Summary
The identification of specific biomarkers will improve the early diagnosis of disease, facilitate the development of targeted therapies, and provide an accurate method to monitor treatment response. A major challenge in the process of verifying biomarker candidates in blood plasma is the complexity and high dynamic range of proteins. This article reviews the current, targeted proteomic strategies that are capable of quantifying biomarker candidates at concentration ranges where biomarkers are expected in plasma (i.e. at the ng/ml level). In addition, a workflow is presented that allows the fast and definitive generation of targeted mass spectrometry-based assays for most biomarker candidate proteins. These assays are stored in publicly accessible databases and have the potential to greatly impact the throughput of biomarker verification studies.
Introduction
A biomarker is a measureable indicator that correlates to a specific biological or disease state. Biomarkers play an important role in various clinical applications [1]. Besides screening for an early diagnosis, biomarkers are measured for the classification and staging of diseases in order to assign patients for targeted treatments, to monitor treatment response and to detect disease recurrence. The process from the discovery of a biomarker to its clinical application can be subdivided into different phases [2,3]. The initial phase, typically referred to as discovery, aims to produce a list of biomarker candidates through various genomic, transcriptomic and proteomic technologies. In the following phase, referred to as verification, the correlation of these candidates to the disease is verified over a large cohort of samples. The candidate markers that perform well through the verification are then selected for the clinical validation phase.
Blood plasma is of particular interest as a source for biomarkers, since it is easily accessible and presumably contains quantifiable molecules that provide information characterizing the physiologic and pathologic state of the human body in the form of proteins shed or secreted from the tissue where a pathologic state is present [4,5]. The major difficulty in finding blood-based biomarkers is the complexity and the dynamic range of protein concentrations in human plasma [6,7]. Tissue-derived proteins, the targets for biomarkers, are found in plasma in the ng/ml concentration range, six orders of magnitude below the classical plasma proteins [6,8]. In order to identify specific, disease-related, changes in the proteome and circumvent the challenges of plasma proteomics, recent biomarker studies have focused on the analysis of tissue or cell lines for the generation of biomarker candidate lists [8–11]. This usually leads to a list from hundreds to thousands of candidate proteins, which subsequently need to be verified in human plasma samples. However, despite the large investment and the effort to generate lists of candidates, only a few protein biomarkers are currently used routinely in the clinical setting. In recent years, the rate of newly-approved diagnostic markers has been steadily decreasing due in part to the demanding technical requirements for the verification of the candidate proteins in plasma samples [11,12]. The technology must be sensitive to allow for the quantification of proteins in the ng/ml concentration range in a highly complex background, and all candidates must be quantified with high reproducibility, accuracy, and in a high throughput manner over large numbers of patient samples.
Currently, the most commonly used approach for verification and clinical validation is the sandwich enzyme-linked immunosorbent assay (ELISA). The advantages of ELISA assays are their high specificity by implementing a pair of antibodies against the candidate protein and their high sensitivity, permitting the quantification of proteins in human plasma at concentrations below the ng/ml range. However, the limiting factors for the ELISA as a technique for serum biomarker verification are the restricted possibility to multiplex assays and the availability of antibodies for novel candidate proteins, combined with the lengthy and expensive development of new assays. Therefore, development of an alternate method for protein quantification with high reproducibility and throughput is needed in order to improve the success rate of approved biomarkers [13]. One solution is a targeted quantitative proteomic concept, such as selected reaction monitoring (SRM) (also referred to as multiple reaction monitoring (MRM)) [14]. This review focuses on the recent advances in targeted mass spectrometric approaches and their impact on biomarker studies.
Selected reaction monitoring mass spectrometry for plasma biomarker verification
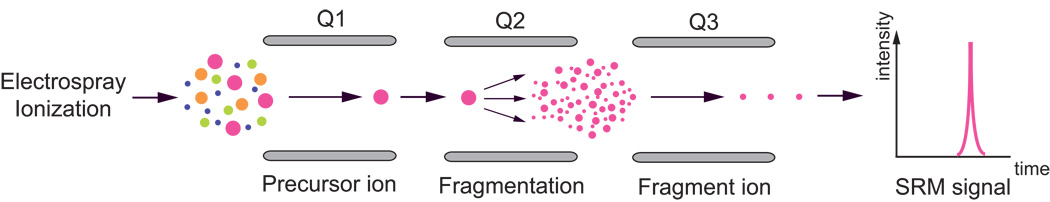
SRM is a mass spectrometric technique for the targeted detection and quantification of selected proteotypic peptides (PTPs) with known fragmentation properties in a complex sample matrix [15,16] (Figure 1). The purpose of PTPs is to serve as surrogates for the candidate protein. They have on the one hand a unique amino acid sequence for the candidate protein and are also easily detectable by MS. To date, SRM is a technology that has been shown to fulfill the requirements that are needed for the verification of biomarker candidates in blood plasma. The high reproducibility of SRM measurements was demonstrated by the Clinical Proteomic Technology Assessment for Cancer Network project (CPTAC) [17]. In this study the precision and reproducibility of SRM-based measurements of proteins spiked in a background of human plasma were assessed over 9 different laboratories with a result of 10–23% interlaboratory coefficient of variance (CV) which includes variations in sample preparation and MS platforms. In addition, recent studies demonstrated the unique capability of SRM to quantify specific sets of proteins consistently and simultaneously over many complex samples [18,19]. Lastly, the major advantage of the SRM assay in comparison to ELISA is the significantly shorter lead-time and reduced costs. The development of a single ELISA assay test usually takes over one year. Meanwhile, the development and optimization of a SRM assay can be performed within weeks and permits multiplexing of assays for several candidates simultaneously. These characteristics allow the verification of most proteins from the biomarker candidate list without previous prioritizing of candidates. Therefore, SRM promises to be a powerful tool for biomarker verification and has the potential to serve as a complementary method to ELISA [3]. One of the major challenges using SRM for candidate biomarker verification in human plasma is the required sensitivity for the quantification of low abundance proteins within a dynamic concentration range of 12 orders of magnitude [6].
Figure 1. Selected Reaction Monitoring.
The figure shows a schematic illustration of the principle of a triple quadrupole mass spectrometer (QQQ). In the first quadrupole (Q1), a specific precursor ion of a PTP is selected based on its mass-to-charge (m/z) ratio. The precursor ion is fragmented in the second quadrupole (Q2) by collision-induced dissociation, which allows for the selection of a specific fragment of the target peptide ion, according to its m/z ratio, in the third quadrupole (Q3). The signal intensity of this fragment is reported over time. The pair of m/z ratios for the precursor and fragment ions is a so-called SRM transition. A series of the best SRM transitions for the target peptide in combination with its retention time and instrument parameters, serve as a fingerprint for a PTP and constitute a definitive SRM assay.
The application of SRM for direct quantification of trypsin-digested plasma proteins without further sample preparation is preferable for many reasons. It is a simple process that is less prone to errors, efficient, reproducible and permits a high sample throughput. In a recent study, Kuzyk et al. reported the development of multiplexed SRM assays for the simultaneous quantification of 45 plasma proteins and tested their accuracy and reproducibility [20]. Measuring separately prepared digests of standard plasma on different days, they achieved a CV below 20% for 94% of the measured analytes. While this demonstrates high reproducibility in the sample preparation as well as in the SRM technique, one limitation of this approach is the lack of sensitivity by applying SRM directly to plasma. For example, the least abundant protein quantified, L-Selectin, had a concentration of 1 ug/ml [21]. While recent technical advances and increasing experience for SRM measurements improved the limit of quantification (LOQ) to 0.3 ug/ml in human plasma [17], this remains 1 to 2 orders of magnitude too high for reliable plasma biomarker measurements.
Therefore, different strategies have been developed with the goal to improve the sensitivity of SRM applied to plasma and to maintain the requirements of (i) high reproducibility over large cohorts of human plasma samples and (ii) high-throughput, permitting the analysis of multiple samples and multiplexed analytes (Table 1).
Table 1.
Comparison of different strategies for the verification of protein biomarkers in plasma
| Assay | Assay development | LOQ | CV | Assay Throughput | Reference |
|---|---|---|---|---|---|
| SRM in plasma | 1. Selection of PTPs 2. SRM assay optimization |
1 ug/ml | 4.6 – 19.7% | +++ | Kuzyk et al. [20] |
| SRM in depleted plasma |
1. Selection of PTPs 2. SRM assay optimization |
25 ng/ml | 8.5 – 19.1% | +++ | Keshishian et al. [22] |
| SRM in depleted and SCX fractionated plasma |
1. Selection of PTPs 2. SRM assay optimization |
2.5 ng/ml | 2.6 – 14.2% | ++ | Keshishian et al. [22] |
| SRM combined with protein antibody enrichment |
1. Generation of the antibody 2. Selection of PTPs 3. SRM assay optimization |
2 ng/ml | <1 – 26% | + | Nicol et al. [26] |
| SRM combined with SISCAPA enrichment |
1. Selection of PTPs 2. Generation of antipeptide antibody 3. SRM assay optimization 4. Test of interference multiplexing assays |
1–10 ng/ml | 9.4 – 20.5% | + | Kuhn et al. [30] |
| SRM combined with isolation of N- glycosites |
1. Selection of PTPs 2. SRM assay optimization |
5 ng/ml | 7 – 27% | +++ | Stahl-Zeng et al. [31] |
| ELISA | Generation of a pair of highly specific monoclonal antibodies |
low pg/ml | <10% | + | Helle et al. [56] Ida et al. [57] |
Sample fractionation for improved sensitivity
Keshishian et al. combined depletion of the 12 highest abundance plasma proteins with minimal fractionation by strong-cation-exchange chromatography (SCX) [22]. The LOQ was improved to 25 ng/ml applying SRM to the depleted plasma. The combination of depletion and SCX fractionation into 6 fractions resulted in further improvement with a reproducible quantification of proteins down to 1–10 ng/ml. The strategy was recently applied for the quantification of cardiovascular biomarkers in human patient plasma [23]. The drawback of this approach is the introduction of additional sample preparation steps, which can introduce variation in the measurement. In addition, this limits the sample throughput since one sample is fractionated into several subsequent fractions that all have to be measured individually. Although Off-Gel™ electrophoresis, a technique based on isoelectric focusing, has been used to fractionate human plasma prior to MS analysis [24,25], to date, it has not been exploited in combination with SRM.
Peptide and protein enrichment using antibodies
In order to reduce the complexity of plasma samples without compromising the capability of high-throughput analysis, protein and peptide enrichment strategies have been combined with the SRM technique.
Nicol et al. introduced an immunoaffinity-SRM approach which implements antibodies to enrich multiple proteins from human sera simultaneously [26]. Using specific antibodies immobilized on hydrazide beads in combination with subsequent SRM analysis, they quantified proteins reproducibly in the low ng/ml range. The comparison of the protein concentrations determined by SRM with results obtained with an ELISA yielded a high correlation of both technologies.
Another immunoaffinity approach was developed using antibodies targeted against tryptic peptides identified as being selective for the protein of interest [27]. Polyclonal rabbit antibodies are immobilized on affinity columns to capture and subsequently elute the target peptide along with its corresponding stable isotope-labeled standard. This method has been termed Stable Isotope Standards for the use with Capture by Anti-Peptide Antibodies (SISCAPA). For automation of the enrichment process, the SISCAPA technology was implemented in a magnetic-bead based platform [28]. Whiteaker et al. demonstrated the potential of SISCAPA combined with SRM for the verification of tissue derived biomarker candidates in plasma without further depletion or fractionation steps [29]. Using the anti-peptide antibody against fibulin-2, they obtained linear quantification over a concentration range of 50 – 8000 ng/ml. However, the analysis of one single peptide per sample preparation is not favorable for the verification of hundreds of biomarker candidates with a high sample throughput. Therefore, Kuhn et al. tested the ability to use a mixture of magnetic beads containing two different anti-peptide antibodies and compared these results with the individual antibody enrichment [30]. Both approaches revealed similar results with a limit of quantification (LOQ) in the low ng/ml range and indicated only minimal deterioration in assay performance using more than one antibody in a single sample preparation. These studies demonstrated the ability of SISCAPA-SRM to achieve high-throughput sample processing due to an automated magnetic bead-based approach and the potential to multiplex the number of peptides measured in one analysis. A limiting factor is that for each peptide an antibody has to be generated, which increases the lead-time and the costs for developing the SRM assay. Additionally, each combination of anti-peptide antibodies multiplexed in a single enrichment has to be tested for interference from cross reactivity of the antibodies. In the end, the sensitivity of the immunoaffinity enrichment methods critically depends on the specificity of the antibody. Therefore, the use of high affinity monoclonal antibodies promises to reach LOQ in the concentration range below ng/ml.
Isolation of a subproteome
Another strategy, which has been applied in combination with SRM for the quantification of low abundance proteins in plasma, focuses on the N-glycosite subproteome [31,32]. This approach is based on the fact that most proteins present at the cell surface or secreted from tissue are glycosylated and are therefore likely to be detected in plasma. In order to isolate the peptides carrying N-glycosylation sites in the intact protein, referred to as N-glycosites, the carbohydrate moieties are oxidized to form aldehydes. Subsequently, these aldehydes are covalently bound to a hydrazide-containing support and then the peptides are specifically released from the solid support in their de-glycosylated form, using peptide N-glycosidase F (PNGaseF) [33–35]. The application of the solid-phase extraction of glycopeptides (SPEG) in combination with SRM was shown to quantify glycoproteins in the 5 – 100 ng/ml concentration range [14,31]. This strategy allows for high-throughput analysis, since the isolation process of N-glycosites can be implemented on a robotic system and, more importantly, since it permits the quantification of multiple analytes in one LC-SRM-MS measurement. A possible complication of this approach is that formerly N-glycosylated peptides of the candidate protein are not appropriate for MS analysis and that the number of peptides per protein is usually low. Additionally, a measured concentration change of the formerly N-glycosylated peptide can arise either from a change in the degree of glycosylation or from a change in the protein concentration itself, which cannot be distinguished without concurrent measurement of the protein abundance. Moreover, some biomarker candidate proteins are not N-glycosylated and are therefore not detectable employing this approach [8]. Additional approaches focusing on sub-proteomes have been reported, for example the enrichment of cysteine-containing peptides [36,37] or the isolation of the serum peptidome [38,39], but they have not been explored yet in combination with the SRM technology.
Generation of SRM assays and their public accessibility
One trait all sample preparation strategies have in common is the development and validation of SRM assays before they can be applied for protein quantification to plasma. Three pieces of information are important for the generation of SRM assays [14]: (i) the target proteins should be selected, which in the case of a biomarker study are usually given by the generated candidate list (ii) for each target protein the PTPs have to be identified and (iii) for each PTP the best transitions and their optimal instrument parameters need to be determined. Lange et al. reviewed comprehensively the selection criteria for PTPs and their transitions as well as provided important considerations for the development of a SRM assay [14]. The information for PTPs is preferentially derived from empirical data of shotgun MS experiments, i.e. from the generation of the candidate list or central repositories of LC-MS data like PeptideAtlas [40–43]. For proteins that have not been detected by MS, computational tools are available that attempt to predict the most likely MS-observable peptides [15,44,45]. After selection of the PTP optimal SRM transitions for this peptide have to be determined, which consist of the most prominent precursor ion charge state and the respective fragment ions that provide the highest and most conclusive fragment ion signals. The information of the most intense fragment ions is usually derived from acquired MS/MS spectra of the PTPs (ideally from fragment ion spectra acquired in a triple quadrupole mass spectrometer (QQQ)). After selection of the SRM transitions, it is necessary to validate that these transitions selectively monitor the target peptide. This task is difficult to perform in the complex background of plasma, especially for low abundance peptides for which MS/MS spectra are mostly of poor quality and cannot be confidentially assigned to a peptide sequence. Moreover, this strategy does not allow the generation of assays for peptides that have not yet been detected by MS.
Therefore, a new strategy was introduced by Picotti et al. to overcome the limitation in the SRM assay generation process (Picotti et al., submitted). It is based on libraries of low-cost, unpurified, synthetic peptides, (e.g. generated by SPOT-synthesis technology [46]), for the best predicted PTPs selected for the protein candidates [15] (Figure 2). The strategy has many advantages, such as the determination of the optimal transitions is fast and can be performed in a multiplexed manner. Therefore, synthetic peptides are pooled before they are analyzed by means of SRM-triggered MS/MS on a QQQ [47]. The most favorable SRM transitions, including their relative signal intensities, are directly extracted from the acquired QQQ MS/MS spectra (or ideally from consensus MS/MS spectra, when multiple spectra are acquired). This allows for the generation of more than 100 assays in a single, one-hour analysis (Picotti et al., submitted). The generated assays are automatically validated by searching the acquired MS/MS spectra against an appropriate protein database. The synthetic peptide library also facilitates further improvements in SRM assay sensitivity by providing transition-specific LC-MS parameters. The optimization of declustering potential and collision energy can result in a measurable intensity gain of the transition of up to 3-fold [14]. Retention time information extracted from the analysis of the synthetic peptides allows performing time-constrained SRM measurements to monitor a higher number of SRM transitions in one single analysis (more than 1000 SRM transitions per hour) [14]. In addition, the fragment ion relative intensities for each PTP, extracted by this approach on a QQQ, can be compared to the transition relative intensities during the SRM measurement of the PTP in a real sample, and serve as a strong validation criterion for the detection of the endogenous peptides.
Figure 2. Impact of SRM in the verification of biomarker candidates.
SRM assays for the candidate proteins are generated using synthetic peptide libraries and stored in a publicly accessible database. The optimized SRM assays are then used to determine the detectability of the candidate proteins in a subset of plasma samples. Isotopically labeled standard peptides for the detectable candidates allow accurate quantification of the target proteins over large cohorts of plasma samples. Scheduled SRM measurements based on the elution time of the peptide permit multiplexing of hundreds of candidates in a single analysis.
SRM assays in publicly available databases
In comparison to ELISA, once the SRM assay is developed, it becomes universally applicable and other researchers can use it for protein quantification. The final SRM transitions and their optimal instrument parameters that constitute a definitive assay for the detection of a targeted peptide can therefore be stored in a centralized, web-accessible database that supports their organization and dissemination [48]. Such a resource of optimized SRM assays would expedite the biomarker verification process, since it offers the scientific community the possibility of retrieving targeted proteomic assays that can be directly applied to the high-throughput verification of biomarker candidates in their own samples of interest.
Of the numerous sample preparation strategies currently applied to biomarker studies, the most likely public resource of SRM assays to emerge is the one for the N-glycosite subproteome, since it is comprised of a limited number of proteins. During the construction of the N-glycosite SRM atlas, unpurified peptides were synthesized for all identified N-glycopeptides from human tissue and plasma.
Furthermore, predicted N-glycosylated peptides were selected based on the presence of the required N×S/T sequence motif (N corresponding to asparagine, × is any amino acid except for proline and S/T corresponds to serine or threonine). This synthetic N-glycosite peptide library is used to develop SRM assays, which are stored in a web-accessible database. In order to use this resource for the biomarker studies focusing on the N-glycosite subproteome, only LC-MS parameters need to be re-optimized for the SRM transitions in order to ensure the highest sensitivity on each instrument platform. The benefit of a public resource, like the N-glycosite SRM atlas, is supported by the study of the CPTAC project. It demonstrated high reproducibility and compatibility using the same transitions in each laboratory even by measuring on different instrument platforms [17].
Until now, the different existing public repositories have made an impact in biomarker studies by speeding up the pipeline. Antibody libraries like the Human Protein Atlas are considered for the selection of candidate proteins, thus, having a positive contribution to the generation of useful candidate lists [49,50]. Public repositories of MS data support the selection of proteotypic peptides, since they contain the expressed and MS-detectable proteome [40–43]. A SRM public database is important for the later steps of the biomarker development process (Figure 2). After generating a biomarker candidate list, the subsequent step is the verification of the candidates by quantification over large cohorts of plasma samples. So far, the most accurate quantification using SRM is achieved by the addition of isotope-labeled reference peptides to the plasma samples at a known concentration and by simultaneously monitoring the transitions for the endogenous and the reference peptides [51,52]. However, the isotope-labeled standard peptides are very expensive ($300–700 per peptide) and prohibit large-scale biomarker studies. The SRM atlas offers the unique possibility to screen for all candidate proteins and to determine those that are detectable in plasma. In order to perform this, SRM assays for each candidate are extracted from the atlas. Next, the scheduled SRM measurements allow for the screening of all candidates in a subset of target plasma samples (ideally samples representing all biological states of the study) in a high-throughput manner in order to determine the detectable candidate proteins. Consequently, isotope-labeled standard peptides can be synthesized for the detectable proteins to subsequently allow for their accurate quantification in a large cohort of plasma samples. This approach is cost effective and helps focus on promising candidates. In the case of N-glycoproteome, it would be conceivable that the N-glycosite SRM atlas replaces the lengthy and expensive generation of previous candidate lists since the number of N-glycosylated proteins is limited. The SRM coordinates for all N-glycosylated proteins could be tested in a subset of the target plasma samples. The detectable candidates would then be selected for quantification using isotope-labeled standard peptides in a large cohort of plasma samples.
Future challenges
Using SRM assays generated by synthetic peptide libraries to determine the detectable range of target proteins in a complex mixture with a high level of sensitivity and selectivity has been demonstrated for a yeast digest [19] and needs to be proven for human plasma samples. However, the high complexity of plasma often leads to the co-elution of unspecific signals with some of the transitions [53,54]. In order to overcome this obstacle and avoid the need to manually inspect all measured transitions, a verification tool is currently being developed that evaluates the correct assignment of the detected transitions to the target peptide and determines probabilities for false positive detection (Reiter L. and Rinner O. et al., in preparation).
Conclusion
Recent advances in the SRM technology show the potential to bridge the gap between the generation of candidate lists and their verification in plasma [55]. The strength of this technology is the ability to develop sensitive and selective assays for proteins in a more cost-effective and time-saving manner compared to standard ELISA assays. Moreover, it has the unique feature to quantify multiple proteins in one analysis with high reproducibility, two factors that play an important role in the verification of hundreds of biomarker candidates. In summary, SRM is a new step to address the challenges in accurately decoding the information residing in the complexity and the dynamic range of blood plasma, which still requires sample preprocessing in order to achieve the sensitivity needed to monitor low-abundance biomarker candidates.
Acknowledgements
We gratefully acknowledge funding from the National Heart, Lung, and Blood Institute of the NIH under contract No. N01-HV-28179 as well as from the Swiss National Science Foundation Grant 3100A0-107679. PP is the recipient of a Marie Curie intra-european fellowship. Dr. Alexander Schmidt from the Institute of Molecular Systems Biology (ETH Zurich) is acknowledged for critical reading of this article.
Abbreviations
- CV
Coefficient of variation
- ELISA
Enzyme-linked immunosorbent assay
- LC
Liquid chromatography
- LOQ
Limit of quantification
- MRM
Multiple reaction monitoring
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- PNGaseF
Peptide N-glycosidase F
- PTP
Proteotypic peptide
- QQQ
Triple quadrupole mass spectrometer
- SCX
Strong cation exchange
- SISCAPA
Stable isotope standards using capture by anti-peptide antibodies
- SPEG
Solid phase extraction of formerly glycosylated peptides
- SRM
Selected reaction monitoring
Contributor Information
Ruth Hüttenhain, Email: huettenhain@imsb.biol.ethz.ch.
Johan Malmström, Email: malmstroem@imsb.biol.ethz.ch.
Paola Picotti, Email: picotti@imsb.biol.ethz.ch.
Ruedi Aebersold, Email: aebersold@imsb.biol.ethz.ch.
References
- 1.Ludwig J, Weinstein J. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 2.Rifai N, Gillette M, Carr S. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 3.Morrison R, Veenstra T, Paulovich A, Whiteaker J, Hoofnagle A, Wang P. The interface between biomarker discovery and clinical validation: The tar pit of the protein biomarker pipeline. Prot. Clin. Appl. 2008;2:1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Liu A, Loriaux P, Wollscheid B, Zhou Y, Watts J, Aebersold R. Mass Spectrometric Detection of Tissue Proteins in Plasma. Molecular & Cellular Proteomics. 2006;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J Clin Invest. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Anderson N. The Human Plasma Proteome: A Nonredundant List Developed by Combination of Four Separate Sources. Molecular & Cellular Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Molecular Oncology. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faça V, Ventura A, Fitzgibbon M, Pereira-Faça S, Pitteri S, Green A, Ireton R, Zhang Q, Wang H, O'briant K, et al. Proteomic Analysis of Ovarian Cancer Cells Reveals Dynamic Processes of Protein Secretion and Shedding of Extra-Cellular Domains. PLoS ONE. 2008;3:e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faca V, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri S, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res. 2005;4:1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- 12.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 13.Anderson N. The Roles of Multiple Proteomic Platforms in a Pipeline for New Diagnostics. Molecular & Cellular Proteomics. 2005;4:1441–1444. doi: 10.1074/mcp.I500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 14. Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:14. doi: 10.1038/msb.2008.61. The authors review comprehensively the principle of the SRM technology, the process of generating SRM assays including current challenges and possible applications.
- 15. Mallick P, Schirle M, Chen S, Flory M, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. For the first time the authors determined physicochemical properties from previously acquired LC-MS/MS data to develop a computational tool for the prediction of the detectability of petides by MS.
- 16.Kuster B, Schirle M, Mallick P. Aebersold R: Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 17. Addona T, Abbatiello S, Schilling B, Skates S, Mani D, Bunk D, Spiegelman C, Zimmerman L, Ham A, Keshishian H, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;12 doi: 10.1038/nbt.1546. This is the first large scale study that shows the reproducibility and precision of SRM measurements in plasma across multiple laboratories and different instrument platforms.
- 18.Lange V, Malmstrom J, Didion J, King N, Johansson B, Schafer J, Rameseder J, Wong C, Deutsch E, Brusniak M, et al. Targeted Quantitative Analysis of Streptococcus pyogenes Virulence Factors by Multiple Reaction Monitoring. Molecular & Cellular Proteomics. 2008;7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full Dynamic Range Proteome Analysis of S. cerevisiae by Targeted Proteomics. Cell. 2009 doi: 10.1016/j.cell.2009.05.051. doi:10.1016/j.cell.2009.1005.1051. In this study the authors demonstrate for the first time the potential of SRM to detect and quantify proteins over the whole range of cellular concentrations in S. cerivisiae
- 20.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NG, Borchers CH. MRM-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson L. Quantitative Mass Spectrometric Multiple Reaction Monitoring Assays for Major Plasma Proteins. Molecular & Cellular Proteomics. 2005;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Keshishian H, Addona T, Burgess M, Kuhn E, Carr S. Quantitative, Multiplexed Assays for Low Abundance Proteins in Plasma by Targeted Mass Spectrometry and Stable Isotope Dilution. Molecular & Cellular Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keshishian H, Addona T, Burgess M, Mani D, Shi X, Kuhn E, Sabatine M, Gerszten R, Carr S. Quantification of Cardiovascular Biomarkers in Patient Plasma by Targeted Mass Spectrometry and Stable Isotope Dilution. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900140-MCP200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller M, Michel PE, Morier P, Crettaz D, Wenz C, Tissot JD, Reymond F, Rossier JS. Two-stage Off-Gel isoelectric focusing: Protein followed by peptide fractionation and application to proteome analysis of human plasma. Electrophoresis. 2005;26:1174–1188. doi: 10.1002/elps.200410106. [DOI] [PubMed] [Google Scholar]
- 25.Stalder A, Haeberli A, Heller M. Evaluation of reproducibility of protein identification results after multidimensional human serum protein separation. Proteomics. 2008;8:414–424. doi: 10.1002/pmic.200700527. [DOI] [PubMed] [Google Scholar]
- 26.Nicol G, Han M, Kim J, Birse C, Brand E, Nguyen A, Mesri M, FitzHugh W, Kaminker P, Moore P, et al. Use of an Immunoaffinity-Mass Spectrometry-based Approach for the Quantification of Protein Biomarkers from Serum Samples of Lung Cancer Patients. Mol Cell Proteomics. 2008;7:1974–1982. doi: 10.1074/mcp.M700476-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 28.Whiteaker J, Zhao L, Zhang HY, Feng L, Piening B, Anderson L, Paulovich A. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteaker J, Zhang H, Zhao L, Wang P, Kelly-Spratt K, Ivey R, Piening B, Feng L, Kasarda E, Gurley K, et al. Integrated Pipeline for Mass Spectrometry-Based Discovery and Confirmation of Biomarkers Demonstrated in a Mouse Model of Breast Cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 30. Kuhn E, Addona T, Keshishian H, Burgess M, Mani D, Lee R, Sabatine M, Gerszten R, Carr S. Developing Multiplexed Assays for Troponin I and Interleukin-33 in Plasma by Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry. Clinical Chemistry. 2009;55:1108–1117. doi: 10.1373/clinchem.2009.123935. This study shows the potential of SISCAPA-SRM for multiplexed quantification of biomarker candidates in plasma in the low ng/ml concentration range.
- 31.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High Sensitivity Detection of Plasma Proteins by Multiple Reaction Monitoring of N-Glycosites. Molecular & Cellular Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Domon B. Glycosylation as means of reducing sample complexity to enable quantitative proteomics. Proteomics. 2009;9:1488–1491. doi: 10.1002/pmic.200800545. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 35.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T, Qian W, Camp D, Smith R. The use of a quantitative cysteinyl-peptide enrichment technology for high-throughput quantitative proteomics. Methods Mol Biol. 2007;359:107–124. doi: 10.1007/978-1-59745-255-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Qian W, Strittmatter E, Camp D, Anderson G, Thrall B, Smith R. High-throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 38.Villanueva JJP, Entenberg D, Chaparro C, Tanwar M, Holland E, Tempst P. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal Chem. 2004;76:1560–1570. doi: 10.1021/ac0352171. [DOI] [PubMed] [Google Scholar]
- 39.Villanueva J, Lawlor K, Toledo-Crow R, Tempst P. Automated serum peptide profiling. Nat Protoc. 2006;1:880–891. doi: 10.1038/nprot.2006.128. [DOI] [PubMed] [Google Scholar]
- 40.Desiere F, Deutsch EW, Nesvizhskii A, Mallick P, King NL, Eng J, Aderem A, Boyle R, Brunner E, Donohoe S. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutsch E, Eng J, Zhang H, King N, Nesvizhskii A, Lin B, Lee H, Yi E, Ossola R, Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005;5:3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 42.Siepen J, Belhajjame K, Selley J, Embury S, Paton N, Goble C, Oliver S, Stevens R, Zamboulis L, Martin N, et al. ISPIDER Central: an integrated database web-server for proteomics. Nucleic Acids Research. 2008;36:W485–W490. doi: 10.1093/nar/gkn196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones P. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Research. 2006;34:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fusaro V, Mani D, Mesirov J, Carr S. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu P, Vogel C, Wang R, Yao X, Marcotte E. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 46.Frank R. The SPOT-synthesis technique: Synthetic peptide arrays on membrane supports--principles and applications. Journal of Immunological Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 47.Unwin RD, Griffiths JR, Whetton AD. A sensitive mass spectrometric method for hypothesis-driven detection of peptide post-translational modifications: multiple reaction monitoring-initiated detection and sequencing (MIDAS) Nat Protoc. 2009;4:870–877. doi: 10.1038/nprot.2009.57. [DOI] [PubMed] [Google Scholar]
- 48.Picotti P, Lam H, Campbell D, Deutsch E, Mirzaei H, Ranish J, Domon B, Aebersold R. A database of mass spectrometric assays for the yeast proteome. Nat Meth. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhlen M. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Molecular & Cellular Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Hober S, Uhlen M. Human protein atlas and the use of microarray technologies. Current Opinion in Biotechnology. 2008;19:30–35. doi: 10.1016/j.copbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi S. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirkpatrick D, Gerber S, Gygi S. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 53.Duncan M, Yergey A, Patterson S. Quantifying proteins by mass spectrometry: The selectivity of SRM is only part of the problem. Proteomics. 2009;9:1124–1127. doi: 10.1002/pmic.200800739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman J, Mckay M, Ashman K, Molloy M. How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics. 2009;9:1120–1123. doi: 10.1002/pmic.200800577. [DOI] [PubMed] [Google Scholar]
- 55.Anderson NL, Anderson NG, Pearson TW, Borchers C, Paulovich A, Patterson S, Gillette M, Aebersold R, Carr SA. A human proteome detection and quantitation project. Mol Cell Proteomics. 2009;8:883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helle M, Boeije L, de Groot E, de Vos A, Aarden L. Sensitive ELISA for interleukin-6: Detection of IL-6 in biological fluids: synovial fluids and sera. Journal of Immunological Methods. 1991;138:47–56. doi: 10.1016/0022-1759(91)90063-l. [DOI] [PubMed] [Google Scholar]
- 57.Ida N, Shingou S, Kazuo H, Tetsunosuke K. A highly sensitive enzyme-linked immunosorbent assay for the measurement of interleukin-8 in biological fluids. Journal of Immunological Methods. 1992;156:27–38. doi: 10.1016/0022-1759(92)90007-g. [DOI] [PubMed] [Google Scholar]