Abstract
The olfactory system of Drosophila melanogaster is one of the best characterized chemosensory systems. Identification of proteins contained in the third antennal segment, the main olfactory organ, has previously relied primarily on immunohistochemistry, and although such studies and in situ hybridization studies are informative, they focus generally on one or few gene products at a time, and quantification is difficult. In addition, purification of native proteins from the antenna is challenging because it is small and encased in a hard cuticle. Here, we describe a simple method for the large-scale detection of soluble proteins from the Drosophila antenna by chromatographic separation of tryptic peptides followed by tandem mass spectrometry with femtomole detection sensitivities. Examination of the identities of these proteins indicates that they originate both from the extracellular perilymph and from the cytoplasm of disrupted cells. We identified enzymes involved with intermediary metabolism, proteins associated with regulation of gene expression, nucleic acid metabolism and protein metabolism, proteins associated with microtubular transport, 8 odorant-binding proteins, protective enzymes associated with antibacterial defense and defense against oxidative damage, cuticular proteins, and proteins of unknown function, which represented about one-third of all soluble proteins. The procedure described here opens the way for precise quantification of any target protein in the Drosophila antenna and should be readily applicable to antennae from other insects.
Keywords: chemosensation, mass spectrometry, odorant-binding proteins, olfaction, proteomics
Introduction
The olfactory system of Drosophila melanogaster has emerged as one of the best characterized chemosensory systems. Odorants are recognized by sensory neurons housed in sensilla of the third antennal segment and the maxillary palps, the main olfactory organs, as well as chemosensory neurons on the tarsi, wing margins, and female reproductive organs. Olfactory sensory neurons in basiconic sensilla of the antenna and maxillary palps express odorant receptors that contain 7 transmembrane domains (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999), but differ from classical G protein–coupled receptors in membrane orientation (Benton et al. 2006) and transduction mechanism, as odorant activation results in the opening of a cation channel formed by a complex between a unique olfactory receptor and the universal Or83b receptor (Sato et al. 2008; Wicher et al. 2008). The molecular receptive fields and activation properties of a large fraction of odorant receptors have been characterized by elegant electrophysiological studies (de Bruyne et al. 1999, 2001; Hallem et al. 2004; Hallem and Carlson 2006), and projections of cells expressing defined receptors have been mapped to individual glomeruli in the antennal lobes (Vosshall et al. 2000). Olfactory sensory neurons in coeloconic sensilla express yet another family of odorant receptors that resemble ionotropic glutamate receptors with distinct ligand specificities, including responses to amines (Benton et al. 2009). In addition to odorant receptors, a large family of odorant-binding proteins (Obps) that are secreted by supporting cells into the antennal perilymph has been characterized (Galindo and Smith 2001; Hekmat-Scafe et al. 2002), and members of this family have been implicated in pheromone detection (Xu et al. 2005; Laughlin et al. 2008), host plant selection (Matsuo et al. 2007), and combinatorial recognition of general odorants (Wang et al. 2007).
Transcriptional profiling studies have shown that expression of the chemosensory repertoire of Drosophila is dynamic and changes under different developmental, environmental, and physiological conditions (Zhou et al. 2009). Clearly, it would be of value to correlate overall protein levels with changes in transcript abundance. Previously, the expression of Obps and odorant receptors, as well as proteins implicated in removal of xenobiotics, including odorants, has been characterized by in situ hybridization (McKenna et al. 1994; Pikielny et al. 1994; Clyne et al. 1999; Vosshall et al. 1999; Rollmann et al. 2005), immunohistochemistry (Hekmat-Scafe et al. 1997), enhancer trap (Riesgo-Escovar et al. 1992; Anholt et al. 1996), and transgenic drivers using the GAL4-UAS binary expression system (Vosshall et al. 2000; Galindo and Smith 2001). Although such studies have been highly informative, they focus generally on one or few gene products at a time and quantification by any of these methods is difficult. Furthermore, biochemical purification of native proteins from the antenna is challenging due to the small size of the Drosophila antenna and because olfactory sensory neurons are encased in a hard cuticle.
Here, we describe a simple method for the large-scale detection of soluble proteins from the Drosophila antenna using chromatographic separation of tryptic peptides by nano-LC followed by tandem mass spectrometry (MS/MS), with femtomole detection sensitivities. Confident protein identifications are obtained when LC retention time data are combined with accurate mass measurements and MS/MS fingerprints. The procedure described here opens the way for precise quantification of any target protein in the Drosophila antenna and should be readily applicable to antennae from other insects, including disease vectors, such as mosquitoes and urban pests, such as cockroaches, which rely on olfactory input for host identification, mating, and oviposition site selection.
Materials and methods
Drosophila stocks and sample preparation
Isogenic D. melanogaster of the Canton S (B) strain were reared on standard cornmeal molasses agar medium at 25 °C and 70% humidity under a 12 h light:dark cycle. Antennae were dissected by hand under a stereomicroscope and immediately placed in microcentrifuge tubes on dry ice. Duplicate pools of 120 antennae from males and females were obtained separately. The antennae were subjected to osmotic lysis by 2 freeze-thaw cycles in 50-μL distilled water followed by homogenization with a small pestle and centrifugation to remove nonsoluble material. The supernatants were recovered, and 32-μL samples were evaporated to dryness and reconstituted in 25.5 μL of 50 mM NH4HCO3 without introduction of detergents. For tryptic digestion of proteins, 1.5 μL of 100 mM aqueous dithiothreitol was added to each sample, and the samples were heated at 95 °C for 5 min. Upon cooling to room temperature, 3 μL of 100 mM aqueous iodoacetamide was added, and the resulting solution incubated at room temperature in the dark for 20 min. This was preceded by the addition of trypsin (1 μL of a 0.1 μg/μL trypsin solution in 1 mM HCl). Digestion was carried out at 37 °C for 3 h, following which another 1 μL of trypsin solution was added and samples were incubated at 30 °C overnight. Trypsin was quenched by addition of 1.5 μL of 5% formic acid. Samples were then evaporated to dryness, reconstituted in 100 μL of LC mobile phase A (98% H2O, 2% acetonitrile, and 0.2% formic acid), and filtered with 10 kDa molecular weight cut off filters (Millipore number 42407) prior to LC/MS/MS analyses. Serial dilutions of the sample solutions up to 1000-fold were performed prior to analysis to determine the optimum concentration for sample introduction into the instrument.
Mass spectrometry
Reversed phase high-performance liquid chromatography separation and MS detection were performed using an Eksigent nano-LC-2D system with an autosampler coupled to a hybrid LTQ-FT Ultra mass spectrometer from Thermo Scientific, Inc. The nano-LC was operated with a “continuous vented column” configuration for in-line trap and elute (Andrews et al. 2009). The analytical column was a self-packed 75-μm inner diameter (i.d.) fused silica PicoFrit capillary with 15 cm of Magic C18AQ stationary phase. The trap and dummy columns were self-packed 75 μm i.d. fused silica IntegraFrit capillaries with 5 cm and 20 cm of Magic C18AQ stationary phase, respectively. LC solvents used are mobile phase A and mobile phase B (acetonitrile/H2O/Hformic acid [98/2/0.2% by volume]). Blank runs were performed after every sample run. Sample injections ranged from 2 to 5 μL on column. Analytical separations were run on the nanoflow pump at 500 nL/min, initially maintaining a composition of 2% B. The MS method consisted of 4 events: a precursor scan followed by 3 data-dependent tandem MS scans of the first, second, and third most abundant peaks in the ion trap. A high resolving power precursor scan of the eluted peptides was obtained using the LTQ-FT with the 3 most abundant ions selected for MS/MS in the ion trap through dynamic exclusion. The instrument was externally calibrated according to the manufacturer's protocol.
Data analysis
The nano-LC/MS/MS data files were processed by Sequest (Bioworks, ThermoFisher Scientific, Inc.; Eng et al. 1994) and Mascot (Matrix Science; Perkins et al. 1999) for protein identifications. These algorithms apply similar general approaches in assigning peptides detected in MS/MS spectra to those in a sequence database. However, the principles behind their mathematical operations are significantly different. Mascot applies a probabilistic metric to determine the likelihood that a fragmented peptide produced an observed MS/MS spectrum. Sequest, on the other hand, applies empirical and correlation measurements to score the alignment between observed and predicted spectra, among other important differences. Batch searching of LC/MS/MS data was performed using the D. melanogaster protein database from InterPro (www.ebi.ac.uk/interpro).
Results and discussion
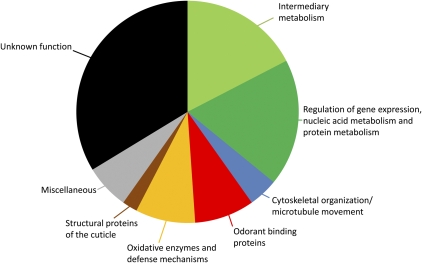
Combining the high mass measurement accuracies of the LTQ-FT mass analyzer with tandem MS fragmentation data and nano-LC retention times allows for confident protein identifications with minimal sample consumption. A representative nano-LC/MS/MS identification of an Obp (PBP2_DROME Pheromone-binding protein-related protein 2 precursor) released from a female antenna is presented in Figure 1. Tandem mass spectrometric analysis of eluted chromatographic peaks allowed for the identification of approximately 100 proteins through Sequest and Mascot database search algorithms. Sequest was able to identify 30 proteins (Table 1), whereas analysis with Mascot resulted in the identification of 92 proteins (Table 2) with high statistical confidence. Examination of the identities of these proteins indicates that they originate both from the extracellular perilymph and from the cytoplasm of disrupted cells. We classified these proteins into 8 major categories (Table 3; Figure 2). These include enzymes involved with intermediary metabolism, primarily the glycolytic pathway, tricarboxylic acid cycle, and oxidative phosphorylation; proteins associated with regulation of gene expression, nucleic acid metabolism and protein metabolism, including regulation of transcription, histone modification and mRNA splicing, protein folding, and degradation; proteins associated with microtubular transport; Obps; protective enzymes associated with antibacterial defense and defense against oxidative damage; cuticular proteins; miscellaneous proteins associated with programmed cell death (Pcs), neuropeptide hormone activity (Nplp2), regulation of mating and courtship behavior (Esterase-6), oxygen transport (glob1), tyrosine kinase activity (Fps85D), and cell adhesion (hig); and proteins of unknown function, which represented the largest group comprising about one-third of all soluble proteins identified (Table 3; Figure 2).
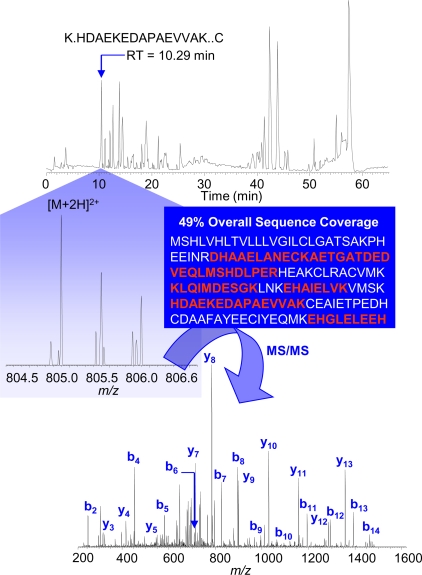
Figure 1.
nano-LC/MS/MS identification of PBP2_DROME Pheromone-binding protein-related protein 2 precursor—Drosophila melanogaster. A tryptic peptide, which eluted from the nano-LC column at a retention time (RT) of 10.29 min (chromatogram shown at the top of the figure), is identified by nano-LC/MS/MS (with database searching) to be derived from PBPRP2 precursor protein. The MS spectrum for this peptide is shown in the center of the figure. The ionized, doubly charged peptide is subjected to dynamic exclusion and fragmentation to generate an MS/MS fingerprint (lower spectrum) that conclusively verifies its molecular structure. As many as 11 peptides derived from PBPRP2 precursor protein were detected in total, providing 49% sequence coverage (shown in red font in the middle of the figure).
Table 1.
Soluble protein identifications (30 total) for Drosophila melanogaster antennae by Sequest (Obps are highlighted in bold font)
| Protein identification | P | Score (XC) |
| ATPB_DROME ATP synthase subunit beta, mitochondrial precursor | 1.6 × 10−13 | 5.42 × 101 |
| PBP2_DROME pheromone-binding protein-related protein 2 precursor | 2.5 × 10−12 | 1.28 × 102 |
| Q7K084_DROME RH04549p | 3.8 × 10−8 | 6.02 × 101 |
| SODC_DROME superoxide dismutase [Cu-Zn] | 7.5 × 10−8 | 5.02 × 101 |
| Q8SY92_DROME RH21971p | 8.6 × 10−8 | 4.02 × 101 |
| O16157_DROME calcium-binding protein | 2.0 × 10−7 | 3.02 × 101 |
| Q9VEB1_DROME CG7998-PA | 3.0 × 10−7 | 3.62 × 101 |
| PRDX1_DROME peroxiredoxin 1 | 3.3 × 10−7 | 1.42 × 101 |
| BNB_DROME protein bangles and beads | 5.1 × 10−7 | 5.03 × 101 |
| ATPA_DROME ATP synthase subunit alpha, mitochondrial precursor | 5.1 × 10−7 | 6.22 × 101 |
| OBP56D_DROME general Obp 56d precursor | 7.7 × 10−7 | 2.02 × 101 |
| G3P1_DROME glyceraldehyde-3-phosphate dehydrogenase | 8.5 × 10−7 | 5.62 × 101 |
| PBP6_DROME pheromone-binding protein-related protein 6 precursor | 1.8 × 10−6 | 6.02 × 101 |
| Q7KUB0_DROME CG7176-PA, isoform A | 2.2 × 10−6 | 6.01 × 101 |
| ENO_DROME isoform 1 of P15007 | 3.4 × 10−6 | 4.22 × 101 |
| Q9VQF7_DROME CG9894-PA, isoform A | 6.1 × 10−6 | 2.02 × 101 |
| Q9VTC3_DROME CG6409-PA | 9.5 × 10−6 | 4.02 × 101 |
| P91941_DROME CG10297-PA | 2.9 × 10−5 | 2.42 × 101 |
| O16043_DROME CG2207-PA, isoform A | 3.0 × 10−5 | 2.02 × 101 |
| PBP5_DROME pheromone-binding protein-related protein 5 precursor | 4.8 × 10−5 | 2.81 × 101 |
| OB10_DROME putative Obp A10 precursor | 4.9 × 10−5 | 4.02 × 101 |
| GSTT1_DROME glutathione S-transferase 1-1 | 6.1 × 10−5 | 2.22 × 101 |
| ALF_DROME isoform Gamma of P07764 | 8.7 × 10−5 | 4.22 × 101 |
| EST6_DROME esterase-6 precursor | 1.0 × 10−4 | 6.02 × 101 |
| APLP_DROME apolipophorins precursor | 1.1 × 10−4 | 7.82 × 101 |
| PBP3_DROME pheromone-binding protein-related protein 3 precursor | 1.1 × 10−4 | 6.62 × 101 |
| NPLP2_DROME neuropeptide-like 2 precursor | 1.4 × 10−4 | 2.02 × 101 |
| ACT1_DROME actin-5C | 3.4 × 10−4 | 3.41 × 101 |
| Q8SXA6_DROME GH20168p | 5.5 × 10−4 | 2.62 × 101 |
| CISY_DROME isoform A of Q9W401 | 1.9 × 10−3 | 6.01 × 101 |
XC is the correlation score.
Table 2.
Soluble protein identifications (94 total) for Drosophila melanogaster antennae by Mascot (Obps are highlighted in bold font; protein mass is in Daltons)
| Accession numbers | Protein identification | Protein score | Protein mass | Protein matches |
| sp|O02649|CH60_DROME | 60 kDa heat shock protein, mitochondrial | 35 | 60 885 | 1 |
| sp|P07487|G3P2_DROME | Glyceraldehyde-3-phosphate dehydrogenase 2 | 212 | 35 518 | 6 |
| sp|P08171|EST6_DROME | Esterase-6 | 168 | 61 486 | 4 |
| sp|P15007-1|ENO_DROME | Isoform B of enolase | 163 | 54 561 | 4 |
| sp|P18106-3|FPS_DROME | Isoform C of tyrosine-protein kinase Fps85D | 23 | 54 963 | 1 |
| sp|P20432|GSTT1_DROME | Glutathione S-transferase 1-1 | 120 | 24 022 | 3 |
| sp|P29746|BNB_DROME | Protein bangles and beads | 305 | 45 902 | 6 |
| sp|P31409|VATB_DROME | V-type proton ATPase subunit B | 34 | 54 800 | 1 |
| sp|P34739|TTF2_DROME | Transcription termination factor 2 | 37 | 118 759 | 2 |
| sp|P35381|ATPA_DROME | ATP synthase subunit alpha, mitochondrial | 183 | 59 612 | 4 |
| sp|P41073-1|PEP_DROME | Isoform B of zinc finger protein on ecdysone puffs | 22 | 78 570 | 1 |
| sp|P46863|KL61_DROME | Bipolar kinesin KRP-130 | 26 | 121 772 | 1 |
| sp|P51123-1|TAF1_DROME | Isoform B of transcription initiation factor TFIID subunit 1 | 25 | 240 307 | 1 |
| sp|P54192|PBP2_DROME | Pheromone-binding protein-related protein 2 | 509 | 17 170 | 11 |
| sp|P54193|PBP3_DROME | Pheromone-binding protein-related protein 3 | 80 | 17 657 | 4 |
| sp|P54195|PBP5_DROME | Pheromone-binding protein-related protein 5 | 39 | 15 484 | 1 |
| sp|P55830|RS3A_DROME | 40S ribosomal protein S3a | 31 | 30 565 | 1 |
| sp|P61851|SODC_DROME | Superoxide dismutase [Cu-Zn] | 333 | 15 974 | 6 |
| sp|P62152|CALM_DROME | Calmodulin | 38 | 16 800 | 2 |
| sp|Q05825|ATPB_DROME | ATP synthase subunit beta, mitochondrial | 186 | 54 074 | 5 |
| sp|Q09101-1|HIG_DROME | Isoform 3 of locomotion-related protein hikaru genki | 26 | 108 557 | 1 |
| sp|Q23970|PBP6_DROME | Pheromone-binding protein-related protein 6 | 137 | 16 500 | 6 |
| sp|Q24120|CAPU_DROME | Protein cappuccino | 23 | 114 705 | 1 |
| sp|Q24407|ATP5J_DROME | ATP synthase-coupling factor 6, mitochondrial | 68 | 11 928 | 1 |
| sp|Q27377|OB10_DROME | Putative Obp A10 | 130 | 18 111 | 2 |
| sp|Q32KD2|SETB1_DROME | Histone-lysine N-methyltransferase eggless | 38 | 143 852 | 1 |
| sp|Q8SY61|OB56D_DROME | General Obp 56d | 181 | 14 452 | 4 |
| sp|Q9NJG9-1|SUZ12_DROME | Isoform 1 of polycomb protein Su(z)12 | 34 | 101 011 | 1 |
| sp|Q9V3P0|PRDX1_DROME | Peroxiredoxin 1 | 118 | 21 952 | 3 |
| sp|Q9V785|3BP5H_DROME | SH3 domain-binding protein 5 homolog | 22 | 54 060 | 1 |
| sp|Q9V7N5-1|VATC_DROME | Isoform D of V-type proton ATPase subunit C | 26 | 79 776 | 1 |
| sp|Q9VPS5|CH60B_DROME | 60 kDa heat shock protein homolog 1, mitochondrial | 34 | 68 992 | 1 |
| sp|Q9VU58|NPLP2_DROME | Neuropeptide-like 2 | 92 | 9463 | 1 |
| sp|Q9W1C9|PEB3_DROME | Ejaculatory bulb-specific protein 3 | 73 | 14 759 | 1 |
| sp|Q9W1R5|VIR_DROME | Protein virilizer | 29 | 210 751 | 1 |
| sp|Q9W401-1|CISY_DROME | Isoform A of probable citrate synthase, mitochondrial | 34 | 51 713 | 3 |
| tr|A1Z6K9|A1Z6K9_DROME | CG17994-PA | 25 | 59 821 | 1 |
| tr|A1Z7Z9|A1Z7Z9_DROME | CG1625-PA | 26 | 129 850 | 2 |
| tr|A1ZA97|A1ZA97_DROME | CG8424-PA | 80 | 62456 | 2 |
| tr|A4V3F9|A4V3F9_DROME | Fructose-bisphosphate aldolase | 193 | 39 251 | 5 |
| tr|A8DZ12|A8DZ12_DROME | CG15140-PA (Fragment) | 21 | 30 108 | 4 |
| tr|A8JV09|A8JV09_DROME | CG4532-PF, isoform F | 21 | 140 127 | 1 |
| tr|A8QI20|A8QI20_DROME | CG41561-PA (Fragment) | 31 | 40 471 | 1 |
| tr|O16043|O16043_DROME | CG2207-PA, isoform A (CG2207-PB, isoform B) (CG2207-PF, isoform F) (LD21289p) (Anon1A4) | 90 | 18 812 | 1 |
| tr|O97102|O97102_DROME | CG4494-PA | 82 | 10 175 | 1 |
| tr|Q0E8L0|Q0E8L0_DROME | CG8486-PC, isoform C | 26 | 305 750 | 1 |
| tr|Q4ABG9|Q4ABG9_DROME | CG33715-PE, isoform E | 22 | 1 064 061 | 2 |
| tr|Q4V5I6|Q4V5I6_DROME | IP07112p (IP06812p) (CG14810-PA) | 26 | 22 874 | 1 |
| tr|Q59DY8|Q59DY8_DROME | CG33552-PA | 24 | 18 848 | 1 |
| tr|Q7JND6|Q7JND6_DROME | Cuticle protein ACP65A | 123 | 10 773 | 1 |
| tr|Q7K084|Q7K084_DROME | RH04549p (CG2297-PA) Obp 44a | 213 | 16 135 | 5 |
| tr|Q7K088|Q7K088_DROME | RH03850p (CG8462-PA) Obp 56e | 44 | 14 505 | 1 |
| tr|Q7K2B0|Q7K2B0_DROME | LD11455p (CG7137-PA) | 26 | 40 605 | 1 |
| tr|Q7KMR7|Q7KMR7_DROME | Thioredoxin-like protein TXL | 25 | 32 140 | 1 |
| tr|Q7KTB7|Q7KTB7_DROME | CG6214-PM, isoform M | 26 | 174 219 | 1 |
| tr|Q7KUB0|Q7KUB0_DROME | CG7176-PA, isoform A (CG7176-PE, isoform E) (CG7176-PF, isoform F) | 192 | 47 030 | 6 |
| tr|Q8I940|Q8I940_DROME | CG1271-PD, isoform D | 22 | 60 539 | 1 |
| tr|Q8IRD3|Q8IRD3_DROME | Glutathione peroxidase | 23 | 26 620 | 1 |
| tr|Q8MLS0|Q8MLS0_DROME | CG13551-PC, isoform C | 65 | 9711 | 1 |
| tr|Q8MSI2|Q8MSI2_DROME | GH15296p (CG15848-PA) | 172 | 21 919 | 4 |
| tr|Q8MSU4|Q8MSU4_DROME | CG2097-PA | 26 | 133 020 | 2 |
| tr|Q8MYW5|Q8MYW5_DROME | CG14667-PA, isoform A | 20 | 30 574 | 1 |
| tr|Q8SWW8|Q8SWW8_DROME | LD18186p (CG12489-PA) | 25 | 74 562 | 1 |
| tr|Q8SX06|Q8SX06_DROME | RH33338p (CG14141-PA) | 30 | 18 788 | 1 |
| tr|Q8SXZ0|Q8SXZ0_DROME | RE47719p | 24 | 56 222 | 1 |
| tr|Q8SY92|Q8SY92_DROME | RH21971p antennal dehydrogenase | 327 | 27 362 | 5 |
| tr|Q8T3H5|Q8T3H5_DROME | AT28279p, CG13382 | 33 | 59 050 | 1 |
| tr|Q8T3Y1|Q8T3Y1_DROME | AT26187p, CG17440 | 25 | 42 763 | 1 |
| tr|Q8T487|Q8T487_DROME | AT10439p, CG1950 | 29 | 39 778 | 2 |
| tr|Q8T8Q5|Q8T8Q5_DROME | SD05887p, CG3493 | 26 | 170 613 | 2 |
| tr|Q8T9I2|Q8T9I2_DROME | GM13608p, Bip1 | 27 | 46 380 | 1 |
| tr|Q95RB2|Q95RB2_DROME | CG8505-PA, Cpr49Ae | 94 | 14 501 | 1 |
| tr|Q95RB2|Q95RB2_DROME | CG8505-PA, Cpr49Ae | 88 | 14 501 | 1 |
| tr|Q961M4|Q961M4_DROME | GH15731p | 28 | 94 143 | 1 |
| tr|Q9NHV6|Q9NHV6_DROME | Dorsal interacting protein 2 | 37 | 26 140 | 1 |
| tr|Q9U1K3|Q9U1K3_DROME | Globin1 | 40 | 17 149 | 1 |
| tr|Q9U3Y5|Q9U3Y5_DROME | Dynein heavy chain | 25 | 529 605 | 1 |
| tr|Q9V3H9|Q9V3H9_DROME | BcDNA.LD27873 | 26 | 113 870 | 2 |
| tr|Q9VCW7|Q9VCW7_DROME | CG6954-PA, CG6954 | 29 | 181 921 | 1 |
| tr|Q9VEB1|Q9VEB1_DROME | CG7998-PA (IP09655p) | 122 | 35 524 | 2 |
| tr|Q9VF51|Q9VF51_DROME | FI04488p | 89 | 139 567 | 3 |
| tr|Q9VGD4|Q9VGD4_DROME | CG14741-PA | 23 | 193 892 | 1 |
| tr|Q9VH97|Q9VH97_DROME | CG9492-PA | 33 | 539 599 | 1 |
| tr|Q9VIK1|Q9VIK1_DROME | CG9318-PA (LD44273p) | 24 | 75 732 | 2 |
| tr|Q9VQF7|Q9VQF7_DROME | CG9894-PA, isoform A (CG9894-PB, isoform B) (RE38782p) | 180 | 15 508 | 3 |
| tr|Q9VQT8|Q9VQT8_DROME | CG16712-PA (RH38008p) (RH05411p) | 75 | 9144 | 1 |
| tr|Q9VSY1|Q9VSY1_DROME | CG4022-PA | 29 | 57 600 | 1 |
| tr|Q9VTC3|Q9VTC3_DROME | CG6409-PA (GH07049p) | 86 | 40 356 | 4 |
| tr|Q9W1A9|Q9W1A9_DROME | CG11290-PA | 27 | 257 089 | 2 |
| tr|Q9W246|Q9W246_DROME | CG4554-PA | 27 | 313 818 | 1 |
| tr|Q9W306|Q9W306_DROME | CG9691-PA, isoform A (CG9691-PB, isoform B) (RH12290p) | 57 | 13 081 | 1 |
| tr|Q9W321|Q9W321_DROME | CG15319-PB | 37 | 343 477 | 2 |
| tr|Q9W3B3|Q9W3B3_DROME | CG1885-PA (GH17465p) | 35 | 31 218 | 2 |
| tr|Q9W4C3|Q9W4C3_DROME | Ubiquitin carboxyl-terminal hydrolase, CG4165 | 38 | 122 838 | 3 |
Table 3.
The soluble proteome of Drosophila melanogaster antennaea
| Intermediary metabolism (17.4%) |
| Glyceraldehyde phosphate dehydrogenase |
| Glycerol kinase |
| Aldolase |
| Enolase |
| L-Malate dehydrogenase |
| Citrate synthase |
| Isocitrate dehydrogenase |
| ATP synthase uncoupling factor 2 |
| Vha44 |
| Vha 55 |
| Blw, ATP synthase |
| ATP synthase subunit B |
| CG6054 |
| CG14741 |
| CG1885 |
| Calmodulin |
| Regulation of gene expression, nucleic acid metabolism, and protein metabolism (17.3%) |
| Taf1 |
| Lds |
| Pep |
| Histone-lysine N-methyltransferase |
| Su(z)12 |
| Cg11290 |
| Vir |
| Nej |
| CG2207 |
| CG33715 |
| Hsp60 |
| Ribosomal protein S3A |
| CG4494 |
| CG2097 |
| CG1950 (ubiquitin thiolesterase) |
| CG4165 (ubiquitin thiolesterase) |
| Cytoskeletal organization/microtubule movement (5.5%) |
| Capu |
| CG4532 |
| Klp61F |
| CG9492 |
| Dynein heavy chain Kl-5 |
| Obps (8.7%) |
| Pbprp2 |
| Pbprp3 |
| Pbprp5 |
| Pbprp6 |
| A10 |
| Obp44a |
| Obp56d |
| Obp56e |
| Oxidative enzymes and defense mechanisms (8.7%) |
| Peroxiredoxin 1 |
| Superoxide dismutase |
| GstD1 |
| Txl |
| CG6214 |
| Glutathione peroxidase |
| CG13551 |
| Dnr1 |
| Unknown function (33.7%) |
| CG17994 |
| CG1625 |
| CG8424 |
| CG41561 |
| CG8486 |
| CG14667 |
| Bnb (gliogenesis) |
| PebIII |
| CG14810 |
| CG33552 |
| CG7137 |
| CG14141 |
| Antdh |
| CG13382 |
| CG17440 |
| CG15296 |
| CG3493 |
| GH15731p |
| BcDNA-LD27873 |
| CG6954 |
| Dip2 |
| CG9318 |
| CG9894 |
| CG16712 |
| CG6409 |
| CG4554 |
| CG9691 |
| CG15140 |
| Smid |
| Bip1 |
| CG4022 |
| Structural proteins of the cuticle (2.2%) |
| Acp65Aa |
| Cpr49Ae |
| Miscellaneous (6.5%) |
| Pcs (programmed cell death) |
| Nplp2 (neuropeptide hormone activity) |
| Esterase-6 (regulation of receptivity, sperm competition, mating, pheromone synthesis, courtship behavior) |
| Globin 1 (oxygen transport) |
| Pfs85D (protein tyrosine kinase activity; photoreceptor cell morphogenesis) |
| Hig (cell adhesion) |
Figure 2.
The number of soluble proteins in different functional categories detected in Drosophila antennae. See also Table 3.
We detected substantially fewer proteins in male antennal extracts than in antennal extracts from females (see Supplementary Tables 1–4). This is most likely due to greater resistance to cell disruption under the conditions used in antennae from males than females, for unknown reasons. However, 6 of 8 Obps and 4 of 8 proteins associated with biotic and abiotic defense were found both in male and female antennae, suggesting that extracellular proteins in the perilymph are released effectively from male antennae and that detection of Obp44a and Obp56e in female but not male samples may reflect true sexual dimorphism in the expression of Obps, in-line with previous observations (Anholt et al. 2003). Pbprp2 (aka Obp19d), Pbprp3 (aka Os-F, Obp83a), Pbprp5 (aka Obp28a), and Pbprp6 (aka Os-E, Obp83b) have been localized previously to antennae (Pikielny et al. 1994; Hekmat-Scafe et al. 1997; Shandbag et al. 2001). In these initial experiments, we identified only 8 members of the Obp family, which comprises more than 50 Obp genes (Hekmat-Scafe et al. 2002). Notably, absent was the Lush protein (Kim et al. 1998), which mediates recognition of the male courtship pheromone 11-cis-vaccenylacetate in T1 trichoid sensilla of the antenna (Xu et al. 2005; Laughlin et al. 2008). It is possible that the amounts of Lush are below our detection limit. The same may be true for other members of the Obp family. Indeed, when the amount of tissue was increased from 120 to 500 antennae, we were able to detect a larger number of proteins with 10 additional Obps, including Lush, A5, Pbprp1 (aka Obp69a), Pbprp4 (aka Obp84a), Obp19a, Obp47b, Obp56a, Obp59a, Obp99b, and Obp99c. Furthermore, some Obps may not be expressed in antennae. For example, some Obps are expressed in the tarsi (Galindo and Smith 2001; Matsuo et al. 2007), fat body (Fujii and Amrein 2002), or male accessory gland (Takemori and Yamamoto 2009) and therefore would not be detected in the antenna.
The vast majority of proteins we detected are widely expressed in many or all cells. Nevertheless, their functions in different cells are essential for enabling distinct physiological functions. For example, intermediary metabolism is necessary to provide energy for olfactory signal transduction, and cytoskeletal organization is essential for maintaining dendritic and axonal structure and function. Two of the categories of soluble proteins indicated in Table 3 are specifically relevant to chemosensation: Obps, which are essential for the transport of hydrophobic odorants in the perilymph, and oxidative enzymes and defense mechanisms which likely contribute to cytochrome P450-mediated inactivation or degradation of odorants and environmental toxins. Obps have been defined based on their structure, including the characteristic positions of disulfide bonds (Hekmat-Scafe et al. 2002). However, olfactory functions have been ascribed to only few members of this family and other possible functions, such as a carrier function for molecules transmitted between males and females during mating have also been noted. Altered regulation of expression of Obp genes has been observed following mating (McGraw et al. 2004; Zhou et al. 2009) and after exposure to starvation stress (Harbison et al. 2005) or alcohol intoxication (Morozova et al. 2006). In addition, changes in expression levels of Obp genes occur as a correlated response to artificial selection for divergent levels of copulation latency (Mackay et al. 2005) and aggression (Edwards et al. 2006), and expression levels change during social crowding and as a result of ageing (Zhou et al. 2009). Systems genetics analyses of 6 X chromosome linked Obp genes showed that their transcripts form part of diverse transcriptional network niches, associated with olfactory behavior, synaptic transmission, detection of signals regulating tissue development and apoptosis, postmating behavior and oviposition, and nutrient sensing (Arya GH, Weber AL, Wang P, Magwire MM, Serrano Negron YL, Mackay TFC, Anholt RRH, unpublished data). Our proteomics analysis has identified Obps that are expressed in the antenna and thus are candidates for contributing directly to chemosensation.
Although the second antennal segment might also contribute to the proteomics profile, the fact that Obps make up a substantial fraction of the soluble proteome (Figure 2) suggests that the major contribution to the mass spectrometric profile is derived from the third antennal segment.
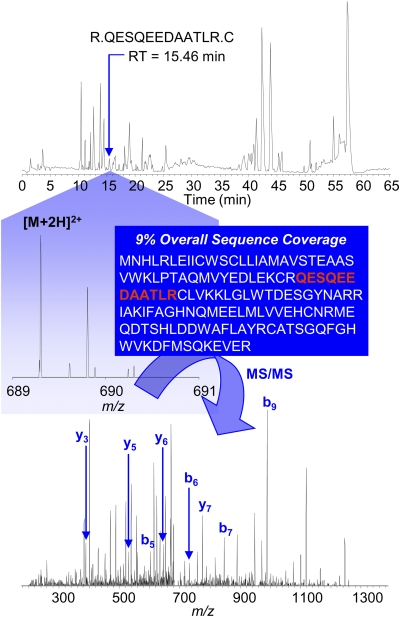
An additional caveat is the notion that protein detection could be limited by masking of peptide fragments in the nano-LC/MS/MS analyses. Comigration on the LC might also have prevented the detection of Lush or other Obps. This problem is likely to be more severe with increasing complexity of the chromatogram and more likely to impact proteins that yield few tryptic fragments, as well as those present in very low concentrations, relative to others. In addition, variations in ionization efficiencies of different peptides can lead to diminished representation in the nano-LC/MS/MS run. Furthermore, it should be noted that the proteins presented in Tables 1 and 2 represent only those that attain a high confidence score with Sequest and Mascot, respectively. Increasing the number of antennae in each sample greatly increases the number of proteins that can be interrogated by nano-LC/MS/MS approaches. This improves the chances for unambiguous identification of proteins such as Obp99d (Figure 3),—previously implicated in responses to benzaldehyde (Wang et al. 2007)—which, while detected by Sequest, did not produce a very confident identification and therefore is not included in Table 1, and Mascot did not allow for the identification of this protein. Nevertheless, information about the presence of Obp99d, which is present in low abundance, can still be extracted from the chromatogram (Figure 3).
Figure 3.
nano-LC/MS/MS identification of tr|Q9VAI7|Q9VAI7_DROME CG15505-PA (Obp 99d). A tryptic peptide, which eluted from the nano-LC column at a retention time (RT) of 15.46 min (chromatogram shown at the top of the figure), is identified by nano-LC/MS/MS (with database searching) to be derived from Obp99d. The MS spectrum for this peptide is shown in the center of the figure. The ionized, doubly charged peptide is subjected to dynamic exclusion and fragmentation to generate an MS/MS fingerprint (lower spectrum) that conclusively verifies its molecular structure. In contrast to PBPRP2 (Figure 1), which is present in abundance and identified by 11 tryptic peptides (Tables 1 and 2), a single peptide (indicated in red font in the middle of the figure) was detected from the Obp99d protein and accounts for only 9% of its sequence but is sufficient to identify this low abundance Obp in the antennal extract.
Finally, whereas integration of chromatographic peaks can establish relative amounts of proteins represented in the sample, precise quantitative determination requires stable isotope labeled internal standard peptides. This approach is now feasible for targeted determination of specific soluble proteins of the antenna.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
National Institutes of Health grant [GM059469].
Supplementary Material
Acknowledgments
We thank Steven A. West for excellent technical assistance and Professor David C. Muddiman (Department of Chemistry, North Carolina State University) for use of the LTQ-FT-MS in his laboratory.
References
- Andrews GL, Shuford CM, Burnett JC, Jr, Hawkridge AM, Muddiman DC. Coupling of a vented column with splitless nanoRPLC-ESI-MS for the improved separation and detection of brain natriuretic peptide-32 and its proteolytic peptides. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:948–954. doi: 10.1016/j.jchromb.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RRH, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TFC. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35:180–184. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- Anholt RRH, Lyman RF, Mackay TFC. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics. 1996;143:293–301. doi: 10.1093/genetics/143.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2006;2:e154. doi: 10.1371/journal.pgen.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fujii S, Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–5363. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TFC. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Steinbrecht RA, Carlson JR. Coexpression of two odorant-binding protein homologs in Drosophila: implications for olfactory coding. J Neurosci. 1997;17:1616–1624. doi: 10.1523/JNEUROSCI.17-05-01616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Heinsohn SL, Lyman RF, Moehring AJ, Morgan TJ, Rollmann SM. Genetics and genomics of Drosophila mating behavior. Proc Natl Acad Sci USA. 2005;102:6622–6629. doi: 10.1073/pnas.0501986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR. Putative Drosophila pheromone- binding proteins expressed in a subregion of the olfactory system. J Biol Chem. 1994;269:16340–16347. [PubMed] [Google Scholar]
- Morozova TV, Anholt RRH, Mackay TFC. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J, Woodard C, Gaines P, Carlson J. Development and organization of the Drosophila olfactory system: an analysis using enhancer traps. J Neurobiol. 1992;23:947–964. doi: 10.1002/neu.480230803. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Mackay TFC, Anholt RRH. Pinocchio, a novel protein expressed in the antenna, contributes to olfactory behavior in Drosophila melanogaster. J Neurobiol. 2005;63:146–158. doi: 10.1002/neu.20123. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Hekmat-Scafe D, Kim MS, Park SK, Carlson JR, Pikielny C, Smith DP, Steinbrecht RA. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc Res Tech. 2001;55:297–306. doi: 10.1002/jemt.1179. [DOI] [PubMed] [Google Scholar]
- Takemori N, Yamamoto MT. Proteome mapping of the Drosophila melanogaster male reproductive system. Proteomics. 2009;9:2484–2493. doi: 10.1002/pmic.200800795. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Shabalina SA, Mackay TFC, Anholt RRH. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics. 2007;177:1655–1665. doi: 10.1534/genetics.107.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Zhou S, Atkinson EA, Mackay TFC, Anholt RRH. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. Forthcoming 2009;5:e1000681. doi: 10.1371/journal.pgen.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.