Abstract
In recent years the development of computational techniques that identify all the likely targets for a given chemical compound, also termed as the problem of Target Fishing, has been an active area of research. Identification of likely targets of a chemical compound helps to understand problems such as toxicity, lack of efficacy in humans, and poor physical properties associated with that compound in the early stages of drug discovery. In this paper we present a set of techniques whose goal is to rank or prioritize targets in the context of a given chemical compound such that most targets that this compound may show activity against appear higher in the ranked list. These methods are based on our extensions to the SVM and Ranking Perceptron algorithms for this problem. Our extensive experimental study shows that the methods developed in this work outperform previous approaches by 2% to 60% under different evaluation criterions.
1 Introduction
Target-based drug discovery, which involves selection of an appropriate target (generally a single protein) implicated in a disease state as the first step, has become the primary approach of drug discovery in pharmaceutical industry.1,2 This was made possible through the advent of High Throughput Screening (HTS) technology in the late 1980s that enabled rapid experimental testing of a large number of chemical compounds against the target of interest (using target-based assays). HTS is now routinely utilized to identify the most promising compounds (called hits) that show desired binding/activity against this target. Some of these compounds then go through the long and expensive process of optimization and eventually one of them goes to clinical trials. If clinical trials are successful then it becomes a drug. HTS technology was considered to usher a new era in drug discovery by reducing time and money taken to find hits that will have a high chance of eventually becoming a drug.
However, the expansion of candidate list of hits via HTS did not result in productivity gains in terms of actual drugs coming out of the drug discovery pipeline. One of the principal reasons ascribed for this failure is that the above approach suffers from a serious drawback of only focusing on the target of interest and therefore taking a very narrow view of the disease. As such, it may lead to unsatisfactory phenotypic effects such as toxicity, promiscuity, and low efficacy in the later stages of drug discovery.2,3 More recently, focus is shifting to directly screen molecules to identify desirable phenotypic effects using cell-based assays. This screening evaluates properties such as toxicity, promiscuity and efficacy from the onset rather than in later stages of drug discovery.2-4 Moreover, toxicity and off-target effects are also a focus of early stages of conventional target-based drug discovery.5,6 But from the drug discovery perspective, target identification and subsequent validation has become the rate limiting step in order to tackle the above issues.7 Targets must be identified for the hits in phenotypic assay experiments and for secondary pharmacology as the activity of hits against all of its potential targets sheds light on the toxicity and promiscuity of these hits.6,8 Therefore, the identification of all likely targets for a given chemical compound, also called Target Fishing,4 has become an important problem in drug discovery.
In this work we focus on the target fishing problem by utilizing the available target-ligand activity data matrix. In this approach, we are given a set of targets and a set of ligands (chemical compounds) and a bipartite activity relation between the targets and the ligands in the two sets. Given a new test chemical compound not in the set, the goal is to correctly predict all the activity relations between the test compound and the targets. We address this problem by formulating it as a category ranking problem. The goal is to learn a model such that for a given test compound it ranks the targets that this compound shows activity against (relevant targets) higher than the rest of the targets (non-relevant targets). In this work, we propose a number of methods that are inspired by research in the area of multiclass/multilabel classification and protein secondary structure prediction. Specifically, we develop four methods based on support vector machines (SVM)9 and ranking perceptrons10 to solve the above ranking problem. Three of these methods try to explicitly capture dependencies between different categories to build models. Our results show that the methods proposed in this work are either competitive or substantially outperform other methods currently employed to solve this problem.
The rest of this paper is organized as follows. Section 2 describes related research in the area of target fishing. Section 3 introduces definition and notations used in this paper. Section 4 describes our methods for target fishing. Section 5 discusses the datasets and experimental methodology used in this work. Section 6 describes our results and finally section 7 has concluding remarks.
2 Related Methods
Computational techniques are becoming increasingly popular for target fishing due to plethora of data from high-throughput screening (HTS), microarrays, and other experiments.4 Given a compound whose targets need to be identified, these techniques initially assign a score to each target based on some measure of likelihood that the compound binds to each target. These techniques then select as the potential compound’s targets either those targets whose score is above a certain cut-off or a small number of the highest scoring targets. Three general classes of methods have been developed for determining the required compound-target scores. The first, referred to as inverse docking, contains methods that score each compound-target pair by using ligand docking approaches.6,11 The second, referred to as nearest-neighbor, contains methods that determine the compound-target scores by exploiting structural similarities between the compound and the target’s known ligands.12 Finally the third, referred to as model-based, contains methods that determine these scores using various machine-learning approaches to learn models for each one of the potential targets based on their known ligands.13-15
The first class of methods to derive a score for each compound-target pair comes from the computational chemistry domain. Specifically, inverse docking process docks a single compound of interest to a set of targets and obtains a score for each target against this compound. The highest scoring targets are then considered as most likely targets that this compound will bind to.6 This approach suffers from a serious drawback that all the targets used in this process must have their three dimensional structure available in order to employ a docking procedure. However, the majority of target proteins do not have such information available.16
The second class of methods, nearest-neighbor based techniques, rely on the principle of structure-activity relationship (SAR)17,18 which suggests that very similar compounds will have a higher chance of overlap between the sets of targets that they show activity against.12 Therefore, identifying targets for a given chemical compound can be solved by utilizing its structural similarity with other chemical compounds that are known to be active or inactive against certain targets. In these approaches, for a given test compound, its nearest-neighbor(s) are identified from a database of compounds with known targets using some notion of structural similarity. The most likely targets for the test compound are then identified as those targets that its nearest neighbors show activity against. A ranking among these targets can be obtained by taking into account the similarity values/rankings of the nearest neighbors that these targets belongs to. In these approaches the solution to the target fishing problem only depends on the underlying descriptor-space representation, the similarity function employed, and the definition of nearest neighbors.
Lastly, a number of methods have been proposed that explicitly build models on the given set of compounds with known targets. These techniques treat the target fishing problem as an instance of a multilabel (multi-category) prediction problem.15,19,20 In this setting, for a given chemical compound, each of the targets is treated as one of the potential labels and the goal is to predict all the labels (i.e., targets) that the compound belongs to (i.e., binds to). One such approach utilizes multi-category bayesian models13 wherein a model is built for every target using the available SAR data. Compounds that show activity against a target are used as positives instances and the rest of the compounds are treated as negatives instances. For a new compound, each of these models is used to compute the compound’s likelihood to be active against the corresponding target and the targets that obtained the highest likelihood scores are considered to be the targets for this compound. In addition, approaches have been developed that build the classification models using one-versus-rest binary support vector machines15 and neural networks.14 Note that even though the underlying machine learning problem is that of multilabel prediction, all of the above model-based methods essentially build one-vs-rest models and then produce a ranking of the possible targets by directly comparing the outputs of these models.
3 Definitions and Notations
The target fishing problem that we consider in this paper is defined as follows:
Definition 1 (Target Fishing Problem). Given a set of compounds (more than one) whose bioactivity is known to be either active or inactive against each of the targets in a given set, learn a model such that it correctly predicts for a test compound a ranking of all the targets according to how likely they are to show activity against the test compound.
Throughout this paper we will use 𝒞 = {𝒸1, … , 𝒸M} to denote a library of chemical compounds, Τ = {τ1, … , τN} to denote a set of protein targets, and will assume that they contain M and N elements, respectively. For each compound 𝒸𝒾, we will use Τ𝒾 to denote the set of all targets that 𝒸𝒾 shows activity against. Note that Τ𝒾 ⊆ Τ. We will use Τ* to denote a total ordering of the targets of Τ. Given two sets A and B such that A ⊂ Τ and B ⊂ Τ, we will use A <Τ* B to denote that every target of A precedes every target of B in Τ*, and A ≮Τ* B otherwise.
Each compound will be represented by a topological descriptor-based representation.18,21 In this representation, each compound is modeled as a frequency vector of certain topological descriptors (e.g., subgraphs) present in its molecular graph. Each dimension’s frequency counts the number of times (i.e., embeddings) the corresponding topological descriptor is present in the compound’s molecular graph. We will use 𝓃 to represent the dimensionality of descriptor-space representation of the chemical compounds in 𝒞. Given a compound 𝒸, and a parameter 𝓀, we define top-𝓀 to be the 𝓀 predicted targets that are most likely to show activity for 𝒸. Lastly, throughout this paper we will use the terms target, category, and labels interchangeably.
4 Methods
Our solution to the Target Fishing problem relies on the principle of Structure Activity Relationship (SAR). Specifically, we develop solutions for the target fishing problem using SAR data by formulating it as a ranking problem. We pursue this approach because in real-world situations a practitioner might want to know the top-𝓀 most likely targets for a given compound so that they can be tested experimentally or further investigated. Therefore, if the relevant (true) targets fall in one of these top-𝓀 predicted targets, they will have a higher chance to be recognized. These methods are described in the rest of this section.
4.1 SVM-based Method
One approach for solving the ranking problem is to build for each target τ𝒾 ∈ Τ a one-versus-rest binary SVM classifier. Given a test chemical compound 𝒸, the classifier for each target τ𝒾 will then be applied to obtain a prediction score 𝒻𝒾(𝒸). The ranking Τ* of the N targets will be obtain by simply sorting the targets based on their prediction scores. That is,
| (1) |
where argsort returns an ordering of the targets in decreasing order of their prediction scores 𝒻𝒾(𝒸). Note that this approach assumes that the prediction scores obtained from the N binary classifiers are directly comparable, which may not necessarily be valid. This is because different classes may be of different sizes and/or less separable from the rest of the dataset indirectly affecting the nature of the binary model that was learned and consequently their prediction scores.22 This SVM-based sorting method is similar to the approach described previously15 in Section 2. We refer to this method as SVMR.
4.2 Cascaded SVM-based Method
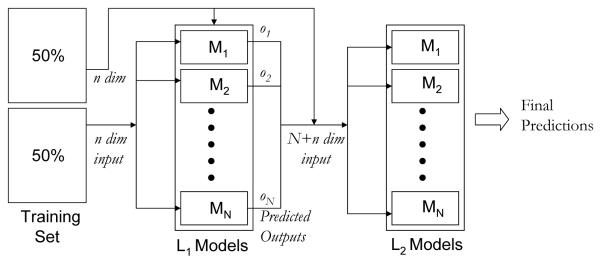
A limitation of the previous approach is that by building a series of one-vs-rest binary classifiers it does not explicitly couple the information on the multiple categories that each compound belongs to during model training and as such it cannot capture dependencies that might exist between the different categories. A promising approach that has been explored to capture such dependencies is to formulate it as a cascaded learning problem.19,23,24 In these approaches, two sets of binary one-vs-rest classification models for each category, referred to as L1 and L2, are connected together in a cascaded fashion. The L1 models are trained on the initial inputs and their outputs are used as input, either by themselves or in conjunction with the initial inputs, to train the L2 models. This cascaded process is illustrated in Figure 1. During prediction time, the L1 models are first used to obtain the required predictions which are used as input to the L2 models from which we obtain the final predictions. Since the L2 models incorporate information about the predictions produced by the L1 models, they can potentially capture inter-category dependencies.
Figure 1.
Cascaded SVM Classifiers.
Motivated by the above observation, we developed a ranking method for the target fishing problem in which both the L1 and L2 models consist of N binary one-vs-rest SVM classifiers, one for each target in Τ. The L1 models correspond exactly to the set of models built by the SVMR method discussed in the previous section. The representation of each compound in the training set for the L2 models consists of its descriptor-space based representation and its output from each of the N L1 models. Thus, each compound 𝒸 corresponds to an 𝓃 + N dimensional vector, where 𝓃 is the dimensionality of the descriptor space. The final ranking Τ* of the targets for a compound 𝒸 will be obtained by sorting the targets based on their prediction scores from the L2 models (). That is,
| (2) |
We will refer to this approach as SVM2R.
A potential problem with such an approach is that the descriptor-space based representation of a chemical compound and the set of its outputs from L1 models are not directly comparable. Therefore, in this work, we also experiment with various kernel functions that combine 𝓃 dimensional descriptor-space representation and the N dimensional L1 model outputs of a chemical compound. Specifically, we experiment with two forms of the kernel function. The first function is given by
| (3) |
where α is a user defined parameter and 𝒦A and 𝒦B are the kernel functions measuring the similarity between compound vector formed using descriptor-space based representation and L1 SVM classifier outputs respectively. The second kernel function is given by
| (4) |
These approaches are motivated by the work on kernel fusion25 and tensor product kernels.26
4.3 Ranking Perceptron Method
We also developed a ranking method that exploits the potential dependencies between the categories based on the ranking perceptron.10 The ranking perceptron extends Rosen-blatt’s linear perceptron classifier27 for the task of learning a ranking function. The perceptron algorithm and its variants have proven to be effective in a broad range of applications in machine learning, information retrieval and bioinformatics.10,20,28
Our approach is based on the online version of the ranking perceptron algorithm proposed to learn a ranking function on a set of categories developed by Crammer and Singer.10 This algorithm takes as input a set of objects and the categories that they belong to and learns a function that for a given object 𝒸 it ranks the different categories based on the likelihood that 𝒸 binds to the corresponding targets. During the learning phase, the distinction between categories is made only via a binary decision function that takes into account whether a category is part of the object’s categories (relevant set) or not (non-relevant set). As a result, even though the output of this algorithm is a total ordering of the categories, the learning is only dependent on the partial orderings induced by the set of relevant and non-relevant categories.
The pseudocode of our ranking perceptron algorithm is shown in Algorithm 1. This algorithm learns a linear model W that corresponds to a N×𝓃 matrix, where N is the number of targets and 𝓃 is the dimensionality of the descriptor space. Using this model, the prediction score for compound 𝒸𝒾 and target τ𝒿 is given by 〈W𝒿, 𝒸𝒾〉, where W𝒿 is the 𝒿th row of W, 𝒸𝒾 is the descriptor-space representation of the compound, and 〈·,·〉 denotes a dot-product operation. Our algorithm extends the work of Crammer and Singer by introducing margin based updates and extending the online version to a batch setting. Specifically, for each training compound 𝒸𝒾 that is active for targets belonging to categories Τ𝒾 ⊆ Τ, our algorithm learns W such that the following constraints as satisfied:
| (5) |
where β is a user-specified non-negative constant that corresponds to the separation margin. The idea behind these constraints is to force the algorithm to learn a model in which the set of relevant categories (Τ𝒾) for a given chemical compound 𝒸𝒾 are well-separated and ranked higher from all the non-relevant categories (T \ Τ𝒾). Therefore, our algorithm tries to satisfy ∑𝒸𝒾∈𝒞 |Τ𝒾| × |Τ \ Τ𝒾| constraints for each of the training set compound 𝒸𝒾 to enforce a degree of acceptable separation between relevant and non-relevant categories that is controlled by β.
Algorithm 1. Learning Category Weight Vectors with the ranking perceptron algorithm.
input:
𝒞: Set of M training compounds.
Τ: Set of N targets (categories).
(𝒸𝒾, Τ𝒾): Compound 𝒸𝒾 and its categories Τ𝒾.
β: User defined margin constraint.
𝓃: Dimensionality of the compound’s descriptor-space representation.
output:
W: N × 𝓃 model matrix.
1: W = 0 {Initial model}2: η = 1/M {Update weight}3: while (STOPPING CRITERION == FALSE) do4: for 𝒾=1 to M do5: Τ* = argsortτ𝒿∈Τ {〈W𝒿, 𝒸𝒾〉}6: τ𝒿 = lowest ranked target of Τ𝒾 in Τ*7: τ𝓀 = highest ranked target of Τ \ Τ𝒾 in Τ*8: if 〈W𝒿, 𝒸𝒾〉 − 〈W𝓀, 𝒸𝒾〉 < β then9: for ∀τq ∈ Τ𝒾: 〈Wq, 𝒸𝒾〉 − 〈W𝓀, 𝒸𝒾〉 < β do10: λ = |{τr ∈ Τ \ Τ𝒾: 〈Wq, 𝒸𝒾〉 − 〈Wr, 𝒸𝒾〉 < β}|11: Wq = Wq + λη𝒸𝒾12: end for13: for ∀τr ∈ Τ \ Τ𝒾: 〈W𝒿, 𝒸𝒾〉 − 〈Wr, 𝒸𝒾〉 < β do14: λ = |{τq ∈ Τ𝒾: 〈Wq, 𝒸𝒾〉 − 〈Wr, 𝒸𝒾〉 < β}|15: Wr = Wr – λη𝒸𝒾16: end for17: end if18: end for19: ∀τ𝒾 ∈ Τ, W𝒾 = W𝒾/||W𝒾||20: end while21: return WDuring each outer iteration (lines 3–20) the algorithm iterates over all the training compounds (lines 4–18) and for each compound 𝒸𝒾 it obtains a ranking Τ* of all the categories (line 5) based on the current model W, and updates the model if any of the constraints in Equation 5 are violated. The check for the constraint violation is done in line 8 by comparing the lowest ranked target τ𝒿 ∈ Τ𝒾 with the highest ranked target τ𝓀 ∈ Τ \ Τ𝒾. If there are any constraint violations, the condition on line 8 will be true and lines 9–16 of the algorithm will be executed. The model W is updated by adding/subtracting a multiple of 𝒸𝒾 from the rows of W involved in the pair of targets of the violated constraints. Instead of updating the model’s vectors by using a constant multiple, which is usually done in perceptron training, our algorithm uses a multiple that is proportional to the number of constraints that each target violates in Τ*. Specifically, for each target τq ∈ Τ𝒾, our algorithm (line 10) finds the number λ of targets τr ∈ Τ \ Τ𝒾 that violate the margin constraint with τq and adds in the qth row of W (which is the portion of the model corresponding to target τq) a λ multiple of η𝒸𝒾, where η is a small constant set to 1/M in our experiments. The motivation behind this proportional update is that if a relevant target τq follows a large number of non-relevant targets in the ordering Τ*, τq’s model (Wq) needs to move towards the direction of 𝒸𝒾 more than the model for another relevant target τq’ which is followed only by a small number of non-relevant targets in Τ*. Note that the term “follows” in the above discussion needs to be considered within the context of the margin β. A similar approach is used to determine the multiple of 𝒸𝒾 to be subtracted from the rows of W corresponding to the non-relevant targets that are involved in violated constraints (lines 13–16). Our experiments (not reported here) showed that this proportional update achieved consistently better results than those achieved by constant update rules.
Since the ranking perceptron algorithm is not guaranteed to converge when the training instances are not β-linearly separable, Algorithm 1 incorporates an explicit stopping criterion. After every pass over the entire training set, it computes the average uninterpolated precision (Section 5.2.4) over all the compounds using the weights W, and terminates when this precision has not improved in N consecutive iterations. The algorithm returns the W that achieved the highest training precision over all iterations. We directly apply the above method on the descriptor-space representation of the training set of chemical compounds.
The predicted ranking for a test chemical compound 𝒸 is given by
| (6) |
We will refer to this approach as RP.
4.4 SVM+Ranking Perceptron-based Method
A limitation of the above ranking perceptron method over the SVM-based methods is that it is a weaker learner as (i) it learns a linear model, and (ii) it does not provide any guarantees that it will converge to a good solution when the dataset is not linearly separable. In order to partially overcome these limitations we developed a scheme that is similar in nature to the cascaded SVM-based approach, but the L2 models are replaced by a ranking perceptron. Specifically, N binary onevs-rest SVM models are trained, which form the set of L1 models. Similar to the SVM2R method, the representation of each compound in the training set for the L2 models consists of its descriptor-space based representation and its output from each of the N L1 models. Thus, each compound corresponds to an 𝓃 + N dimensional vector, where 𝓃 is the dimensionality of the descriptor space. Finally, a ranking model W learned using the ranking perceptron of Algorithm 1. Since the L2 model is based on the descriptor-space based representation and the outputs of the L1 models, the size of W is N × (𝓃 + N). A recent study within the context of remote homology prediction and fold recognition has shown that this way of coupling SVM and ranking perceptrons improves the overall performance.28 We will refer to this approach as SVMRP.
5 Materials
5.1 Datasets
We evaluated the methods proposed in this work using a set of assays derived from a wide variety of databases that store the bioactivity relationship between a target and a set of small chemical molecules or ligands. In particular, these databases provide us target-ligand activity relationship pairs.
We use the PubChem29 database to extract target-specific dose-response confirmatory assays. For each assay we choose compounds that show the desired activity and confirmed as active by the database curators. We filter compounds that show different activity signals in different experiments against the same targets and they are deemed to be inconclusive and so not used in the study. Duplicate compound entries are removed by comparing the canonical SMILES30 representations of these molecules. We also incorporate target-ligand pairs from the following databases: BindingDB,31 Drug-Bank,32 PDSP K𝒾 database,33 KEGG BRITE database,34 and an evaluation sample of the WOMBAT database.35 All the protein targets that can be mapped to an identifier in PDB database16 are extracted from this set. We then eliminate all the targets that have < 10 actives to ensure that there is some amount of activity information available for each target in our database. Note that a minority of databases report binding affinity between compound and targets (that can be converted to binds to or does not bind to a target) instead of activity. In this work we do not distinguish between the two. It should be noted that the datasets used in our study consists of mostly confirmatory dose-response assays. Therefore it is expected to have lower level of noise than either the single point or the High Throughput Screening data.
After the above integration and filtering steps our final dataset contains 231 targets, and 27,205 compounds or ligands with a total of 40,170 target-ligand active pairs. Note that certain compounds may show activity in relation with two or more targets as well. Table 1 shows the number of compounds that belong to one or more categories. Out of the 27,205 compounds, 19,154 belong to a single category, 5,363 belong to two categories, 1,697 belong to three categories and the rest of the compounds belong to greater than three categories. It should also be noted that most of these compounds have been experimentally tested for activity against a very small subset of the 231 targets. Thus, if a compound belongs to a very small number of categories, it may be simply due to the fact that it has not been experimentally tested against many targets. Figure 2 shows the distribution of actives against the 231 targets in this dataset. The dataset has a skewed distribution wherein most targets have few active compounds (less than hundred) but a small number of targets have a large number active compounds (in thousands).
Table 1.
Multi-target activity of the compounds.
| # compounds | 19,154 | 5,363 | 1,697 | 648 | 129 | 139 | 29 | 14 | 7 | 25 |
| # targets | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10–41 |
The number of compounds that are active against different number of targets. The last entry indicates that there are a total of 25 compounds that are active against at least 10 different targets and that there are no compounds that are active against more that 41 targets.
Figure 2.
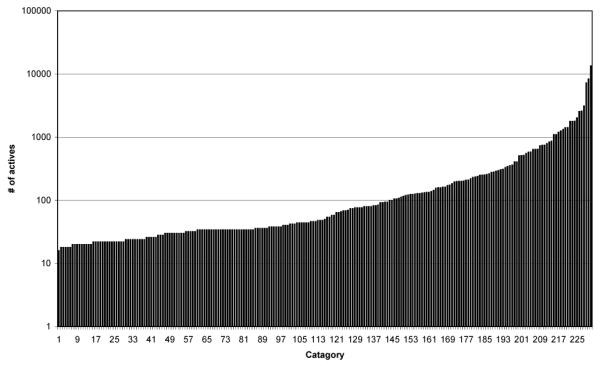
Distribution of Actives in various categories (using log-scale).
The filtered dataset consists of many clinically important enzymes such as PDEs as well as many kinase proteins. It also consists of many targets of clinically important virulent bacterial and viral strains such as tuberculosis, cholera, HIV, and herpes. In particular the dataset contains 165 human targets, 25 non-human mammalian targets and the remaining 41 consists mostly of bacterial, viral, and fungal targets. Figure 3 describes the distribution of the 231 targets in various classes. (The dataset with smiles representation of compounds and all the PDB ids of targets it binds to can be found onlinea).
Figure 3.
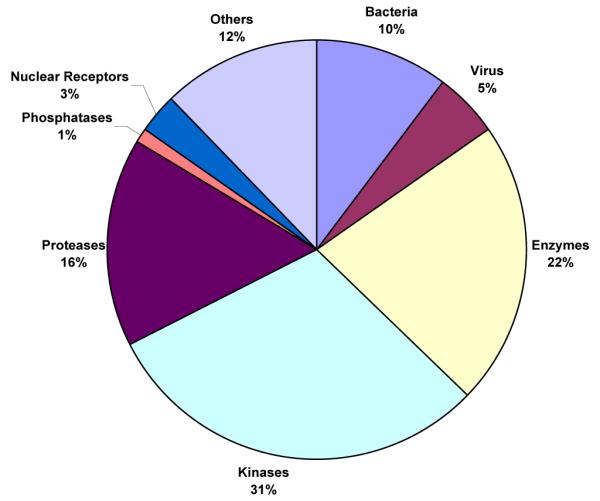
Distribution of targets in various classes.
5.2 Experimental Methodology
All the experiments were performed on dual core AMD Opterons with 4 GB of memory. The following sections will review our experimental methodology to assess the performance of various methods proposed in Section 4.
5.2.1 Descriptor Spaces and Similarity Measures
The similarity between chemical compounds is usually computed by first transforming them into a suitable descriptor-space representation.18,21 A number of different approaches have been developed to represent each compound by a set of descriptors. These descriptors can be based on physiochemical properties as well as topological and geometric substructures (fragments).30,36-40 In this study we use the ECFP descriptors37,38 as it has been shown to be very effective in the context of classification, ranked-retrieval, and scaffold-hopping.39,41 We utilize Scitegic’s Pipeline Pilot42 to generate ECFP4, a variation of ECFP descriptor-space for our dataset.
We use the Tanimoto coefficient40 (extended Jacquard similarity coefficient) to measure the similarity of chemical compounds based on their descriptor-space representation. This similarity measure was used to form the kernel function in SVMR and the kernel 𝒦A in SVM2R (Equations 3 and 4). We also utilize the cosine similarity to measure the similarity between two compounds represented by the L1 SVM outputs (𝒦B in Equations 3 and 4). Tanimoto coefficient was selected because it has been shown to be an effective way of measuring the similarity between chemical compounds using sparse descriptor-space representations,39,40,43 whereas the cosine function was selected because it achieved slightly better performance as compared to Tanimoto coefficient and variants of Eucledian distance for this task.
5.2.2 Multi-Category Bayesian Predictor
We implemented the multi-category bayesian models as described in13 and will call this approach BAYESIAN as the original authors call their approach Laplacian-corrected multi-category bayesian models.13 We compare this scheme to our methods developed in this paper. We have experimented with nearest-neighbor based scheme12 as well and found the overall results to be comparable to the BAYESIAN method. Therefore, we do not report the results for the nearest neighbor scheme in this paper.
5.2.3 Training Methodology
For each dataset we separated the compounds into test and training sets, ensuring that the test set is never used during any parts of the learning phase. We split the entire data randomly into ten parts and use nine parts for training and one part for test. We will refer to the nine parts training set as 𝒞r and the one part test set as 𝒞s.
In order to learn models using BAYESIAN, SVMR, and RP, we utilize the entire set 𝒞r for training. To learn the models for the cascaded methods (SVM2R and SVMRP) we experiment with an approach that allows us to use the entire training set (𝒞r) to build both L1 as well as L2 models. This approach is motivated by the cross-validation methodology and is similar to that used in previous works.20,28 In this approach we further partition the entire training set 𝒞r into ten equal-size parts. Nine out of these ten parts are used to train N first level (L1) binary classifiers (one for each target). A total of N prediction values are then obtained for each compound in the remaining part. This process is repeated for each of the ten parts. At the end of this process, each training instance in 𝒞r has been predicted by a set of N binary classifiers, and these predicted values serve as descriptors of the training samples (𝒞r) for the second-level learning (using the SVMRP or the SVM2R algorithm). Having learned the second-level (L2) models, we take the entire training set 𝒞r and retrain the L1 models. These L1 models are then used to obtain output values (that form the input representation for L2 models) for the independent test set 𝒞s.
In our setup we use each part (𝒞s) from the initial ten way split as the test set exactly once. Therefore we have ten different variants of 𝒞r and 𝒞s. In order to be consistent with the methodology described in13 for learning BAYESIAN models, we assumed all the compound-target pairs with unknown activity as inactives. Moreover, we did not have inactivity information for many targets in our dataset so it was impossible to model most of our targets using true actives and inactives.
5.2.4 Evaluation Metrics
During the evaluation stage, we compute the prediction for our untouched test dataset. These predictions are then evaluated using the uninterpolated precision46 and precision/recall values in top-𝓀.47
To calculate uninterpolated precision for each test compound we utilize the following methodology. For a test compound we obtain top-𝓀 ranked targets (using one of the five schemes described in this paper), where 𝓀 is equal to the number of targets that this test compound is active against. Now, for every correctly predicted target that appears at a position 𝒿 the top-𝓀 predictions we compute precision value at that position. This precision value is defined as the ratio of the number of correct targets identified up to that position 𝒿 over the total number of targets seen thus far (which is the same as the number 𝒿). We calculate this value for every one of the positions in the top-𝓀 ranking that corresponds to a correctly predicted target for the given test compound. The final un-interpolated precision value is given by summing up all the precision values obtained above and dividing it by 𝓀.
We also calculate precision and recall values in the top one to fifteen ranks of the target for each test compound. This precision value for a test compound is defined as the fraction of correct targets in the top-𝓀 ranked targets (where 𝓀 ranges from one to fifteen). Note that this precision is different from the uninterpolated precision described above. The recall value is defined as the number of correctly predicted targets in the top-𝓀 ranked predictions divided by the total number of true targets for a test compound. Note that a high recall value indicates the ability of a scheme to identify a high fraction of true targets for a given compound in top-𝓀 ranks. The final values of uninterpolated precision, precision and recall reported in this work are averaged over all the compounds in the test set.
We used the box plots to compare the relative performance of the different methods. These box plots were derived from the performance achieved in each one of the ten independent test sets in the ten-fold cross validation experiments. Using these box plots, the relative performance of two methods was assessed by comparing the first (lower) quantile (q1) of one method against the median of the other. Specifically, we will consider that method A performs relatively better than another method B if the first quantile of A is higher than the median of B. Note that this approach only provides a qualitative way by which to compare the relative performance of two schemes and does not convey any indication of statistical significance. We resorted to this approach because the results of the ten-fold cross validation are not entirely independent from each other (training compounds overlap), and as such the traditional statistical significance tests cannot be used.48
5.2.5 Model Selection
The performance of SVM depends on the parameter that controls the trade-off between the margin and the misclassification cost (“C” parameter in SV Mlight49), whereas the performance of ranking perceptron depends on the margin β in Algorithm 1.
We perform a model selection or parameter selection step. To perform this exercise fairly, we split our test set into two equal halves of similar distributions, namely sets A and B. Using set A, we vary the controlling parameters and select the best performing model for set A. We use this selected model and compute the accuracy for set B. We repeat the above steps by switching the roles of A and B. The final results are the average of the two runs. While using the SV Mlight program we let C take values from the set {0.0001, 0.001, 0.01, 0.1, 0.1, 0.5, 1.0, 2.0, 5.0, 10.0, 15.0, 25.0}. While using the perceptron algorithm we let the margin β take values in the set {0.00001, 0.0001, 0.0005, 0.001, 0.005, 0.01, 0.05, 0.1, 1.0, 2.0, 5.0, 10.0}.
For SVM2R, we experiment with kernel functions described by Equations 3 and 4. For the kernel function described by equation 4 no parameter tuning is required. For the kernel function described by Equation 3 we experiment with five different values of the parameter α (α = 0.2, 0.4, 0.5, 0.6, and 0.8). Since Equation 3 with parameter α = 0.5 showed the best performance, we only present results for SVM2R using this kernel and parameter value.
6 Results
In this section, we will evaluate the performance of the BAYESIAN scheme as well as the four methods described in Section 4: SVMR, SVM2R, RP, and SVMRP.
We compare these methods along three directions. The first direction compares the overall performance of these methods with each other on the entire dataset. The second direction compares the effect of the number of categories that a compound belongs to on the results obtained. We utilize un-interpolated precision described in Section 5.2.4 as our metric for these comparisons. The third direction compares these five methods on their ability to retrieve all the relevant categories in the top-𝓀 ranks. In order to evaluate and compare the performance of these category ranking algorithms for this task we utilize two measures - precision in top-𝓀 and recall in top-𝓀 (also described in Section 5.2.4). Finally, we also compare each of these methods for the time they take to train the final ranking model.
6.1 Overall Comparison
Table 2 compares the uninterpolated precision of the five methods using the ECFP4 descriptor-space for each of the ten tests sets. These results are derived using compounds that bind to at least one target (L ≥ 1) and therefore include all the compounds in the dataset. Figure 4(a) shows the box plot results corresponding to L ≥ 1.
Table 2.
Performance of various schemes on ECFP4 descriptor-space
| Test Set # | BAYESIAN | SVMR | SVM2R | RP | SVMRP |
|---|---|---|---|---|---|
| 1 | 0.461 | 0.710 | 0.725 | 0.716 | 0.730 |
| 2 | 0.467 | 0.728 | 0.739 | 0.724 | 0.734 |
| 3 | 0.474 | 0.718 | 0.736 | 0.726 | 0.730 |
| 4 | 0.471 | 0.728 | 0.739 | 0.728 | 0.733 |
| 5 | 0.465 | 0.724 | 0.735 | 0.716 | 0.728 |
| 6 | 0.473 | 0.731 | 0.739 | 0.723 | 0.736 |
| 7 | 0.473 | 0.714 | 0.728 | 0.707 | 0.725 |
| 8 | 0.465 | 0.735 | 0.747 | 0.731 | 0.739 |
| 9 | 0.468 | 0.717 | 0.733 | 0.723 | 0.733 |
| 10 | 0.471 | 0.740 | 0.749 | 0.731 | 0.748 |
| Avg | 0.469 | 0.725 | 0.737 | 0.723 | 0.734 |
The entry in each cell corresponds to the uninterpolated precision of the column method for a given test set. Entries in bold represent winners for every test set.
Figure 4.
Box Plots comparing the five methods using average uninterpolated precision values of the 10 test sets.
From Table 2 it can be observed that the best performing schemes over the ten test sets are the SVM2R and SVMRP. The two schemes are better than all the other methods tested in our work over these ten splits. Comparing SVM2R and SVMRP shows that SVM2R performs better than SVMRP. However, from figure 4(a) it can be observed that this difference is not substantial as the variance in the results of SVM2R and SVMRP overlap considerably. Similarly, the next two methods, SVMR and RP show equivalent performance among each other as observed through the box plots. Note that the variance in the results of SVM2R and SVMRP (Figure 4(a)) is much smaller than their single stage counterparts, SVMR and RP respectively. Finally, all of the above four methods are significantly better than the BAYESIAN approach.
Table 2 also indicates that the absolute gain in performance achieved by SVM2R over the other methods is not very high (with the exception of its performance over the BAYESIAN scheme). The gains are relatively modest with SVM2R gaining 0.4% over SVMRP, 2.0% over RP and nearly 1.5% over SVMR on an average. Similarly, SVMRP achieves a gain of only about 1.5% over RP as well as about 1.2% over SVMR. However, the gains are consistent across all the ten test sets highlighting the power of SVM2R and SVMRP in capturing the interclass dependencies.
Finally, it can be observed from Table 2 and Figure 4(a) that RP performs as well as SVMR (a insignificant difference of 0.6% between the two methods). RP is a simple linear learning method that does not guarantee convergence. However, it tries to capture dependencies between the different categories by explicitly trying to rank them in the context of a given test compound. SVMR on the other hand is based on the powerful SVM methodology which employs a sophisticated Tanimoto kernel function shown to be the most effective kernel function for chemical compounds.40 However, it builds independent one-vs-rest classifiers that fail to capture dependencies among different categories and therefore does not perform better than RP.
6.2 Effect of the number of categories
We also investigated the effect of the number of categories (L) that a compound belongs to on the performance of the different methods. This type of evaluation can provide insights on the ability of these methods to identify compounds that hit more than one target and may pose problems in terms of toxicity or promiscuity.
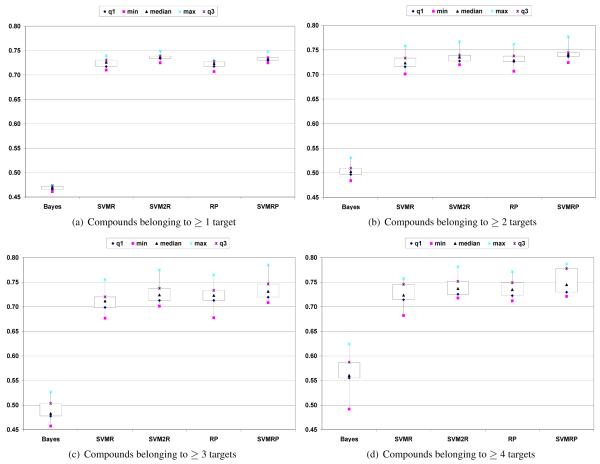
Table 3 summarizes the average performance of the five different methods over subsets of compounds that belong to L or more targets in the ten test sets utilizing the uninterpolated precision metric. The first row in this table corresponds to L ≥ 1, which is the set of all the compounds in the dataset. Therefore the average results in the first row of Table 3 are identical to the averages reported in Table 2. The subsequent rows show the performance of different methods over compounds that belong to two or more categories. We also utilize Figures 4(a)-4(d) to compare the performance across the methods for different values of L and the variability within each one of them.
Table 3.
Performance of various schemes on ECFP4 descriptor-space with compound that belong to L or more categories (targets).
| L | # of compounds | BAYESIAN | SVMR | SVM2R | RP | SVMRP | Comparisons using Figures 4(a)-4(d) |
|---|---|---|---|---|---|---|---|
| ≥ 1 | 2720 | 0.469 | 0.725 | 0.737 | 0.723 | 0.734 | BAYESIAN ≪ (RP,SVMR) ≪ (SVMRP, SVM2R) |
| ≥ 2 | 805 | 0.504 | 0.726 | 0.736 | 0.731 | 0.743 | BAYESIAN ≪ SVMR ≪ (RP, SVM2R) ≪ SVMRP |
| ≥ 3 | 269 | 0.488 | 0.713 | 0.725 | 0.724 | 0.737 | BAYESIAN ≪ (SVMR, RP) ≪ (RP, SVM2R, SVMRP) |
| ≥ 4 | 99 | 0.565 | 0.723 | 0.735 | 0.727 | 0.746 | BAYESIAN ≪ (SVMR, RP) ≪ (RP, SVM2R, SVMRP) |
The entry in each cell corresponds to the uninterpolated precision of the column method averaged over compounds belonging to L or more targets in all the ten test set. The second column (# of compounds) shows the total number of compounds that satisfy the inequality given by L in the test set over which these statistics are calculated. We do not show statistics beyond L ≥ 4 as the corresponding number of compounds in the test set is far too small to make any reliable conclusion using these results. Entries in bold represent winners for a given L. The different entries were compared using the box plots in Figures 4(a)-4(d), using the approach described in Section 5.2.4. The symbol ≪ indicates that methods on the right are relatively better than the methods on the left, and “( )” indicates that they are similar. The order of the methods within parentheses represent the order of the weak relationship comparing the average value in this table. Finally, if a method appears in multiple parentheses (such as RP), the comparison with other methods (such as SVMR,SVM2R) within the parentheses overrides other comparison.
From this table it can be observed that, in general, as the number of targets (L) that a compound belongs to increases, the methods that try to capture dependencies among the different categories (SVM2R, RP, and SVMRP) perform better than the SVMR method, which does not.
Moreover, as L increases from one to four, the edge of SVM2R over the two ranking perceptron based schemes disappears, and SVMRP achieves the best results (Table 3 and Figures 4(a)-4(d)). However, as seen from Figures 4(a) to 4(d), the variability of these methods within themselves increases considerably when L is ≥ 3 and 4. Finally, simple sorting based scheme SVMR which performs slightly better than RP in terms of absolute performance for L ≥ 1 performs worse than RP for L ≥ 2, 3, and 4.
Overall, these results indicate that the schemes that capture interclass dependencies tend to outperform schemes that do not for compounds that belong to more than one category.
6.3 Ability to retrieve all relevant categories
In this section we compare the ability of the five methods to identify relevant categories in the top-𝓀 ranks. We analyze the results along this direction because this directly corresponds to the use case scenario where a user may want to look at top-𝓀 predicted targets for a test compound and further study or analyze them for toxicity, promiscuity, off-target effects, pathway analysis etc. If all the true targets fall in the top-𝓀 ranks, there is a high likelihood of successfully recognizing them for the above analysis.
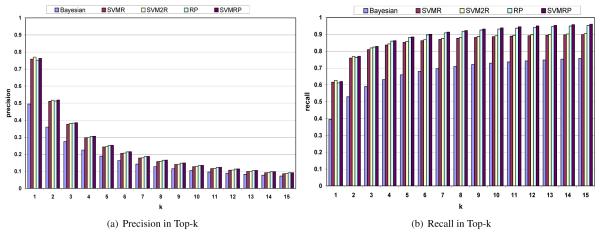
For this comparison we utilize precision and recall metric in top-𝓀 for each of the five schemes as shown in Figures 5(a) and 5(b). These figures show the actual precision and recall values in top-𝓀 by varying 𝓀 from one to fifteen. These figures are obtained by averaging precision and recall results over all the test sets used in this study. Therefore, these results are averaged over all the 27,205 compounds present in the dataset.
Figure 5.
Precision and Recall results
A number of trends can be observed from these figures. First, for identifying one of the correct categories or targets in the top 1 predictions, SVM2R outperforms all the other schemes in terms of both precision and recall. This is followed by SVMRP, RP and SVMR in that order. Second, BAYESIAN is still the worst performing method among the five compared and the performance order of these schemes is exactly the same as the performance order for the uninterpolated results in Table 2.
However, as 𝓀 increases from one to fifteen, the precision and recall results indicate that the best performing scheme is now SVMRP and it outperforms all other schemes for both precision as well as recall. This is followed by RP, which outperforms the other three schemes (BAYESIAN, SVMR, SVM2R) for both precision and recall in the top fifteen. The performance of RP is followed by SVM2R, SVMR, and finally BAYESIAN. The ranking perceptron based methods achieve an average recall of approximately 0.96 and 0.95 for SVMRP and RP respectively for 𝓀 = 15 and is better than the other schemes for the ten test sets (with average recall rates of 0.90, 0.89, and 0.76 respectively for SVM2R, SVMR, and BAYESIAN). Moreover, these values in figure 5(b) show that as 𝓀 increases from one to fifteen, ranking perceptron based schemes start performing consistently better that others in identifying all the correct categories. The two ranking perceptron based schemes also achieve average precision values that are significantly better than other schemes in the top fifteen (Figure 5(a)).
In summary, these result indicate that ranking perceptron based methods because they have a higher recall will tend to find more of the correct categories in top ranks than other schemes. Thus, these methods present a better chance of finding and analyzing targets in the context of a chemical compound.
6.4 Performance on Dissimilar Compounds
To evaluate how well the models being learned by the different methods can generalize to a set of compounds that are structurally different from those used for training, we partitioned the compounds into ten clusters and then used a leave-one-cluster-out cross validation approach44 to assess the performance of the different methods. The clustering was computed using the direct 𝓀-way clustering algorithm of CLUTO45 with cosine similarity and its default clustering criterion function. The size and various characteristics of the resulting clusters as they relate to the inter- and intra-cluster similarities are shown in Table 4.
Table 4.
Distribution of compounds in the ten clusters.
| Cluster # | Size | ISIM | ISDEV | ESIM | ESDEV |
|---|---|---|---|---|---|
| 1 | 320 | 0.571 | 0.123 | 0.046 | 0.026 |
| 2 | 1348 | 0.306 | 0.077 | 0.080 | 0.016 |
| 3 | 5061 | 0.280 | 0.066 | 0.074 | 0.018 |
| 4 | 1900 | 0.243 | 0.067 | 0.066 | 0.027 |
| 5 | 2840 | 0.224 | 0.055 | 0.063 | 0.019 |
| 6 | 4225 | 0.199 | 0.046 | 0.066 | 0.024 |
| 7 | 2070 | 0.199 | 0.043 | 0.071 | 0.019 |
| 8 | 1033 | 0.133 | 0.047 | 0.033 | 0.015 |
| 9 | 3946 | 0.139 | 0.052 | 0.073 | 0.024 |
| 10 | 4462 | 0.069 | 0.023 | 0.035 | 0.013 |
The column ‘Size’ shows the total number of compounds in each cluster. Column ISIM shows the average pairwise similarity of compounds that belong to the same cluster (within cluster similarity). Column ESIM shows the average pairwise similarity between compounds that belong to a cluster and compounds in all other clusters (external similarity). Column’s ISDEV and ESDEV show the standard deviations associated with the ISIM and ESIM values, respectively.
Table 5 shows the average uninterpolated precision that was achieved by the different methods on the ten clusters. From these results we can see that even though the absolute performance achieved by the methods developed in this paper is lower than the corresponding performance reported in Table 2, it is still considerably higher than what would have been achieved by a random predictor (which for the number of classes involved, is close to zero) and they are still much better than the performance achieved by the Bayesian approach. In fact the relative performance gains achieved by the SVM- and ranking perceptron-based methods over the Bayesian approach are higher in this experiment than in the earlier one. These results indicate that the methods that we developed are not only able to effectively predict the targets of the compounds when these compounds are drawn from the same overall distribution as those used for training (Table 2) but they can also generalized to compounds that are structurally different from those whose activity information is already known. Finally, comparing the relative performance of the different methods, the results of Table 5 show that SVM2R performs better than the rest of the schemes and that the approaches based on ranking perceptron do not perform as well as they did in the earlier experiments.
Table 5.
Performance of various schemes on ECFP4 descriptor-space
| Cluster # | BAYESIAN | SVMR | SVM2R | RP | SVMRP |
|---|---|---|---|---|---|
| 1 | 0.602 | 0.789 | 0.815 | 0.809 | 0.812 |
| 2 | 0.252 | 0.612 | 0.628 | 0.656 | 0.616 |
| 3 | 0.237 | 0.593 | 0.601 | 0.554 | 0.578 |
| 4 | 0.338 | 0.568 | 0.591 | 0.553 | 0.526 |
| 5 | 0.280 | 0.608 | 0.611 | 0.592 | 0.605 |
| 6 | 0.460 | 0.655 | 0.671 | 0.664 | 0.608 |
| 7 | 0.362 | 0.634 | 0.637 | 0.589 | 0.595 |
| 8 | 0.117 | 0.515 | 0.522 | 0.441 | 0.458 |
| 9 | 0.376 | 0.637 | 0.636 | 0.610 | 0.632 |
| 10 | 0.296 | 0.496 | 0.512 | 0.495 | 0.476 |
| Avg | 0.332 | 0.611 | 0.622 | 0.596 | 0.590 |
The entry in each cell corresponds to the uninterpolated precision of the column method for a given test set. Entries in bold represent winners for every test set.
6.5 Computational Complexity
The complexity of Ranking Perceptron based methods depend on the number of compounds as well as the number of targets. If the number of compounds is M and the number of targets is N, our ranking perceptron iterates over the dataset until the stopping criterion is satisfied (Algorithm 1). The complexity of the outer loop on line 4 in Algorithm 1 is O(M). Each compound in the worst case has to resolve O(N2) constraints in the inner loops on lines 8 and 13. Therefore, for every iteration the algorithm has a complexity of O(MN2).
BAYESIAN approach has a linear complexity with respect to the number of compounds and targets as for each feature it computes the probability of occurrence in every target using all the compounds in the dataset. This can be efficiently computed in O(M) time. Lastly, computational complexity of the SVM algorithm depends on the implementation as well as the kernel employed. For our setting, we use SV Mlight implementation49 in a one-vs-rest setting and use the Tanimoto kernel. The computational complexity is quadratic with respect to the number of compounds49 and linear with respect to the number of targets. Therefore, the final complexity of the SVM algorithm is O(M2N).
We also compare the actual run-time performance of different methods we developed and the BAYESIAN method on 64 bit AMD Opetron Machines. We report both the training and test set run-times for each method. Table 6 summarizes the run-time results for one of the 10 cross-validation folds for each of the five methods. The run-times for other cross-validation folds were found to be very similar.
Table 6.
Computational Performance of various schemes
| Method | BAYESIAN | SVMR | SVM2R | RP | SVMRP | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | |
| Total Run Time (sec) | 3 | 0.87 | 9802 | 173 | 47077 | 1123 | 7892 | 0.69 | 44098 | 175 |
The entry in cell corresponds to either the time taken for training or testing the first test set.
It can be observed from this table that two stage methods, particularly SVM2R, has the longest training time among all of the methods. This is followed by SVMRP, SVMR, RP, and BAYESIAN in the decreasing order of training times. Therefore, for training models, BAYESIAN is the fastest among all the methods. This is not surprising as BAYESIAN scheme does not perform any direct optimization. However, as seen in the preceding sections, it is also the worst performing scheme to others by a significant margin.
Looking at the time taking for the testing 2720 compounds from Table 6 it can again be observed that SVM2R has the longest run-time. This is followed by SVMR, SVMRP, BAYESIAN, and RP in that order. Thus, the fastest methods are RP and BAYESIAN as they only requires dot-product of each test compound with the weight vectors of each of 231 classes.
7 Discussion and Conclusion
SVM based methods have been extensively employed for the task of virtual screening, classification, as well as selectivity analysis of chemical compounds and their performance has been assessed for these tasks.39,50-52 For example, Wassermann and co-workers52 showed that the SVM results in better predictions for selectivity analysis than either nearest neighbor based or BAYESIAN method. Similarly, Glick and co-workers50 showed that the SVM outperformed BAYESIAN scheme when performance was measured as enrichment of actives in top 1% of the high throughput screening data. However, they also found that the performance of SVM and BAYESIAN was near identical when the noise level was significantly increased. In this work, we proposed methods based on the SVM and ranking perceptrons for the task of Target Fishing. Extensive experiments and analysis comparing our methods showed that our methods are either as good as or superior to the current state-of-the-art.
However, a number of issues still need to be addressed. First, in this work we assumed the compound with no activity information against a target to be inactive against that target. The primary reason for such assumption was almost no inactive data available for many of the targets in our dataset collected from publicly available sources. Similar assumption has been made by previous studies.13 However, this is not a very satisfactory assumption from the point of view of drug discovery as it may result in the method missing some rare compound-target activity. Therefore, in our future work, we will try to address the issue of unknown compound-target activity explicitly in order to come up with practical methods for drug discovery.
Second, in this work we have not utilized target category information (for example target sequence, structure, family) in building these ranking perceptron or SVM based models. Identification of key characteristics common across two target themselves (similar geometry of binding sites or similar biochemical characteristics of binding residues) might identify two targets that will likely bind to the same compound. Therefore, effective solutions can be devised using both SAR data and target information. A number of recent approaches for chemogenomics utilize SAR data as well as target information to build predictive models on the target-ligand graph.53,54 Our initial studies in trying to include target information did not yield promising results for the problem of target fishing. They were found to be no better as compared to only SAR data based approaches proposed in this paper. So we did not pursue approaches that include target information in this work. However, we believe that exploring a good way of including target information is a worthwhile effort and plan to investigate it rigorously as a part of our future work.
Finally, recent approaches have shown that the interclass dependencies could be learned within structural learning framework that utilizes structural SVM.20,28,55 We also experimented with this approach in our work using the SVM-struct algorithm.55 However, our preliminary results showed that this approach did not yield any significant gains over the ranking perceptron based approach over the large number of categories in our domain. Moreover, the computational cost of employing structural learning was much higher than that of utilizing ranking perceptrons. Therefore we did not pursue structural SVM based approach in the present work.
Acknowledgements
This work was supported by NSF ACI-0133464, IIS-0431135, NIH RLM008713A, and by the Digital Technology Center at the University of Minnesota.
Footnotes
References
- 1.Eglen RM, Schneider G, Bohm HJ. High Throughput Screening and Virtual Screening: Entry Points to Drug Discovery. In: Bohm HJ, Schneider G, editors. Virtual Screening for Bioactive Molecules. Vol 10. Wiley-VCH; Weinheim Germany: 2000. pp. 1–14. [Google Scholar]
- 2.Terstappen GC, Schlopen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discovery. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 3.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discovery Today. 2005;10:139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins JL, Bender A, Davies JW. In silico target fishing: Predicting biological targets from chemical structure. Drug Discovery Today: Technologies. 2006;3:413–421. [Google Scholar]
- 5.Azzaoui K, Hamon J, Faller B, Whitebread S, Jacoby E, Bender A, Jenkins JL, Urban L. Modeling promiscuity based on in vitro safety pharmacology profiling data. ChemMedChem. 2007;2:874–880. doi: 10.1002/cmdc.200700036. [DOI] [PubMed] [Google Scholar]
- 6.Chen YZ, Ung CY. Prediction of potential toxicity and side effect protein targets of a small molecule by a ligand-protein inverse docking approach. J. Mol. Graphics Modell. 2001;20:199–218. doi: 10.1016/s1093-3263(01)00109-7. [DOI] [PubMed] [Google Scholar]
- 7.Eggert US, Mitchison TJ. Small molecule screening by imaging. Curr. Opin. Chem. Biol. 2006;10:232–237. doi: 10.1016/j.cbpa.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Hart CP. Finding the target after screening the phenotype. Drug Discovery Today. 2005;10:513–519. doi: 10.1016/S1359-6446(05)03415-X. [DOI] [PubMed] [Google Scholar]
- 9.Vapnik V. Statistical learning theory. John Wiley; New York U.S.A: 1998. Support Vector Methods for Estimating Indicator Function. [Google Scholar]
- 10.Crammer K, Singer Y. A new family of online algorithms for category ranking. Journal of Machine Learning Research. 2003;3:1025–1058. [Google Scholar]
- 11.Chen YZ, Zhi DG. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins. 2001;43:217–226. doi: 10.1002/1097-0134(20010501)43:2<217::aid-prot1032>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.Nettles JH, Jenkins JL, Bender A, Deng Z, Davies JW, Glick M. Bridging chemical and biological space: “target fishing” using 2d and 3d molecular descriptors. J. Med. Chem. 2006;49:6802–6810. doi: 10.1021/jm060902w. [DOI] [PubMed] [Google Scholar]
- 13.Nidhi, Glick M, Davies JW, Jenkins JL. Prediction of biological targets for compounds using multiple-category bayesian models trained on chemogenomics databases. J. Chem. Inf. Model. 2006;46:1124–1133. doi: 10.1021/ci060003g. [DOI] [PubMed] [Google Scholar]
- 14.Niwa T. Prediction of biological targets using probabilistic neural networks and atom-type descriptors. J Med Chem. 2004;47:2645–2650. doi: 10.1021/jm0302795. [DOI] [PubMed] [Google Scholar]
- 15.Kawai K, Fujishima S, Takahashi Y. Predictive activity profiling of drugs by topological-fragment-spectra-based support vector machines. J. Chem. Inf. Model. 2008;48:1152–1160. doi: 10.1021/ci7004753. [DOI] [PubMed] [Google Scholar]
- 16.RCSB Protein Data Bank [accessed January 28, 2008]; http://www.pdb.org/
- 17.Hansch C, Fujitai T, Maolney PP, Muir RM. Correlation of biological activity of phenoxyacetic acids with hammett substituent constants and partition coefficients. Nature. 1962;194:178–180. [Google Scholar]
- 18.Bravi G, Gancia E, Green D, Hann VS, Mike M. Modelling structure-activity relationship. In: Bohm HJ, Schneider G, editors. Virtual Screening for Bioactive Molecules. Vol 10. Wiley-VCH; Weinheim Germany: 2000. pp. 81–113. [Google Scholar]
- 19.Godbole S, Sarawagi S. Discriminative methods for multi-labeled classification. PAKDD., Proceedings of the 8th Pacific Asia Conference in Knowledge Discovery and Data Mining; Sydney. May 2004.pp. 22–30. [Google Scholar]
- 20.Ie E, Weston J, Noble WS, Leslie C. Multi-class protein fold recognition using adaptive codes. ICML, Proceedings of the 22st Annual International Conference on Machine Learning; Bonn Germeny. August 2005.pp. 329–336. [Google Scholar]
- 21.Barnard JM, Downs GM, Willett P. Descriptor-Based Similarity Measures for Screening Chemical Databases. In: Bohm HJ, Schneider G, editors. Virtual Screening for Bioactive Molecules. Vol 10. Wiley-VCH; Weinheim Germany: 2000. pp. 59–79. [Google Scholar]
- 22.Platt JC. Fast training of Support Vector Machines Using Sequential Minimal Optimization. In: Scholkopf B, Burges C, Smola A, editors. Advances in Kernel Methods: Support Vector Learning. MIT-Press; Boston USA: 1998. pp. 185–208. [Google Scholar]
- 23.Jones DT. Protein secondary structure prediction based on position-specific scoring matricies. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 24.Karypis G. Yasspp: Better kernels and coding schemes lead to improvements in svm-based secondary structure prediction. Proteins: Structure, Function and Bioinformatics. 2006;64:575–586. doi: 10.1002/prot.21036. [DOI] [PubMed] [Google Scholar]
- 25.Lanckriet GRG, Cristianini N, Bartlett P, Ghaoui LE, Jordan MI. Learning the kernel matrix with semi-definite programming. ICML, Proceedings of the 19th Annual International Conference on Machine Learning; Sydney Australia. July 2002.pp. 323–330. [Google Scholar]
- 26.Basilico J, Hofmann T. Unifying collaborative and content-based filtering. ICML, Proceedings of the 21st Annual International Conference on Machine Learning; Banff. Alberta Canada. July 2004.pp. 9–17. [Google Scholar]
- 27.Rosenblatt F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychological Review. 1958;65:386–407. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 28.Rangwala H, Karypis G. Building multiclass classifiers for remote homology detection and fold recognition. BMC Bioinformatics. 2006;7:455–471. doi: 10.1186/1471-2105-7-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The PubChem Project [accessed November 15, 2007]; http://pubchem.ncbi.nlm.nih.gov
- 30.Daylight Toolkit. version 4.73 Daylight Chemical Information System; Aliso Viejo, CA: 2007. [Google Scholar]
- 31.Liu T, Lin Y, Wen X, Jorrisen R, Gilson MK. Bindingdb: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res (Database Issue) 2007;35:D198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. Drug-Bank: a comprehensive resource for in silico drug discovery and exploration. Nucl. Acids Res. 2006;34:668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth BL, Kroeze WK, Patel S, Lopez E. The multiplicity of serotonin receptors: Uselessly diverse molecules or an embarrasment of riches? The Neuro-scientist. 2000;6:252–262. [Google Scholar]
- 34. [accessed Aug 20, 2007];BRITE Database. http://www.genome.jp/kegg/brite.html
- 35.Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, Oprea T. Wombat: World of molecular bioactivity. In: Oprea TI, editor. Chemoinformatics in Drug Discovery. Wiley-VCH; New York: 2004. pp. 223–239. [Google Scholar]
- 36.Integrated Scientific Information System. version 3.1 MDL Information Systems; San Ramon, CA: 2007. [Google Scholar]
- 37.Hert J, Willett P, Wilton D, Acklin P, Azzaoui K, Jacoby E, Schuffenhauer A. Comparison of topological descriptors for similarity-based virtual screening using multiple bioactive reference structures. Org. Biomol. Chem. 2004;2:3256–3266. doi: 10.1039/B409865J. [DOI] [PubMed] [Google Scholar]
- 38.Rogers D, Brown R, Hahn M. Using extended-connectivity fingerprints with laplacian-modified bayesian analysis in high-throughput screening. J. Biomol. Screen. 2005;10:682–686. doi: 10.1177/1087057105281365. [DOI] [PubMed] [Google Scholar]
- 39.Wale N, Watson IA, Karypis G. Comparison of descriptor spaces for chemical compound retrieval and classification. Journal of Knowledge and Information Systems. 2008;14:347–375. [Google Scholar]
- 40.Willett P. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 41.Wale N, Watson IA, Karypis G. Indirect similarity based methods for effective scaffold-hopping in chemical compounds. J. Chem. Inf. Model. 2008;48:730–741. doi: 10.1021/ci700369e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pipeline Pilot. version 6.1 Accelrys Inc; San Diego CA: 2008. [Google Scholar]
- 43.Whittle M, Gillet VJ, Willett P. Enhancing the effectiveness of virtual screening by fusing nearest neighbor list: A comparison of similarity coefficients. J. Chem. Inf. Comput. Sci. 2004;44:1840–1848. doi: 10.1021/ci049867x. [DOI] [PubMed] [Google Scholar]
- 44.Saeh JC, Lyne PD, Takasaki BK, Cosgrove DA. Lead Hopping Using SVM and 3D Pharmacophore Fingerprints. J. Chem. Inf. Model. 2005;45:1122–1133. doi: 10.1021/ci049732r. [DOI] [PubMed] [Google Scholar]
- 45. [accessed April 25, 2009];Cluto: Clustering Toolkit. (version 2.1.2). http://glaros.dtc.umn.edu/gkhome/views/cluto
- 46.Hearst M, Pedersen J. Reexamining the cluster hypothesis: Scatter/gather on retrieval results. ACM SIGIR; Proceedings of the 19th Annual International Conference; Zurich. June 1996.pp. 76–84. [Google Scholar]
- 47.Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. 1st ed Addison Wesley; New York U.S.A: 1999. Modeling. [Google Scholar]
- 48.Bland JM. An introduction to medical statistics. 3rd edn Oxford University Press; New York U.S.A: 2000. Significance tests. [Google Scholar]
- 49.Joachims T. Making large-scale svm learning practical. In: Scholkopf B, Burges C, Smola A, editors. Advances in Kernel Methods: Support Vector Learning. MIT-Press; Boston USA: 1998. pp. 169–184. [Google Scholar]
- 50.Glick M, Jenkins JL, Nettles JH, Hitchings H, Davies JW. Enrichment of High-Throughput Screening Data with Increasing Levels of Noise Using Support Vector Machines, Recursive Partitioning, and Laplacian-Modified Naive Bayesian Classifiers. J. Chem. Inf. Model. 2006;46:193–200. doi: 10.1021/ci050374h. [DOI] [PubMed] [Google Scholar]
- 51.Geppert H, Horvath T, Gartner T, Wrobel S, Bajorath J. Support-vector-machine-based ranking significantly improves the effectiveness of similarity searching using 2D fingerprints and multiple reference compounds. J. Chem. Inf. Model. 2008;48:742–746. doi: 10.1021/ci700461s. [DOI] [PubMed] [Google Scholar]
- 52.Wassermann AM, Geppert H, Bajorath J. Searching for target-selective compounds using different combinations of multiclass support vector machine ranking methods, kernel functions, and fingerprint descriptors. J. Chem. Inf. Model. 2009;49:582–592. doi: 10.1021/ci800441c. [DOI] [PubMed] [Google Scholar]
- 53.Park K, Kim D. Binding network similarity of ligand. Proteins. 2008;71:960–971. doi: 10.1002/prot.21780. [DOI] [PubMed] [Google Scholar]
- 54.Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M. Prediction of drug-target interaction networks from integration of chemical and genomics spaces. Bioinformatics. 2008;24:232–240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsochantaridis I, Hofmann T, Joachims T, Altun Y. Support vector machine learning for interdependent and structured output spaces. ICML, Proceedings of the 21st Annual International Conference on Machine Learning; Banff. Alberta Canada. July 2004.pp. 9–17. [Google Scholar]