M. Chen, L. Yi, J. W. de Gier, A. Kuhn, and R. E. Dalbey. 2003. Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278:48965-48972. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, F., L. Yi, M. Moore, M. Chen, T. Rohl, K. J. Van Wijk, J. W. de Gier, R. Henry, and R. E. Dalbey. 2002. Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J. Biol. Chem. 277:19281-19288. [DOI] [PubMed] [Google Scholar]
- 19.Kaufmann, A., E. H. Manting, A. K. Veenendaal, A. J. Driessen, and C. van der Does. 1999. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38:9115-9125. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer, D., and A. Kuhn. 2007. YidC as an essential and multifunctional component in membrane protein assembly. Int. Rev. Cytol. 259:113-138. [DOI] [PubMed] [Google Scholar]
- 21.Kim, R. G., and C. Y. Choi. 2001. Expression-independent consumption of substrates in cell-free expression system from Escherichia coli. J. Biotechnol. 84:27-32. [DOI] [PubMed] [Google Scholar]
- 22.Kohler, R., D. Boehringer, B. Greber, R. Bingel-Erlenmeyer, I. Collinson, C. Schaffitzel, and N. Ban. 2009. YidC and OxaI form dimeric insertion pores on the translating ribosome. Mol. Cell 34:344-353. [DOI] [PubMed] [Google Scholar]
- 23.Kol, S., W. Majczak, R. Heerlien, J. P. van der Berg, N. Nouwen, and A. J. M. Driessen. 2009. Subunit a of the ATP synthase requires YidC and SecYEG for membrane insertion. J. Mol. Biol. 390:893-901. [DOI] [PubMed] [Google Scholar]
- 24.Kol, S., N. Nouwen, and A. J. Driessen. 2008. Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J. Biol. Chem. 283:31269-31273. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 27.Matsushita, K., L. Patel, and H. R. Kaback. 1984. Cytochrome o type oxidase from Escherichia coli. Characterization of the enzyme and mechanism of electrochemical proton gradient generation. Biochemistry 23:4703-4714. [DOI] [PubMed] [Google Scholar]
- 28.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, T., K. Haga, M. Takeuchi, and T. Sato. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J. Bacteriol. 184:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagamori, S., I. N. Smirnova, and H. R. Kaback. 2004. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preuss, M., M. Ott, S. Funes, J. Luirink, and J. M. Herrmann. 2005. Evolution of mitochondrial oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 280:13004-13011. [DOI] [PubMed] [Google Scholar]
- 32.Price, C. E., and A. J. M. Driessen. 2008. YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J. Biol. Chem. 283:26921-26927. [DOI] [PubMed] [Google Scholar]
- 33.Saaf, A., M. Monne, J. W. de Gier, and G. von Heijne. 1998. Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J. Biol. Chem. 273:30415-30418. [DOI] [PubMed] [Google Scholar]
- 34.Samuelson, J. C., F. Jiang, L. Yi, M. Chen, J. W. de Gier, A. Kuhn, and R. E. Dalbey. 2001. Function of YidC for the insertion of M13 procoat protein in Escherichia coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276:34847-34852. [DOI] [PubMed] [Google Scholar]
- 35.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 36.Serrano, M., L. Corte, J. Opdyke, C. P. Moran, Jr., and A. O. Henriques. 2003. Expression of spoIIIJ in the prespore is sufficient for activation of sigma G and for sporulation in Bacillus subtilis. J. Bacteriol. 185:3905-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano, M., F. Vieira, C. P. Moran, Jr., and A. O. Henriques. 2008. Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J. Bacteriol. 190:7786-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilov, I. V., S. L. Seymour, A. A. Patel, A. Loboda, W. H. Tang, S. P. Keating, C. L. Hunter, L. M. Nuwaysir, and D. A. Schaeffer. 2007. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6:1638-1655. [DOI] [PubMed] [Google Scholar]
- 39.Stuart, R. 2002. Insertion of proteins into the inner membrane of mitochondria: the role of the OxaI complex. Biochim. Biophys. Acta 1592:79-87. [DOI] [PubMed] [Google Scholar]
- 40.The UniProt Consortium. 2007. The Universal Protein Resource (UniProt). Nucleic Acids Res. 35:D193-D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tjalsma, H., S. Bron, and J. M. van Dijl. 2003. Complementary impact of paralogous OxaI-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278:15622-15632. [DOI] [PubMed] [Google Scholar]
- 42.Tzagoloff, A., A. Barrientos, W. Neupert, and J. M. Herrmann. 2004. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J. Biol. Chem. 279:19775-19780. [DOI] [PubMed] [Google Scholar]
- 43.van Bloois, E., H. L. Dekker, L. Froderberg, E. N. Houben, M. L. Urbanus, C. G. de Koster, J. W. de Gier, and J. Luirink. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419-1424. [DOI] [PubMed] [Google Scholar]
- 44.van Bloois, E., G. J. Haan, J. W. de Gier, B. Oudega, and J. Luirink. 2006. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281:10002-10009. [DOI] [PubMed] [Google Scholar]
- 45.van Bloois, E., G. Koningstein, H. Bauerschmitt, J. M. Herrmann, and J. Luirink. 2007. Saccharomyces cerevisiae Cox18 complements the essential Sec-independent function of Escherichia coli YidC. FEBS J. 274:5704-5713. [DOI] [PubMed] [Google Scholar]
- 46.van der Does, C., J. de Keyzer, M. van der Laan, and A. J. M. Driessen. 2003. Reconstitution of purified bacterial preprotein translocase in liposomes. Methods Enzymol. 372:86-98. [DOI] [PubMed] [Google Scholar]
- 47.van der Laan, M., P. Bechtluft, S. Kol, N. Nouwen, and A. J. M. Driessen. 2004. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Laan, M., M. L. Urbanus, C. M. ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A. J. M. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vik, S. B., and R. D. Simoni. 1987. F1F0-ATPase from Escherichia coli with mutant F0 subunits. Partial purification and immunoprecipitation of F1F0 complexes. J. Biol. Chem. 262:8340-8346. [PubMed] [Google Scholar]
- 50.Wagner, S., O. Pop, G. J. Haan, L. Baars, G. Koningstein, M. M. Klepsch, P. Genevaux, J. Luirink, and J. W. de Gier. 2008. Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283:17881-17890. [DOI] [PubMed] [Google Scholar]
- 51.Xie, K., D. Kiefer, G. Nagler, R. E. Dalbey, and A. Kuhn. 2006. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45:13401-13408. [DOI] [PubMed] [Google Scholar]
- 52.Yen, M. R., K. T. Harley, Y. H. Tseng, and M. H. Saier, Jr. 2001. Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol. Lett. 204:223-231. [DOI] [PubMed] [Google Scholar]
- 53.Yi, L., N. Celebi, M. Chen, and R. E. Dalbey. 2004. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279:39260-39267. [DOI] [PubMed] [Google Scholar]
- 54.Yi, L., F. Jiang, M. Chen, B. Cain, A. Bolhuis, and R. E. Dalbey. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F1F0 ATP synthase and SecE of the SecYEG translocase. Biochemistry 42:10537-10544. [DOI] [PubMed] [Google Scholar]