Abstract
Clubfoot is a common birth defect that affects 135,000 newborns each year worldwide. It is characterized by equinus deformity of one or both feet and hypoplastic calf muscles. Despite numerous study approaches, the cause(s) remains poorly understood although a multifactorial etiology is generally accepted. We considered the HOXA and HOXD gene clusters and insulin-like growth factor binding protein 3 (IGFBP3) as candidate genes because of their important roles in limb and muscle morphogenesis. Twenty SNPs from the HOXA and HOXD gene clusters and 12 SNPs in IGFBP3 were genotyped in a sample composed of nonHispanic white and Hispanic multiplex and simplex families (discovery samples) and a second sample of nonHispanic white simplex trios (validation sample). Four SNPs (rs6668, rs2428431, rs3801776 and rs3779456) in the HOXA cluster demonstrated altered transmission in the discovery sample, but only rs3801776, located in the HOXA basal promoter region, showed altered transmission in both the discovery and validation samples (p=0.004 and p=0.028). Interestingly, HOXA9 is expressed in muscle during development. A SNP in IGFBP3, rs13223993, also showed altered transmission (p=0.003) in the discovery sample. Gene-gene interactions were identified between variants in HOXA, HOXD and IGFBP3 and with previously associated SNPs in mitochondrial-mediated apoptotic genes. The most significant interactions were found between CASP3 SNPS and variants in HOXA, HOXD and IGFBP3. These results suggest a biologic model for clubfoot in which perturbation of HOX and apoptotic genes together affect muscle and limb development, which may cause the downstream failure of limb rotation into a plantar grade position.
Keywords: association study, genotyping, complex disease, clubfoot, HOXA, IGFBP3
INTRODUCTION
Clubfoot, or talipes equinovarus (TEV), is a common birth defect which is easily recognized and diagnosed because of the characteristic forefoot adduction, hindfoot varus and equinus, midfoot cavus, and hypoplastic lower limb musculature [Dietz, 1985]. Isolated, or idiopathic, cases occur in the absence of any other anomaly and/or known cause(s) [Moorthi et al., 2005]. The birth prevalence varies among ethnic groups from 1/150-2500, with a worldwide rate of approximately 1/1000 [Moorthi et al., 2005]. A variety of approaches including genome scans and candidate gene testing have been used to identify the underlying causes of complex genetic disorders. Interrogation of minimal overlapping chromosomal deletions in syndromic clubfoot led to the identification of a group of candidate genes, the HOXD gene cluster (UCSC Genome Bioinformatics) [Brewer et al., 1998].
The HOX genes encode transcription factors that direct patterning of the axial skeleton and muscle and limb development [Mark et al., 1997; McGinnis and Krumlauf, 1992]. This family consists of 39 genes distributed in four paralogous clusters (A, B, C and D). Each of the HOX clusters share common genomic structure and are temporally and spatially expressed during embryogenesis. The 5’ HOXA genes are expressed in the muscles of both the fore and hind limbs, and are involved in patterning and differentiation of muscles in embryonic limbs and during muscle repair of adult limbs [Houghton and Rosenthal, 1999]. Mutations in HOX genes perturb normal limb development; for example, the HOXD10 956T>A/319M>K mutation causes autosomal dominant congenital vertical talus (CVT) [Dobbs et al., 2006; Shrimpton et al., 2004].
Conditional HOXA13 knockout mice show decreased expression of insulin-like growth factor binding protein 3 (IGFBP3) in the limbs (Dr. S. Stadler, personal communication), which has been shown to have a role in apoptosis [Baxter and Martin, 1989]. This observation is important because we previously reported an association between genetic variants in Caspase genes and clubfoot [Ester et al., 2007; Heck et al., 2005].
Given the role of HOXA, HOXD and IGFBP3 in limb development, this study was undertaken to determine whether variation in the HOXA, HOXD or IGFBP3 genes is associated with isolated clubfoot and to further evaluate whether the 956T>A/319M>K HOXD10 mutation causes clubfoot.
MATERIALS AND METHODS
Clubfoot study samples and sample preparation
SNPs from the HOXA and HOXD gene clusters and IGFBP3 were genotyped in the discovery population composed of 1783 genotyped individuals from 598 singly ascertained families as described previously [Ester et al., 2007; Heck et al., 2005]. This included a total of 179 extended families (122 nonHispanic white (NHW) and 57 Hispanic) and 331 simplex families (130 NHW and 201 Hispanic) and 88 trios with a positive family history (42 NHW and 46 Hispanic). The validation population consisted of 144 NHW simplex trios.
Families in the discovery dataset were identified through a proband with clubfoot from one of six orthopedic centers: Shriners Hospital for Children of Houston, Los Angeles and Shreveport, Texas Scottish Rite Hospital for Children of Dallas, University of Iowa, Department of Orthopaedics and Rehabilitation, and other collection sites, as previously described [Ester et al., 2007; Heck et al., 2005]. Individuals from the validation population were ascertained in the Department of Orthopedics at Washington University in St. Louis (144 probands). All affected individuals were diagnosed with clubfoot using standardized clinical criteria and radiographic examination as previously described [Lochmiller et al., 1998]. Age of ascertainment for probands generally ranged from birth to 18 years of age. Cases were excluded if they were caused by a chromosomal abnormality or were syndromic. Ethnicity was self-reported and informed consent was obtained.
Blood or saliva samples were collected from the affected individuals and family members. DNA was extracted using either the Roche DNA Isolation Kit for Mammalian Blood (Roche, Switzerland) or the Oragene Purifier for saliva (DNA Genotek, Inc., Ontario, Canada) following the manufacturer’s protocol.
956T>A/319M>K HOXD10 mutation detection
To determine whether the 956T>A/319M>K HOXD10 mutation played a causal role in isolated clubfoot, 494 clubfoot probands (267 NHW and 227 Hispanic) and 595 unrelated, unaffected controls (380 NHW and 215 Hispanic) were genotyped using a custom TaqMan assay designed using FileBuilder 3.0 (ABI, Foster City, CA). One hundred forty-four probands ascertained later were not genotyped for this mutation. The positive control in all assays was DNA from two individuals with CVT and the HOXD10 956T>A/319M>K mutation.
Genotyping
SNPs were identified using NCBI and Ensembl websites and the software program Haploview [Barrett et al., 2005]. Final SNP selection was based on heterozygosity (>0.3), inter- and intragenic positions, coverage of the gene, and tagging ability. Using these criteria, nine HOXA, 11 HOXD and 12 IGFBP3 SNPs were identified (Table I). SNPs were genotyped using TaqMan® Genotyping Assays (Applied Biosystems, Foster City, CA) and detected on a 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA).
Table I.
Allele frequencies for HoxA, HoxD and IGFBP3 SNPs
| Gene | dbSNP | Pos. | Alleles | MAFa | HMAFb,c | Location |
|---|---|---|---|---|---|---|
| HoxD | rs6749771 | SNP1 | A/G | 0.391 | 0.685 | 3' of HoxD cluster |
| rs1446575 | SNP2 | C/A | 0.385 | 0.697 | HoxD3-HoxD1 | |
| rs1318778 | SNP3 | G/C | 0.289 | 0.145 | 3' of HoxD3 | |
| rs1542180 | SNP4 | G/A | 0.378 | 0.692 | 5' of HoxD3 | |
| rs1867863 | SNP5 | C/A | 0.341 | 0.606 | 5' of HoxD4 | |
| rs2113563 | SNP6 | T/C | 0.306 | 0.257 | HoxD8-HoxD4 | |
| rs2592394 | SNP7 | C/T | 0.305 | 0.182 | HoxD9-HoxD8 | |
| rs847146 | SNP8 | G/T | 0.354 | 0.361 | HoxD11 (intronic) | |
| rs741610 | SNP9 | G/A | 0.316 | 0.443 | HoxD12-HoxD11 | |
| rs711812 | SNP10 | T/G | 0.325 | 0.438 | 5' of HoxD12 | |
| rs6758117 | SNP11 | T/C | 0.302 | 0.594 | 5' of HoxD cluster | |
| HoxA | rs2462907 | SNP12 | C/T | 0.491 | 0.461 | 3' of HoxA cluster |
| rs6668 | SNP13 | C/T | 0.375 | 0.413 | 3' of HoxA2 | |
| rs2428431 | SNP14 | C/G | 0.367 | 0.409 | HoxA2 (intronic) | |
| rs3757640 | SNP15 | C/T | 0.267 | 0.415 | HoxA5 (intronic) | |
| rs3801776 | SNP16 | G/A | 0.226 | 0.315 | 5' of HoxA9 | |
| rs3779456 | SNP17 | T/C | 0.442 | 0.540 | HoxA10 (intronic) | |
| rs1859164 | SNP18 | T/C | 0.479 | 0.386 | HoxA10 (intronic) | |
| rs6968828 | SNP19 | G/T | 0.462 | 0.445 | HoxA11 (intronic) | |
| rs3807598 | SNP20 | C/G | 0.451 | 0.305 | 5' of HoxA cluster | |
| IGFBP3 | rs2132571 | SNP21 | A/G | 0.306 | 0.218 | upstream |
| rs2132572 | SNP22 | A/G | 0.227 | 0.478 | upstream | |
| rs2854744 | SNP23 | A/C | 0.472 | 0.316 | upstream | |
| rs2854746 | SNP24 | C/G | 0.415 | 0.264 | exon 1 (missense) | |
| rs2854747 | SNP25 | C/T | 0.415 | 0.631 | intron 1–2 | |
| rs3793345 | SNP26 | C/T | 0.199 | 0.150 | intron 1–2 | |
| rs2471551 | SNP27 | C/G | 0.192 | 0.154 | intron 1–2 | |
| rs3110697 | SNP28 | A/G | 0.422 | 0.629 | intron 3–4 | |
| rs10255707 | SNP29 | C/T | 0.244 | 0.545 | intron 3–4 | |
| rs2453839 | SNP30 | C/T | 0.197 | 0.130 | intron 4–5 | |
| rs6670 | SNP31 | A/T | 0.216 | 0.129 | 3' UTR | |
| rs13223993 | SNP32 | A/G | 0.248 | 0.289 | downstream |
MAF = Minor allele frequency in nonHispanic white sample
HMAF = Frequency of NHW minor allele in Hispanics
HMAF signficiantly different from MAF (p<0.001) in bold
Statistical analysis
Allele frequencies and Hardy-Weinberg Equilibrium (HWE) were calculated using SAS (v9.1). The SNPs not in HWE (p<0.001) were identified and appropriate methods for correction were used in the analyses. Pair-wise linkage disequilibrium values (D′ and r2) were calculated using GOLD [Abecasis and Cookson, 2000]. Multiple methods for assessing linkage and/or association were performed to extract the greatest amount of information from each dataset. Parametric and non-parametric linkage analyses were conducted using Merlin [Abecasis et al., 2002]. Two parametric models based on segregation analysis were tested on the 179 extended families [de Andrade et al., 1998; Rebbeck et al., 1993; Wang et al., 1988]. Model I used penetrances of 0.000, 0.020, 0.494 and 0.0, 0.008, 0.358 while model II used 0.0, 0.002, 0.067 and of 0.0, 0.008, 0.358 for males and females, respectively. The rsq option (rsq=0.16) of Merlin was used to remove the effects of LD.
Association in the Presence of Linkage (APL) was used to analyze association because this program incorporates data even when a parental genotype is missing [Chung et al., 2006]. The APL software was also used to assess transmission of 2-SNP haplotypes within a gene or gene clusters. Generalized Estimating Equations (GEE) were used to detect gene-gene interactions for all SNPs [Hancock et al., 2007].
Identification of transcription factor binding sites
In silico analyses of all the over-transmitted SNP sequences in regulatory regions were performed using three online algorithms: Alibaba2, Patch and Transcription Element Search Software (TESS) [Grabe 2002; Matys et al., 2006; Schug 2003]. Ancestral and alternate (www.ncbi.nlm.nih.gov/) allele sequences were inputted into the program and the output was compared.
This study was reviewed and approved by the Committee for the Protection of Human Subjects at the University of Texas Heath Science Center (HSC-MS-04-239).
RESULTS
The 956T>A/319M>K HOXD10 mutation was not detected in any of the 494 probands (267 NHW and 227 Hispanic) or the 595 unrelated controls (380 NHW and 215 Hispanic). This excluded a maximum carrier frequency 0.005 with over 80% power in both ethnic groups.
We next interrogated the HOXA and HOXD gene clusters and IGFBP3 in the discovery population. Only rs10255707 was not in HWE (p<0.0001) in either ethnic group (data not shown). Comparisons of SNP allele frequencies between NHW and Hispanics detected significant differences in the majority of SNPs in all genes even after Bonferroni correction (p<0.001) (Table I). Therefore, the data were stratified by ethnicity and examined separately. The data was combined across ethnicities for SNPs showing no differences in frequency.
Affected individuals had slightly higher levels of linkage disequilibrium (LD) than unaffected individuals in both ethnic groups (Supplemental Tables I–IV). Parametric and nonparametric multipoint linkage analysis detected suggestive evidence (LOD>1.5) for linkage to IGFBP3 in the Hispanic subset (Table II). No evidence of linkage was found for HOXA or HOXD with clubfoot (data not shown).
Table II.
IGFBP3 multipoint linkage results for discovery Hispanic sample
| Nonparametric | Parametric | ||||
|---|---|---|---|---|---|
| Gene | dbSNP | Position | LOD | p-value | HLODa |
| IGFBP3 | rs2132571 | SNP21 | 2.06 | 0.001 | 1.93 |
| rs2854746 | SNP24 | 2.13 | 0.0009 | 2.01 | |
| rs2453839 | SNP30 | 2.22 | 0.0007 | 2.12 | |
| rs6670 | SNP31 | 2.29 | 0.0006 | 2.19 | |
| rs13223993 | SNP32 | 2.29 | 0.0006 | 2.19 | |
All families linked under model II
The APL analysis in the discovery population found significant evidence for association with SNPs in the HOXA and HOXD gene clusters and IGFBP3 (Table III). The strongest associations were seen for rs3801776 in the combined discovery NHW multiplex and simplex group (p=0.004) and when stratified by family history, and for rs13223993 in IGFBP3 in the simplex families (p=0.003) (Table III). Additionally, a strong association for rs6749771 in HOXD was found in the NHW simplex families (p=0.009). In contrast, in the Hispanic sample, the strongest association was with rs2428431 (p=0.008) in the HOXA gene cluster, with no evidence for association found with SNPs in HOXD or IGFBP3. Interestingly, when the ethnicities were combined, rs2428431 in HOXA was still significant (Table III).
Table III.
| All | Multiplex | Simplex | |||||||
|---|---|---|---|---|---|---|---|---|---|
| dbSNP | Pos | All Ethn. |
NHW | Hisp | NHW | Hisp | NHW | Hisp | |
| HoxD | rs6749771 | SNP1 | — | 0.015 | 0.899 | 0.505 | 0.699 | 0.009 | 0.754 |
| rs1446575 | SNP2 | — | 0.021 | 0.334 | 0.026 | 0.440 | 0.224 | 0.134 | |
| HoxA | rs6668 | SNP13 | 0.101 | 0.802 | 0.025 | 0.968 | 0.119 | 0.872 | 0.073 |
| rs2428431 | SNP14 | 0.006 | 0.345 | 0.008 | 0.158 | 0.081 | 0.876 | 0.043 | |
| rs3801776 | SNP16 | — | 0.004 | 0.143 | 0.032 | 0.114 | 0.046 | 0.449 | |
| r3779456 | SP17 | — | 0.179 | 0.845 | 0.042 | 0.459 | 0.980 | 0.702 | |
| IGFBP3 | rs3793345 | SNP26 | — | 0.015 | 0.307 | 0.066 | 0.497 | 0.101 | 0.393 |
| rs2471551 | SNP27 | 0.045 | 0.032 | 0.556 | 0.041 | 0.489 | 0.311 | 0.769 | |
| rs13223993 | SNP32 | — | 0.047 | 0.320 | 0.852 | 0.398 | 0.003 | 0.552 | |
SNPs with p<0.05 shown in bold,
p-values uncorrected for multiple testing,
NHW=nonHispanic white, Hisp.=Hispanic
Significantly over-transmitted haplotypes for HOXA and HOXD were seen in NHW and only for HOXA in Hispanics (Table IV and Supplemental Tables 5 – 7). These haplotypes primarily involve the SNPs identified in the single SNP association analyses (Table III). Among the NHW, the most significant haplotype for HOXA involved rs3801776 (HOXA9) and rs1859164 (HOXA10), which were located in the 5’ region of the HOXA cluster; both HOXA9 and HOXA10 are expressed during limb development (Table IV). In Hispanics, haplotypes involving rs2428431 or rs2428431 (HOXA) were also over-transmitted (0.006≤p≤0.009, Table IV and Supplemental Table V).
Table IV.
| A. nonHispanic White | ||
|---|---|---|
| Haplotype | p-value | |
| HoxD | SNP1/SNP2 | 0.007 |
| SNP1/SNP6 | 0.007 | |
| SNP1/SNP7 | 0.006 | |
| HoxA | SNP16/SNP18 | 0.004 |
| B. Hispanic | ||
|---|---|---|
| Haplotype | p-value | |
| HoxA | SNP14/SNP15 | 0.009 |
| SNP14/SNP19 | 0.006 | |
| SNP16/SNP12 | 0.006 | |
Haplotypes with p<0.01 shown,
p-values uncorrected for multiple testing
There was no evidence for interactions between HOXA and HOXD in the discovery NHW group. However for the discovery Hispanic dataset, there was evidence for interactions between HOXA and HOXD for rs6758117/ rs2428431 and rs1867863/ rs3801776 (p=0.006 and p=0.007, respectively, data not shown). Interactions were found in the discovery sample for SNPs in HOXA/HOXD and IGFBP3 (Table V). The strongest interaction was between rs3801776 (HOXA) and rs13223993 (IGFBP3) in the NHW subset (p=0.0001, Table V and Supplemental Table IX and X). Interestingly, significant HOXA interactions were found for rs13223993 and 5’ HOX SNPs expressed in limb development in both NHW and Hispanic groups.
Table V.
| A. Discovery | |||||
|---|---|---|---|---|---|
| Gene1 | SNP1 | Gene2 | SNP2 | p-value | Pop |
| HOXD | rs2113563 (SNP6) | IGFBP3 | rs3793345 (SNP26) | 0.008 | NHW |
| rs2113563 (SNP6) | rs2471551 (SNP27) | 0.007 | NHW | ||
| rs2592394 (SNP7) | rs3793345 (SNP26) | 0.003 | NHW | ||
| rs2592394 (SNP7) | rs2471551 (SNP27) | 0.001 | NHW | ||
| rs741610 (SNP9) | rs13223993 (SNP32) | 0.002 | H | ||
| rs711812 (SNP10) | rs13223993 (SNP32) | 0.003 | H | ||
| HOXA | rs3801776 (SNP16) | rs13223993 (SNP32) | 0.0001 | NHW | |
| rs6968828 (SNP19) | rs2854747 (SNP25) | 0.003 | NHW | ||
| rs6968828 (SNP19) | rs3110697 (SNP28) | 0.001 | NHW | ||
| B. Validation | ||||
|---|---|---|---|---|
| Gene1 | SNP1 | Gene2 | SNP2 | p-value |
| HOXD | rs1542180 (SNP4) | IGFBP3 | rs2854747 (SNP25) | 0.002 |
| rs1542180 (SNP4) | rs3110697 (SNP28) | 0.0003 | ||
| rs1542180 (SNP4) | rs2453839 (SNP30) | 0.002 | ||
| rs2592394 (SNP7) | rs3110697 (SNP28) | <0.0001 | ||
| rs2592394 (SNP7) | rs2453839 (SNP30) | 0.004 | ||
| rs2592394 (SNP7) | rs2854747 (SNP25) | 0.0006 | ||
| rs847146 (SNP8) | rs2854747 (SNP25) | 0.004 | ||
| rs847146 (SNP8) | rs3110697 (SNP28) | 0.001 | ||
| HOXA | rs2462907 (SNP12) | rs2471551 (SNP27) | 0.007 | |
Only p<0.01 shown, p<0.001 in bold,
NHW=nonHispanic white H=Hispanic,
p-values uncorrected for multiple testing
All SNPs were then tested in the validation dataset. There was evidence for association with rs3801776 in HOXA (p=0.028). Analysis of 2-SNP haplotypes in this sample identified seven HOXA haplotypes with altered transmission, none of which were identified in the discovery population (Table IV and Supplemental Table VIII). The most significant haplotypes in the validation population were rs3779456/ rs3807598 (p=0.007) and rs1859164/ rs3807598 (p=0.0003, Supplemental Table VIII).
There was no evidence for an interaction between SNPs in HOXA and HOXD in the validation dataset (data not shown). In contrast, multiple interactions between SNPs in HOXA and IGFBP3 and HOXD and IGFBP3 were identified in the validation population; the majority of these interactions involved SNPs in HOXD and SNPs in IGFBP3 (Table VI and Supplemental Table XI) but are different from those found in the discovery population (Supplemental Tables IX and XI).
Table VI.
Gene-gene interactions for Hox IGFBP3 and mitochondrial-mediated apoptotic variants in discovery dataseta,b
| A. nonHispanic White | ||||
|---|---|---|---|---|
| Gene 1 | SNP 1 | Gene 2 | SNP 2 | p-value |
| Casp3 | rs1049216 | HoxD | rs6749771 (SNP1) | 0.002 |
| Casp3 | rs1405937 | rs6749771 (SNP1) | 0.005 | |
| Casp3 | rs1049253 | rs2592394 (SNP7) | 0.001 | |
| Casp3 | rs1049216 | rs2592394 (SNP7) | 0.008 | |
| Bid | rs8919 | rs6758117 (SNP11) | 0.007 | |
| Casp3 | rs2720378 | HoxA | rs6668 (SNP13) | 0.004 |
| Casp3 | rs1049216 | rs1859164 (SNP18) | 0.004 | |
| Casp3 | rs2720378 | rs1859164 (SNP18) | 0.004 | |
| Casp10 | rs3900115 | rs3779456 (SNP17) | 0.008 | |
| Bid | rs181405 | rs3779456 (SNP17) | 0.001 | |
| Bid | rs181405 | rs1859164 (SNP18) | 0.005 | |
| Apaf1 | rs2132572 | IGFBP3 | rs2132572 (SNP22) | 0.008 |
| Casp3 | rs1049210 | rs2132572 (SNP22) | <.0001 | |
| Casp3 | rs1049210 | rs2854744 (SNP23) | 0.006 | |
| Casp3 | rs1049210 | rs10255707 (SNP29) | 0.002 | |
| Casp3 | rs1049210 | rs2854746 (SNP24) | 0.005 | |
| Casp8 | rs3769825 | rs10255707 (SNP29) | 0.003 | |
| Casp10 | rs3900115 | rs2854747 (SNP25) | 0.008 | |
| B. Hispanic | ||||
|---|---|---|---|---|
| Gene 1 | SNP 1 | Gene 2 | SNP 2 | p-value |
| Casp3 | rs4647602 | HoxD | rs1318778 (SNP3) | 0.0005 |
| Casp3 | rs4647602 | rs2113563 (SNP6) | 0.009 | |
| Casp3 | rs2696057 | rs741610 (SNP9) | 0.008 | |
| Bcl2 | rs1564483 | HoxA | rs3801776 (SNP16) | 0.002 |
| Casp9 | rs4233533 | rs3779456 (SNP17) | 0.001 | |
| Apaf1 | rs1866477 | rs6968828 (SNP19) | 0.009 | |
| Apaf1 | rs2278361 | IGFBP3 | rs2132571 (SNP21) | 0.007 |
| Apaf1 | rs2278361 | rs2453839 (SNP30) | 0.002 | |
| Apaf1 | rs7968661 | rs2453839 (SNP30) | 0.006 | |
only p<0.01 shown,
p-values uncorrected for multiple testing
We have previously reported an association of variants in mitochondrial-mediated apoptotic genes with clubfoot in our discovery dataset [Ester et al., 2007]. Since the genes in the current study were involved in both apoptosis and limb formation, we combined the data from the previous study with that of the current study to evaluate potential gene-gene interactions in the discovery sample. The results are presented in Table VI and Supplemental Tables XII and XIII. While multiple interactions between HOX and IGFBP3 genes with mitochondrial-mediated genes were found in both ethnicities, none of these interactions having a p<0.01 were the same in both ethnicities. There was, however, strong evidence for interaction between SNPs in HOXA and HOXD and SNPs in both CASP3 and Bid in NHW (Table VIA). The strongest interaction was between a SNP in CASP3 and one in IGFBP3 in NHW (Table VIA). In Hispanics, the strongest interactions were between SNPs in CASP3 and HOXD; two SNPs in IGFBP3 interacted with two SNPs in Apaf1 (Table VIB).
Because the HOX clusters were controlled by many different regulatory factors, SNPs in the intergenic regions of the clusters may be in DNA enhancer or suppressor binding sites. Therefore, each of the over-transmitted SNPs in these regions was assessed using three transcription factor binding site prediction algorithms and the results are shown in Table VII. rs2428431 was predicted to change ancestral binding sites by all three programs, but predicted binding factors differed (Table VII). In contrast, the ancestral rs3801776 allele was not identified as a potential binding site by any of the programs, while the alternative allele was identified as a potential binding site by two of the programs (Table VII). However, rs6749771 was not predicted to change transcription factor binding sites (data not shown).
Table VII.
| Alibaba2 | Patch | TESS | |||||
|---|---|---|---|---|---|---|---|
| Alleles | Alleles | Alleles | |||||
| Ancestral | Alternate | Ancestral | Alternate | Ancestral | Alternate | ||
| HoxA |
rs2428431 (SNP14) |
MIG1, Sp1 | None | Sp1, AML1 | NF-E | TCF2α, MIG1, AML1 |
NF-E, RAF |
|
rs3801776 (SNP16) |
None | 1-Oct | None | None | None | RAF | |
Only SNPs with p<0.01 in regulatory regions shown,
Abbreviations: AML1: acute myeloid leukemia 1, MIG1: MIG1, NF-E: nuclear factor E, Oct-1: octomer-binding transcription factor 1, RAF: RAF, Sp1: simian virus 40 protein 1, TCF2α: transcription factor 2 α
DISCUSSION
This study was undertaken to assess the roles of limb and muscle morphogenesis genes in clubfoot. The 956T>A/319M>K HOXD10 mutation that causes syndromic vertical talus was not found in any cases or controls. This result confirms previous findings and provides further evidence that this mutation does not cause isolated clubfoot [Gurnett et al., 2007]. Next, we evaluated whether variants in the HOXA and HOXD gene clusters and IGFBP3 are associated with clubfoot in our family-based dataset. This was accomplished by genotyping SNPs spanning the HOXA, HOXD and IGFBP3 genes first in our discovery sample with follow-up in our validation sample. Located 3’ of the HOXD cluster, SNP1 showed altered transmission and may regulate expression of genes within the cluster. In the HOXA gene cluster, rs2428431 and rs3801776 were significantly over-transmitted. In intron 1 of HOXA2, rs2428431 is changes predicted DNA binding sites (Table VII). The HOXA2 gene is not expressed in the limb, but we cannot exclude the possibility that rs2428431 may regulate expression levels of other upstream 5’ HOXA genes or is in linkage disequilibrium with SNPs that do [Sasaki et al., 2007; Tarchini and Duboule, 2006]. Located in the HOXA9 basal promoter region, rs3801776 showed altered transmission in NHW in the discovery sample and was confirmed in the validation group. Furthermore, pair-wise haplotypes involving rs3801776 were significantly over-transmitted and gene-gene interactions involving this SNP were found. The HOXA9 gene is expressed in both the proximal and distal limb and the variant allele is predicted to introduce a novel DNA binding site (Table VII). This change could alter expression levels of HOXA9 and affect normal limb or muscle development. Future studies are aimed at determining whether rs3801776 is functional or if it is in linkage disequilibrium with a functional change.
Three SNPs (rs3793345, rs2471551 and rs13223993) in IGFBP3 were over-transmitted in the discovery sample. Interestingly, rs3793345 and rs2471551 are located in a potential enhancer/regulatory region in intron 1 and the SNP variants are predicted to change DNA binding sites (data not shown). Located in the 3’ UTR of IGFBP3, rs13223993 showed the most significant results. Previously, SNPs in 3’ UTRs are have been shown to regulate protein expression through microRNAs or proteins that can stabilize or destabilize the half-life of mRNA [Wickens et al., 2002]. Additionally, rs2132572 and rs2854747 in IGFBP3 were only marginally significant in the validation group but not the discovery dataset and do not change DNA binding sites (data not shown). Functional studies are necessary to further assess these SNP variants that could potentially change DNA binding and protein expression and thus affect limb development [Wolters and Wijmenga, 2008].
Because clubfoot is a complex disorder, multiple genes are likely involved although a major gene effect cannot be excluded. This allows us to postulate a model in which multiple genes interact and together are necessary but individually are not sufficient to cause disease. Moreover, we suggest that multiple variants within a gene may cause perturbation of gene function, rather than a single common variant, and thus any one SNP may not demonstrate significant evidence (very small p-values) for association. Therefore association within a gene is considered replication, even if different SNPs are over-transmitted in the two datasets. This standard of replication also was applied to gene-gene interactions. Here, we find that rs3801776 is associated with clubfoot, and in addition, we find that variation in apoptotic genes interact with several variants in HOXA and HOXD. These results are consistent with a recent study demonstrating that apoptosis is important for muscle morphogenesis [Rodriguez-Guzman et al., 2007].
Apoptosis is associated with interdigital removal of excess skin; however, it has recently been shown to play a far more subtle role in later limb development by shaping muscle and tendons [Rodriguez-Guzman et al., 2007]. Specifically, CASP3 has been shown to modulate apoptosis in later muscle and tendon development. While we did not find a strong association with CASP3 and clubfoot alone [Ester et al., 2007], we found evidence for interactions between CASP3 and variants in HOX genes and IGFBP3. This implies that individual variation in CASP3 may not be sufficient to cause clubfoot but that the interaction with genes in other limb and muscle developmental pathways, such as HOXA and HOXD, are necessary in order to disrupt the developmental process. The interactions observed between the many mitochondrial-mediated apoptotic genes and HOXA and HOXD genes suggest that there may be a common mechanism controlling these two pathways. Perturbations of this regulatory pathway may also contribute to clubfoot.
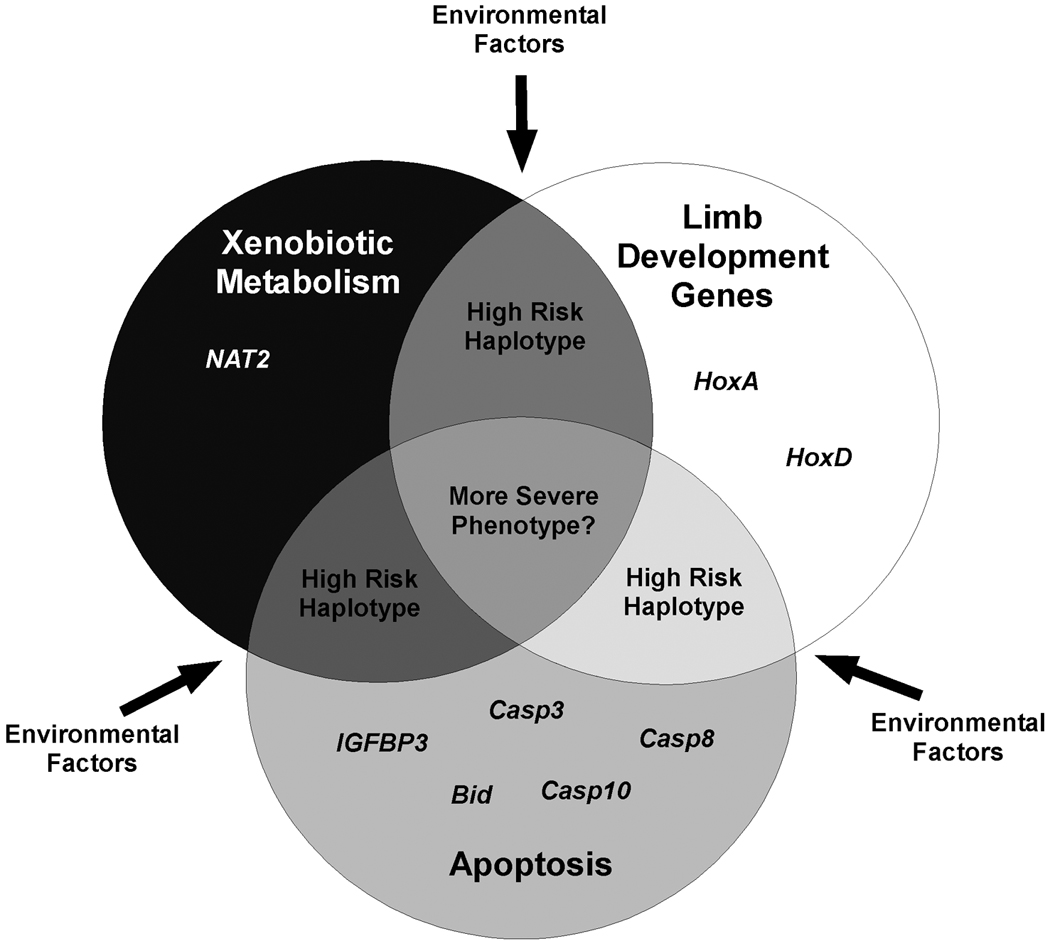
The goal of this study was to identify high-risk haplotypes that would contribute to clubfoot. The first step in this process is to identify genetic variants that are associated with clubfoot and to determine whether they interact. The results of our studies suggest a model for clubfoot that is shown in Figure 1 and includes the interactions between different genes. It is possible that environmental exposures alone can cause clubfoot, but it is more likely that an individual’s genotype confers an increased susceptibility to these exposures. This model further suggests that severity of phenotype may relate to the number of genetic variants and/or genetic pathways affected by the variants (Fig. 1). In this study, we found evidence for an association between a basal promoter SNP in HOXA9 and clubfoot, and provide further evidence that interactions between genes involved in limb development play a role in clubfoot.
Figure 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria Elena Serna and Dr. S. Shahrukh Hashmi for sample collection and database management and Dr. Scott Stadler for helpful discussions. This work was supported by grants from Shriners Hospital for Children [to JTH and MBD]; National Institutes of Health [5R01HD043342 to JTH; Texas Scottish Rite Hospital Research Fund [04-96-353 to CAW]; Pediatric Orthopaedic Society of North America, and St. Louis Children's Hospital to MBD. The project described was supported by Award Number TL1RR024147 from the National Center For Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baxter RC, Martin JL. Binding proteins for the insulin-like growth factors: structure, regulation and function. Prog Growth Factor Res. 1989;1:49–68. doi: 10.1016/0955-2235(89)90041-0. [DOI] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am J Hum Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61(4):189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- de Andrade M, Barnholtz JS, Amos CI, Lochmiller C, Scott A, Risman M, Hecht JT. Segregation analysis of idiopathic talipes equinovarus in a Texan population. Am J Med Genet. 1998;79:97–102. doi: 10.1002/(sici)1096-8628(19980901)79:2<97::aid-ajmg4>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Dietz FR. On the pathogenesis of clubfoot. Lancet. 1985;1:388–390. doi: 10.1016/s0140-6736(85)91400-x. [DOI] [PubMed] [Google Scholar]
- Dobbs MB, Gurnett CA, Pierce B, Exner GU, Robarge J, Morcuende JA, Cole WG, Templeton PA, Foster B, Bowcock AM. HOXD10 M319K mutation in a family with isolated congenital vertical talus. J Orthop Res. 2006;24:448–453. doi: 10.1002/jor.20052. [DOI] [PubMed] [Google Scholar]
- Ester AR, Tyerman G, Wise CA, Blanton SH, Hecht JT. Apoptotic gene analysis in idiopathic talipes equinovarus (clubfoot) Clin Orthop Relat Res. 2007;462:32–37. doi: 10.1097/BLO.0b013e318073c2d9. [DOI] [PubMed] [Google Scholar]
- Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2:S1–S15. [PubMed] [Google Scholar]
- Gurnett CA, Keppel C, Bick J, Bowcock AM, Dobbs MB. Absence of HOXD10 mutations in idiopathic clubfoot and sporadic vertical talus. Clinical Orthopaedics & Related Research. 2007;462:27–31. doi: 10.1097/BLO.0b013e31805d8649. [DOI] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Li YJ, Scott WK. Methods for interaction analyses using family-based case-control data: conditional logistic regression versus generalized estimating equations. Genet Epidemiol. 2007;31:883–893. doi: 10.1002/gepi.20249. [DOI] [PubMed] [Google Scholar]
- Heck AL, Bray MS, Scott A, Blanton SH, Hecht JT. Variation in CASP10 gene is associated with idiopathic talipes equinovarus. J Pediatr Orthop. 2005;25:598–602. doi: 10.1097/01.bpo.0000173248.96936.90. [DOI] [PubMed] [Google Scholar]
- Houghton L, Rosenthal N. Regulation of a muscle-specific transgene by persistent expression of Hox genes in postnatal murine limb muscle. Dev Dyn. 1999;216:385–397. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<385::AID-DVDY7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Lochmiller C, Johnston D, Scott A, Risman M, Hecht JT. Genetic epidemiology study of idiopathic talipes equinovarus. Am J Med Genet. 1998;79:90–96. [PubMed] [Google Scholar]
- Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Research. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Moorthi RN, Hashmi SS, Langois P, Canfield M, Waller DK, Hecht JT. Idiopathic talipes equinovarus (ITEV) (clubfeet) in Texas. Am J Med Genet Part A. 2005;132A:376–380. doi: 10.1002/ajmg.a.30505. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Dietz FR, Murray JC, Buetow KH. A single-gene explanation for the probability of having idiopathic talipes equinovarus. Am J Hum Genet. 1993;53:1051–1063. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Guzman M, Montero JA, Santesteban E, Ganan Y, Macias D, Hurle JM. Tendon-muscle crosstalk controls muscle bellies morphogenesis, which is mediated by cell death and retinoic acid signaling. Dev Biol. 2007;302:267–280. doi: 10.1016/j.ydbio.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Sano M, Kin T, Asai K, Hirose T. Coordinated expression of ncRNAs and HOX mRNAs in the human HOXA locus. Biochem Biophys Res Commun. 2007;357:724–730. doi: 10.1016/j.bbrc.2007.03.200. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. John Wiley and Sons, Inc.; 2003. pp. 2.6.1–2.6.15. [DOI] [PubMed] [Google Scholar]
- Shrimpton AE, Levinsohn EM, Yozawitz JM, Packard DS, Jr, Cady RB, Middleton FA, Persico AM, Hootnick DR. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. Am J Hum Genet. 2004;75:92–96. doi: 10.1086/422015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes' collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Wang JH, Palmer RM, Chung CS. The role of major gene in clubfoot. Am J Hum Genet. 1988;42:772–776. [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wolters VM, Wijmenga C. Genetic background of celiac disease and its clinical implications. Am J Gastroenterol. 2008;103:190–195. doi: 10.1111/j.1572-0241.2007.01471.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


