Abstract
We sought to develop a cancer-targeted, infectivity-enhanced oncolytic adenovirus that embodies a capsid-labeling fusion for noninvasive dual-modality imaging of ovarian cancer virotherapy. A functional fusion protein composed of fluorescent and nuclear imaging tags was genetically incorporated into the capsid of an infectivity-enhanced conditionally replicative adenovirus. Incorporation of herpes simplex virus thymidine kinase (HSV-tk) and monomeric red fluorescent protein 1 (mRFP1) into the viral capsid and its genomic stability were verified by molecular analyses. Replication and oncolysis were evaluated in ovarian cancer cells. Fusion functionality was confirmed by in vitro gamma camera and fluorescent microscopy imaging. Comparison of tk-mRFP virus to single-modality controls revealed similar replication efficiency and oncolytic potency. Molecular fusion did not abolish enzymatic activity of HSV-tk as the virus effectively phosphorylated thymidine both ex vivo and in vitro. In vitro fluorescence imaging demonstrated a strong correlation between the intensity of fluorescent signal and cytopathic effect in infected ovarian cancer cells, suggesting that fluorescence can be used to monitor viral replication. We have in vitro validated a new infectivity-enhanced oncolytic adenovirus with a dual-imaging modality-labeled capsid, optimized for ovarian cancer virotherapy. The new agent could provide incremental gains toward climbing the barriers for achieving conditionally replicated adenovirus efficacy in human trials.
OVARIAN CANCER remains the deadliest malignancy of the female reproductive tract, with over 14,600 deaths predicted in the United States for 2009. The 10-year relative survival rate for all women diagnosed with ovarian cancer is approximately 39%.1 It is evident that new therapeutic approaches are greatly needed for the treatment of women with this deadly disease. Conditionally replicative adenovirus (CRAd)-based virotherapy represents a promising option to treat malignancy through viral amplification, oncolysis, and lateralization—a mechanism that is distinct from other forms of cancer therapy.2 Although CRAd-based therapies have emerged as novel strategies for the treatment of ovarian cancer, clinical efficacy has been fleeting and information derived from clinical trials has yielded limited insight into factors limiting success.3 A flexible, dynamic, noninvasive CRAd imaging system remains clinically relevant and a promising prospect for future clinical evaluation of ovarian cancer virotherapy.
Various imaging strategies have been employed in the context of adenoviral (Ad) gene therapy vectors. We and others have reported adenoviral labeling systems based on incorporation of an imaging reporter into the E1A, E2B, and the fiber genes of Ad5.4–7 Use of different types of reporter genes allowed exploiting a variety of signal detection techniques. Imaging motifs such as mutated herpes simplex virus thymidine kinase (HSV-tk)8 allow for use of common and clinically relevant imaging techniques such as computed tomography (CT), single-photon emission computed tomography (SPECT), or positron emission tomography (PET) using radioactive tracers, whereas monomeric red fluorescent protein (mRFP1) offers more economical optical fluorescence imaging options for in vitro and in vivo investigations. Given that no single modality is capable of addressing all aspects of molecular imaging, construction of fusion reporters that combine the positive attributes of different modalities has recently been attempted for multimodality imaging applications.7
Existing CRAd imaging strategies are based on detection of virus-encoded transgene expression as opposed to direct visualization of viral progeny particles. Being dependent on the viability of the infected cells to produce an imaging signal, these strategies are not compatible with the cell-killing functions of oncolytic agents and are therefore inadequate for assessment of CRAd biodistribution, progeny production, and accumulation in tumors.
A genetic technology of Ad capsid labeling involving fusing imaging motifs to the minor capsid protein IX (pIX) allows direct visualization of viral particles and overcomes the CRAd imaging limitation. Feasibility of Ad capsid labeling by imaging reporter fusions has been established in several recent proof-of-principle studies.9–14 Matthews and colleagues fashioned a pIX fusion incorporating both HSV-tk and firefly luciferase (luc) reporters for use in the context of a nonreplicative Ad. Light signal produced by luc, however, requires administration of potentially toxic substrate, which makes this reporter not practical for human uses.13 In contrast, use of fluorescent reporters such as mRFP1 that require light activation (excitation), but no substrates for activity represents a better alternative for dynamic optical imaging approach. Capsid incorporation of both HSV-tk and mRFP1 moieties in a dual-modality reporter fusion could result in combined capacity for SPECT/PET and highly sensitive fluorescent imaging, respectively. Expression timing of the imaging fusion from the native promoter of the Ad gene IX could allow an accurate tracking of CRAd replication by imaging of the reporter in CRAd-infected cells and whole tumors.11
One of the most effective CRAds in ovarian cancer as it pertains to therapeutic efficacy, replication ability, oncolysis, and in vivo tumor resolution has been a chimeric, infectivity-enhanced, conditionally replicating, serotype 5 adenovirus: Ad5Δ24-F5/3.15,16 In this study, we sought to develop a capsid-labeled variant of this powerful CRAd with tk-mRFP1 dual-imaging modality fusion and validate its utility for multimodality imaging applications in various ovarian cancer cell lines.
Materials and Methods
Cell Culture
The ovarian carcinoma cell lines HEY, OV4, SKOV3.ip1 and the normal human epidermal fibroblast cell line VH-10 were used for in vitro experiments. HEY, SKOV3.ip1 and OV4 cell lines were generous gifts from Dr. Judith Wolf and Dr Janet Price (The University of Texas M.D. Anderson Cancer Center, Houston, TX) and Dr. Timothy Eberlein (Harvard University, Cambridge, MA), respectively. The VH-10 cell line was obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Both human embryonic kidney (HEK) 293 and human embryonic retinoblasts 91117 cells were purchased from the ATCC. All cell lines were maintained according to the manufacturers' protocols in recommended media in the presence of 10% fetal bovine serum (FBS), 5 mM glutamine (Cellgro, Mediatech), and 1% penicillin and streptomycin (Cellgro, Mediatech). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Recombinant Ad Construction
All viruses were constructed by previously described methods of homologous recombination in Escherichia coli.18 To generate the double-imaging CRAd Ad5Δ24pIX-tk-mRFP1, F5/3 (heretofore referred to as CRAd pIX-tk-mRFP), we used a shuttle vector pSl Δ24, pIX-tk-mRFP1 and a modified AdEasy-1 vector, AdEz F5/3, which contains a modified F5/3 chimera fiber (knob domain from Ad3 fiber). To generate the AdEz F5/3 backbone we first generated AdEasy-1 with fiber gene, replaced by a unique SwaI site (AdEz ΔF/SwaI), which, following SwaI-linearization, was subsequently recombined with the shuttle vector pKAN3.1F5/3 (Invitrogen, Carlsbad, CA), containing a 3.1 kb XbaI–KpnI fragment of Ad5.
The resulting recombinant was selected and validated via a recombination-specific polymerase chain reaction (PCR) assay of our own design (Borovjagin and colleagues unpublished data, 2007), linearized with PacI and transfected into 293HEK helper cells.
AdEz, ΔF/SwaI was constructed by recombination of the commercial AdEasy-1 vector (Quantum Biotechnologies, Montreal, QC) with pZero2 ΔE3.6.9 shuttle plasmid, which, in turn, was derived from the parental pZero2E3.6.9 plasmid by deletion of the E3 region using a two-step PCR approach.19 The resulting E3 deletion (nts 28,133–30,817) contained three extra nucleotides at each boundary of the deletion as compared with the one in the commercial AdEasy-1 (nts 28,130–30,820).
All viruses carried a 24 bp deletion in the E1A gene identical to the one in the “delta 24” (Δ24) oncolytic adenovirus,20 which made them conditionally replicative (replication competent in cancer cells with Rb-defective pathway).
The rescue vectors for control viruses Ad5Δ24pIX-tk-F5/3 and Ad5Δ24pIX-mRFP1-F5/3 were constructed by the same approach, using AdEz F5/3 and the corresponding shuttle vectors: pSlΔ24pIX-tk and pSlΔ24pIX-mRFP1.
The pSlΔ24pIX-tk shuttle vector was generated from the previously described shuttle vectors pshuttle-wt-pIX-tk12 and pshuttle-d24-pIX-EGFP14 by their digestion with EcoR1 and Hind III and ligation of the 4,165 bp fragment from pshuttle-wt-pIX-sr39tk with the 6,702 bp fragment from pshuttle-d24-pIX-EGFP. The E1-shuttle vector pSl Δ24, pIX-tk-mRFP1 was constructed on the basis of the above--described pSlΔ24pIX-tk shuttle vector by replacing the tk sequence with a PCR-generated fusion tkmRFP1 product between the Nhe I sites. To generate the tk-mRFP1 fusion, the HSV-tk and mRFP1 genes were amplified by PCR using the corresponding pSlΔ24pIX-tk and pSlΔ24pIX-mRFP1 vectors as templates. The following primers were used for tk gene amplification: 5′-GACAAGCTAGCCGGATCCGGTTCAGGCAGTGCTTC-3′ (sense primer) and 5′-GGCGACC--GGTGGATCCCAATTAGCCTCCCCCATCTCCCGGGCA-3′ (antisense primer). The sense primer contained an Nhe I site (underlined) 5 bp away from its 5′ terminus, whereas the antisense primer contained an Age I site (shown in underlined italics), unique in the resulting vector. For mRFP1 amplification, the sense and antisense PCR primers: 5′-ATCCACCGGTCGCCACCATGGCCTCCTCCGAGGACGTCA-3′ and 5′-TA-TGACTAGTCGACTTAGGCGCCGGTGGAGTGG-3′ were used, respectively. The sense primer contained Age I site and the antisense primer contained two overlapping sites: SpeI (underlined) and Sal I (shown in bold). Following purification from 2% agarose gel with a deoxyribonucleic acid (DNA) extraction kit (Qiagen, Valencia, CA), the resulting PCR fragments were digested with Age I and ligated together using the Fast-Link DNA Ligation Kit (Epicentre Technologies, Madison, WI). The ligation was used as a template for another PCR amplification with flanking sense (Nhe I) and antisense (Spe I/Sal I) primers used for amplification of each of the two fragments at the first step. The resulting 1.8 kb–long tk-mRFP1 product was simultaneously digested with Nhe I and Spe I and cloned into the Nhe I–digested and gel-purified pSl Δ24, pIX-tk vector with the tk gene removed because digestion at Spe I and Nhe I sites produces compatible cohesive ends. As a result, the downstream Nhe I and Spe I sites disappeared. All shuttle vectors used in this study carried a linker sequence, encoding 10–amino acid peptide (SADYKDDDDK) with an octapeptide flag motif (shown in bold) at the C-terminus of polypeptide IX coding sequence in place of its “stop” codon followed by a unique Nhe I restriction site. Both pSlΔ24pIX-tk and pSlΔ24pIX-tk-mRFP1 constructs contained an additional 19 bp linker between the Nhe I site and the tk coding sequence, placing tk or tk-mRFP1 fusion in frame with pIX. The size of the linker peptide separating pIX and tk in the fusions was 18 amino acids.
To generate nonreplicative control vector Ad5ΔE1/luc-pIX-mRFP1-F5/3 we used a two-step procedure. First, we generated Ad5ΔE1/luc–pIX-tk-mRFP1-F5/3 rescue vector by using the same procedure as for the construction of its “Δ24” variant except that the tk-mRFP1 fragment was cloned into an Nhe I site of an earlier described parental shuttle vector AdlucpIXpK21 bearing the cytomegalovirus promoter-driven firefly luc gene in place of E1. To generate pSlΔE1/luc-pIX-mRFP1, the tk sequence was excised from the pSlΔE1/luc-pIX-tk-mRFP1 shuttle by Nhe I-Age I digestion and religated with a short-adaptor double-stranded oligonucleotide with Nhe I and Age I overhangs, generated by annealing of two complementary single-stranded oligonucleotides, 5′-CTAGCCTGGGATCCA-3′ and 5′-CCGGTGGATCCCAGG-3′, which restored the 19 bp linker between pIX and mRFP1 sequences, placing the latter in frame with pIX.
The pSlΔ24pIX-mRFP1 construct was derived from the previously described vector pShuttle wt-IX-mRFP1,11 which was digested by Hind III and Pme I to remove the wild-type E1A portion of the Ad sequence and replace it with the Δ24E1A-containing Hind III-Pme I fragment, derived from the previously described pScs Δ24.22 In contrast to the other shuttle vectors, pSlΔ24pIX-mRFP1 contained BamH1 site instead of Nhe I and was missing the last 19 bp of the linker while carrying the same flag peptide sequence (Figure 1).
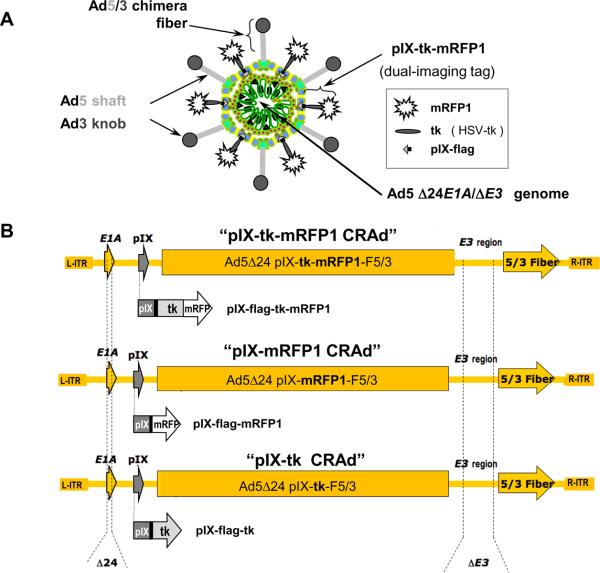
Figure 1.
Schematic representation of the capsid-labeled CRAd genomes. A, pIX-tk-mRFP1 CRAd structure. B, Recombinant genomes of pIX-tk-mRFP1 CRAd (top), pIX-mRFP1 CRAd (middle), and pIX-tk CRAd (bottom) with the corresponding pIX fusions in place of native pIX. All viruses contain Δ24 mutation in E1A, deletion in E3 (ΔE3), and chimera F5/3 fiber, carrying the knob domain of serotype 3 Ad (Ad3).
Virus Recue, Propagation, and Purification
All of the CRAds were rescued in 293HEK cells and then plaque-purified in A549 adenocarcinoma cells to prevent formation and propagation of the wild-type E1-recombinants (also known as “RCA”), which could spontaneously occur in the 293HEK helper cells, carrying the Ad5 E1 genes in the genome. CRAds were upscaled and purified from approximately 3 × 108 infected A549 cells. For virus purification, we used two sequential cesium chloride gradient ultracentrifugation rounds (AdEasy system, QBiogene, Montreal, QC) and two-step dialysis against buffer containing 10 mM Tris HCl, pH 8.0, 50 mM NaCl, 2 mM MgCl, and 10% (v/v) glycerol. Final aliquots of each virus were analyzed for viral particle titer by measuring nucleic acid concentration at A260 and protein concentration by “DC assay” (BioRad, Hercules, CA). The Spearman-Karber method for tissue culture infective dose of 50% of cells (TCID50) was used to determine infection titers. Genetic identity of the genomes isolated from purified viral particles was verified by various PCR-based assays. The RCA-free status of all viral preparations was confirmed by in-house-developed PCR assay of our own design (Borovjagin and colleagues unpublished data, 2007), which uses an E1A sequence-specific PCR primer, complementary to the 24 bp sequence, deleted in the Δ24 Ad mutants. The absence of visible PCR product after 37 PCR cycles was considered evidence for the absence of wild-type (WT) E1 recombinants in the preparations.
Validation of Functionality of the Imaging Fusion Components In Vitro
Tritium (3H)-labeled thymidine conversion assay was performed using a previously reported protocol23 to confirm functionality of the tk moiety on the virus capsid. Reaction mixtures included 50 mM Tris (pH 7.2), 5 mM adeno-sine triphosphate, 5 mM MgCl, 2 mg/mL bovine serum albumin (BSA), and 1 mM of 3H-thymidine (Amersham Biosciences, Westborough, MA). The experiment was performed in triplicate for each purified virus, including CRAd pIX-tk-mRFP1, CRAd pIX-tk, and the previously reported Ad5WTpIX, ΔE3-tk12 virus with tk transgene cloned in place of the Ad5 E3 region as a negative control (for the tk capsid inclusion-associated background). In addition, a CRAd with no tk gene (Ad5Δ24WTpIX) or pIX modification, but otherwise the same genome, served as a negative control. The reaction mixtures were incubated at 37°C for 5 minutes and inactivated by heating to 95°C for 3 minutes to stop the reaction. To quantify the amount of phosphorylated 3H-thymidine, 10 μL from each reaction mixture was mixed with 200 μL of diethylaminoethyl (DEAE)-cellulose and the slurry was centrifuged down and washed three times with 4 mM ammonium acetate and once with methanol. The amount of DEAE-bound radioactivity (phosphorylated 3H-thymidine) was quantified in a scintillation counter using 8 mL of scintillation mixture per sample. The results were plotted as a percentage of the input radioactive substrate.
Validation of functionality of the mRFP1 moiety was carried out by fluorescent microscopy of CsCl-purified imaging viruses (1 × 1010 vp) using an inverted IX-70 stereomicroscope (Olympus, Melville, NY) equipped with a Magnifire digital charge-coupled device camera (Optronics, Golets, CA) at 10x or 40x magnification. Fluorescence signal quantification was carried out by Nuance 2.4.2 spectral imaging software (CRI Inc., Woburn, MA).
Evaluation of CRAd Replication Efficiency
To quantitatively assess CRAd DNA replication in infected cells, we analyzed changes in each virus genome copy number over time by TaqMan quantitative real-time polymerase chain reaction (qPCR) (LightCycler System, Roche Applied Science, Indianapolis, IN) using E4-specific primers as described previously.24 The readings were normalized by copy number of β-actin as a cellular housekeeping gene. For each assay, a known amount of E4 template DNA (108, 106, 104, and 102 copies/μL) was amplified to generate a standard curve for quantification of the E4 copy number in each sample. Thermal cycling conditions were as follows: 10 minutes at 95°C and 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Data were analyzed with LightCycler software.
Ovarian cancer cell lines SKOV3.ip.1, OV4, and HEY and normal human fibroblasts VH-10 as a non-permissive control were infected with a panel of five viruses, including a nonreplicative control, and total cellular DNA was harvested on days 1 and 3 postinfection. The experiment was performed in triplicate. DNA isolation was carried out using the QIAamp DNA Blood Mini kit (Qiagen) according to the manufacturer's instructions.
Validation of CRAd Oncolytic Potency In Vitro
Cytopathic potential of CRAds was assessed by a Cell Titer 96Aqueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) based on the MTS ([3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenol)-2-(4-sulfophenyl)-2H-tetrazolium]) reagent reduction, supported by enzymatic activity of live but not dead cells. Ovarian cancer cells SKOV3.ip.1, OV4, and HEY and VH-10 fibroblasts were plated at 1 × 104 cells/well in 96-well plates in recommended media and allowed to adhere overnight. Cells were infected with varying multiplicities of infection (MOI) (0, 0.01, 0.1, 1, 10, 100 plaque-forming units [PFU]/cell) for each of the five viruses: CRAd pIX-tk-mRFP1, CRAd pIX-tk, CRAd pIX-mRFP1, CRAd WTpIX and Ad5ΔE1/lucpIX-mRFP1. All experiments were performed in triplicate. Four days postinfection, 20 μL of MTS solution was added to each well per each 100 μL of media and cells were incubated for 4 hours at 37°C in 5% CO2-humidified atmosphere. Light absorbance of each sample at 490 nm (A490) was analyzed with a colorimetric plate reader, and the results were quantified and plotted against a previously derived standard curve and plotted as percentage of cell viability relative to uninfected control.
The cell-killing (cytotoxic) potency of CRAds was verified by crystal violet staining assay. SKOV3.ip.1, OV4, HEY, and VH-10 cell lines were plated in 24-well plates in 2% FBS-supplemented Dulbecco's Modified Eagle's Medium: Nutrient Mixture F12 (DMEM/F12) medium and grown to 80% confluency. Cells were infected in 2% FBS medium at an MOI of 100, 10, 1, and 0 PFU/cell. Eight hours after infection, the virus-containing medium was removed and replaced with fresh 2% FBS medium. When the wild-type control virus showed cytopathic effect (CPE) at 1 PFU/cell, the cells were fixed in 10% formaldehyde and stained with 1% crystal violet to visualize live but not dead cells.
Evaluation of CRAd In Vitro Imaging Capabilities
To evaluate imaging capabilities of thymidine kinase, VH-10, OV4, and SKOV3.ip.1 cell lines were plated in separate 24-well plates and 24 hours later infected with pIX-tk-mRFP1, pIX-tk, and pIX-mRFP1 CRAds. No virus was added in the control samples. The virus-containing medium was replaced approximately 8 hours postinfection. Seventy-two hours postinfection, culture medium was aspirated and replaced with internalization buffer. Half of the samples were treated with an excess of unlabeled 1-(2-deoxy-2-fluoro-1-D-arabinofuranosyl)-5-iodouracil (FIAU) substrate to serve as a competitive inhibitor. All wells were treated with 0.2 mL of 10 μCi/mL of 125I-FIAU. The samples were immediately imaged via gamma camera with 1-minute exposure followed by incubation for 2 hours at 37°C, after which medium was aspirated and cells were washed twice with Hank's balanced salt solution (HBSS) (pH 7.2) and reimaged. Cells were lysed with 1M NaOH and imaged for 20 minutes. The resulting lysate was collected and the percentage of labeled FIAU per microgram of protein was calculated for each sample and plotted.
To test in vitro imaging capabilities of the mRFP1 moiety VH-10, OV4, and SKOV3.ip.1, cell lines were plated in recommended 2% FBS medium on 24-well plates and infected with the panel of viruses or mock-infected. Both phase contrast and fluorescent microscopy at 40x magnification were performed using the above-mentioned equipment. Imaging was performed every 24 hours for 4 days.
Results
Genomic Construction and Generation of Capsid-Labeled CRAds
We constructed a new-generation conditionally replicative “delta 24” (Δ24) oncolytic adenovirus carrying a tk-mRFP1 dual-imaging reporter on the capsid, which provides utility of multimodality imaging. This imaging reporter fusion can provide an important benefit of flexibility in imaging method selection, which is useful for clinical applications of ovarian cancer virotherapy.
The new CRAd genomes (see Figure 1), constructed by the conventional method of homologous recombination in bacteria, carried the following modifications: (1) a 24 bp deletion in the E1A gene, rendering Ad replication cancer selective; (2) a genetic fiber modification, providing an optimal infectivity enhancement for ovarian cancer, both in vitro and in vivo; (3) C-terminal fusion of the minor capsid protein pIX with a dual-imaging reporter tk-mRFP1, capable of labeling adenoviral particles via capsid incorporation of its structural component pIX; and (4) deletion of the genomic E3 region to avoid DNA packaging constraints and retain genomic stability of the virus. Owing to the presence of a large number of modifications in the genome, including large pIX modification, it was important to validate genomic stability and biologic properties of the new CRAd. This study represents biologic validation of the new CRAd in ovarian cancer cells and characterizes its imaging capacities in vitro.
Genomic stability of the tk-mRFP1 CRAd was confirmed by PCR-based analysis of the purified virus genomic DNA. PCR size verification of tk-mRFP1 in the packaged genomes suggested that it remained intact and was not removed or shortened by spontaneous deletions in the fusion (data not shown), which could result from biologic selection during virus propagation in culture.
To determine if the biologic characteristics of the virus are hampered by the large size of the tk-RFP1 fusion protein, we constructed capsid-modified control viruses carrying each component of the fusion separately. We observed no reduction in assembly or yields of the tk-mRFP1 virus particles (2.0E+12 viral particles (vp)/mL) compared with those of pIX-tk (1.7E+12 vp/mL) and pIX mRFP1 (2.6E+ 12 vp/mL) CRAds, carrying significantly shorter pIX extensions or even nonmodified pIX, as in the control WTpIX CRAd (6.0E+11 vp/mL). This was evidenced by similar physical titer (vp/mL) and overall yields of each CRAd in large-scale preparations as well as a low amount of defective particles (empty capsids) seen on the cesium chloride gradients of the viral preparations (data not shown).
Comparison of infectious titers of all capsid-modified CRAds with that of the unlabeled but otherwise isogenic control WTpIX CRAd (8.0E+10 TCID50/mL) showed some mild reduction in infectivity for tk-mRFP1 CRAd (2.2E+10 TCID50/mL) and a singly modified pIX-tk CRAd (1.8E+10 TCID50/mL), but not for pIX-mRFP1 control virus (4.0E+10 TCID50/mL). These data suggest that a C-terminal pIX modification with a large fusion (615 amino acids) reporter does not abolish viral infectivity and assembly in infected cells; thus, the new CRAd might have practical utility as a virotherapy agent.
Both Imaging Moieties of the CRAd Capsid-Associated Fusion Are Functional
Activity of the tk enzyme in the context of its fusion with both pIX and mRFP1 as analyzed by a test tube thymidine conversion assay using a (3H)-thymidine substrate revealed no significant difference in the ability of purified viral particles, carrying the pIX-tk-mRFP1 or the pIX-tk fusions, to catalyze thymidine phosphorylation. The enzymatic activity of the capsid-attached tk-mRFP1 and tk moieties was approximately 10-fold higher compared with that of the negative control viruses, carrying no tk gene or the tk transgene cloned in the E3 region of the genome (Ad5-WTpIX-ΔE3-tk). The observed differences were statistically significant (p < .05) (data not shown).
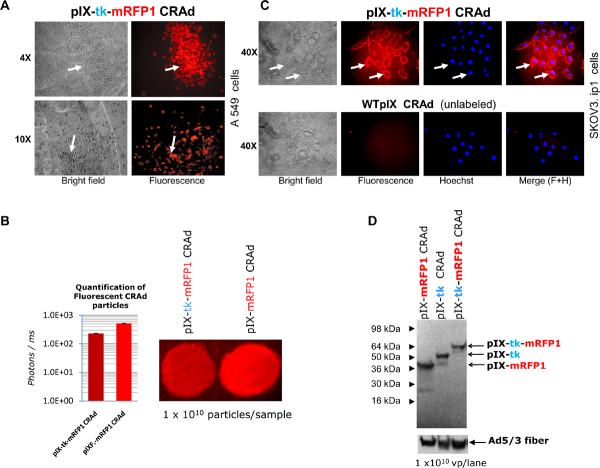
Functionality of mRFP1 reporter protein in the context of its C-terminal fusion with pIX or pIX-tk was evidenced by red fluorescence signals, visualized by fluorescence microscopy of various culture cells on infection with the pIX-tk-mRFP1- or the pIX-mRFP1-labeled CRAds (Figure 2A) and fluorescent imaging of CsCl-purified particles in solutions. The latter provided direct evidence of mRFP1 incorporation in the CRAd capsids, showing only about a twofold but clinically insignificant reduction in pIX incorporation efficiency compared with the pIX-mRFP1 control CRAd (Figure 2B).
Figure 2.
Molecular and biologic validation of fluorescent labeling of the CRAd capsids with mRFP1. A, Visualization of red fluorescence in A549 cells during initial propagation of plaque-purified pIX-tk-mRFP1 CRAd. Arrows point to the infected cells/plaques. B, Quantification of fluorescence. Left bar and sample: pIX-tk-mRFP1 CRAd. Right bar and sample: pIX-mRFP1 CRAd. C, Viral particle tracking in SKOV3.ip.1 cells. Top frames: pIX-tk-mRFP1 CRAd. Bottom frames: WTpIX CRAd. All images 40x objective. Arrows point to individual cells. From left to right: brightfield, fluorescence imaging of virus, Hoechst stain of cellular DNA and Adobe Photoshop 5.5-merged images. D, Western blot of pIX fusion moieties with ANTI-FLAG® M2 Ab confirms appropriate sizes.
Biologic activity of capsid-labeled viral particles was demonstrated by an in vitro cell-binding assay, in which a large excess of purified viral particles was incubated with target SKOV3.ip.1 ovarian cancer cells at 4°C to block virus internalization and synchronize infection at the cell-binding step. Using fluorescence microscopy, we were able to detect an efficient labeling of the cell surface with red fluorescence (Figure 2C). This evidenced the ability of the capsid-labeled CRAd particles to bind to their cognate (Ad3) receptors on the target cells. Furthermore, on switching of cell incubation temperature to 37°C, we could easily track infection of the labeled tk-mRFP1 CRAd from cell membrane to the cytoplasm and later to the nuclei via the capsid-shed pIX-tk-mRFP1 fluorescent protein (data not shown).
Incorporation of the pIX fusion in the capsids and its stability were verified in Western blot by using an equal number of viral particles for each CRAd and antiflag monoclonal antibody for detection of the flag tag at the C-terminus of pIX in each fusion protein. Equal loading of each lane on the gel and fiber incorporation in the capsids were controlled by reprobing of the same blots with antifiber tail (4D2) antibodies (Figure 2D). Staining of an identical gel with Coomassie blue confirmed normal incorporation of other capsid proteins (data not shown).
In Vitro Replication of the Capsid-Labeled CRAds Is Efficient and Cancer Specific
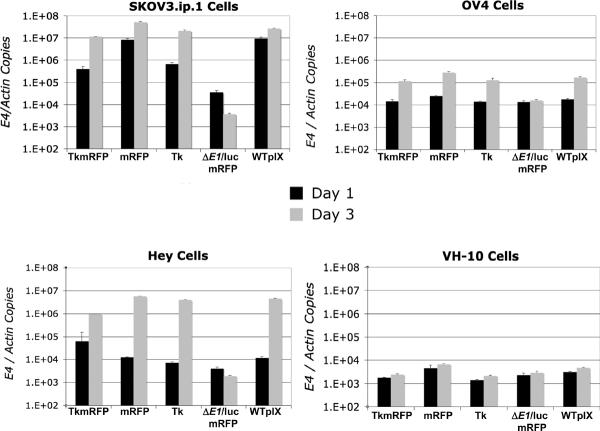
Analysis of viral replication in ovarian cancer cell lines by qPCR demonstrated that incorporation of pIX-tk-mRFP1 fusion in the CRAd capsid caused negligible reduction in the CRAd replication efficiency in most ovarian cancer cell lines compared with WTpIX or pIX-mRFP1 controls. Retention of cancer selectivity was evidenced by minimal replication of the pIX-tk-mRFP CRAd in normal human fibroblasts (Figure 3). Replication of pIX-tk CRAd relative to the above controls was reduced as well but to a lesser extent than for the pIX-tk-mRFP CRAd. The relative rates of the capsid-modified CRAd's replication in different ovarian cancer cell lines remained essentially unchanged, whereas their absolute levels varied dramatically between different cell lines, being the highest in SKOV3.ip and lowest in OV4. All CRAds consistently showed a time-dependent progression in their replication in all cells, as opposed to a nonreplicative pIX-mRFP1 control virus (Ad5ΔE1/luc pIX-mRFP1-F5/3) (Figure 4).
Figure 3.
Replication on analysis. Replication activity in normal human fibroblast and ovarian cancer cell lines. Bar graphs represent the amount of viral replication by quantitative polymerase chain reaction measured at 1 and 3 days postinfection with viruses as housekeeping gene beta-actin-normalized Ad5 E4 DNA copy number. Error bars represent standard deviation of triplicate. TkmRFP = CRAd pIX-tk-mRFP1; mRFP = CRAd pIX-mRFP1; Tk = CRAd pIX-tk; ΔE1/luc mRFP1 = Ad5 ΔEq/luc pIX-mRFP1-F5/3; WTpIX = Ad5 Δ24 WTpIX-F5/3.
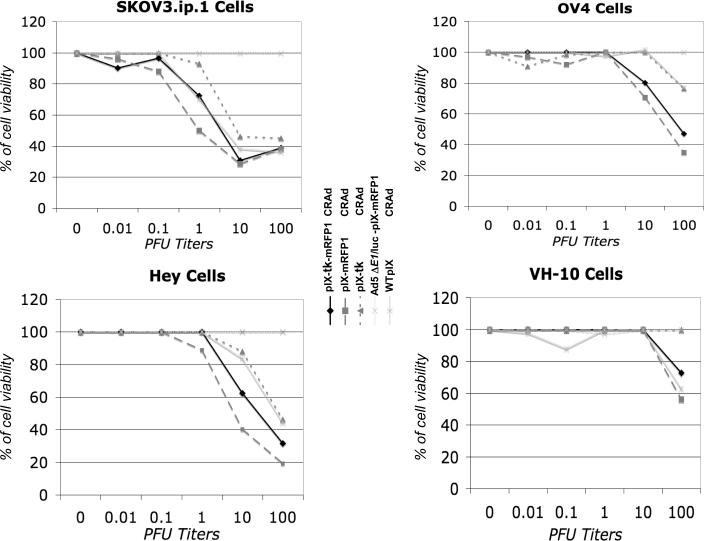
Figure 4.
MTS cell proliferation assay. Cytopathic effect and spread of investigated viruses in normal human fibro-blasts and ovarian cancer cell lines 4 days postinfection at varying multiplicities of infection expressed as % of live cells.
Oncolysis Rates of Tk-mRFP CRAd In Vitro Are Not Diminished
To further investigate the replication-dependent oncolytic capacity of the pIX-tk-mRFP1 CRAd, infected SKOV3.ip.1, OV4, HEY, and VH-10 control cells were analyzed with MTS and crystal violet cell proliferation assays. In MTS cell proliferation assays, the CPE and spread of the pIX-tk-mRFP1 CRAd in ovarian cancer cell lines was not significantly different from that of the WTpIX and the single-moiety controls. This confirmed minimal hindrance of the pIX-tk-mRFP1 fusion on the cell-killing (oncolytic) potency of the virus. SKOV3.ip.1 cells appear to be the most sensitive to both the dual-imaging modality CRAd and the single-modality controls (see Figure 4). In crystal violet assays the efficiency of CPE induction varied with cell type such that SKO3.ip.1 and HEY cell assays were completed by day 7, OV4 by day 8, and VH-10 by day 12. In ovarian cancer lines, CRAd pIX-tk-mRFP1 was indistinguishable from the single-modality CRAds in its oncolytic activity and comparable to the WTpIX control virus. As expected, in normal human VH-10 fibroblasts none of the CRAds exhibited cell killing at an MOI below 100 PFU/cell by day 12, as opposed to in cancer cells, where CPE was observed at an MOI of 1 PFU/cell even at earlier times. Almost a 100-fold lower cell-killing potency in VH-10, as compared with WTpIX control CRAd, demonstrated cancer selectivity of all of the capsid-modified CRAds (data not shown).
Evaluation of Gamma Imaging Capabilities of the tk-mRFP CRAd
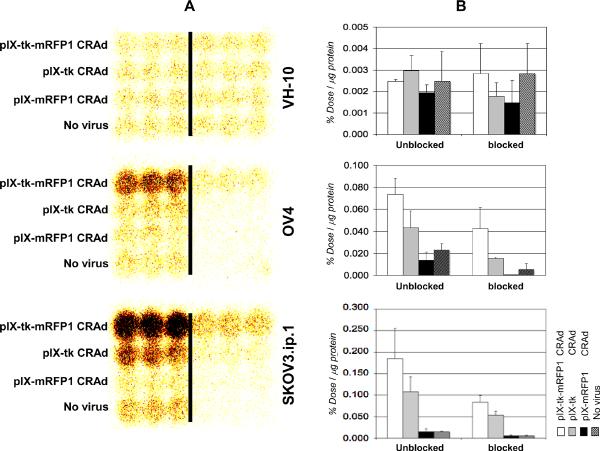
In vitro evaluation of tk enzymatic activity via gamma--camera imaging in the presence of 125I-FIAU as a tracer was performed in SKOV3.ip.1 and OV4 ovarian cancer cell lines as well as control VH-10 normal fibroblasts. We were able to effectively demonstrate minimal tk activity in normal human fibroblasts owing to their nonpermissiveness for CRAd replication. In contrast, we were able to visualize conversion of the radioactive substrate in both ovarian cancer cell lines, which was efficiently blocked by an unlabeled substrate, used as a competitor. Furthermore, gamma-camera imaging in both ovarian cancer cell lines revealed significantly stronger signals in cells infected with the pIX-tk-mRFP1 CRAd compared with a singly-modified pIX-tk control CRAd (Figure 5). This difference is likely to be secondary to the somewhat lower oncolytic potency of the latter CRAd, observed in our MTS and crystal violet experiments (see Figure 4); therefore, more intracellular tracer was converted.
Figure 5.
A, Evaluation of dynamic in vitro thymidine kinase gamma-imaging capabilities. Figure reveals evidence of both replication and function that is at least as active as controls and significantly improved over “no virus” and competitively inhibited controls. Samples to the right of the bar: in the presence of excess unlabeled FIAU. Columns to the left of the bar: in the absence of unlabeled competitor; all cells were labeled with 125I-FIAU. Darkened areas represent tk-converted substrate. B, Quantification of signals shown in A as percentage of labeled FIAU up-take by different ovarian cells per microgram of total protein. Input tracer radioactivity is 100%. Error bars represent standard deviation of three samples for each bar.
Evaluation of the Fluorescent Imaging Capacity of the pIX-tk-mRFP1 CRAd In Vitro
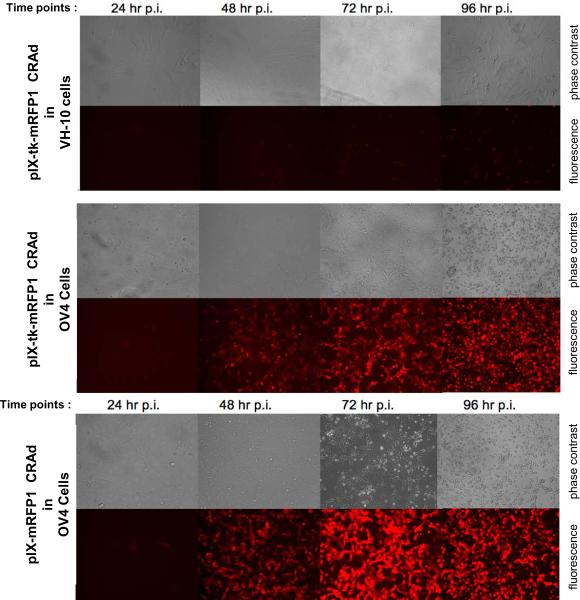
Fluorescence imaging capacity of the capsid-modified CRAds, containing mRFP1 imaging modality on the capsid, was evaluated in SKOV3.ip.1 and OV4 ovarian cancer cell lines by a conventional fluorescence microscopy approach at different time points of infection. We demonstrated proportional fluorescence associated with cytopathic evidence of CRAd replication, revealed by parallel visualization of CPE with phase contrast imaging (Figure 6). At all time points, cells infected with the pIX-tk-mRFP1 CRAd demonstrated slightly weaker fluorescence compared with the single-moiety pIX-mRFP1 CRAd, which was consistent with the difference in replication efficiency observed between these two viruses by qPCR. Moreover, our fluorescence imaging experiments were able to confirm cancer selectivity of CRAd replication and cell killing, revealed by MTS and crystal violet assays.
Figure 6.
In vitro fluorescence imaging. Top panel: CRAd pIX-tk-mRFP1 in VH-10 normal fibroblasts cells. Middle panel: CRAd pIX-tk-mRFP1 in OV4 ovarian cancer cells. Bottom panel: CRAd pIX-mRFP1 in OV4 ovarian cancer cells.
Discussion
Noninvasive imaging reporter categories that have been incorporated into adenoviral genomes include chemical labeling markers,4 bioluminescent,13 fluorescent6,11,25 and nuclear imaging reporter genes,7,12 or their various combinations.7,13 Each reporter category has certain disadvantages. For example, soluble chemical markers are in general not suitable for molecular localization studies. Bioluminescent markers, although useful for increasingly sensitive imaging applications in contrast to fluorescent and nuclear techniques, require reaction with substrate luciferin and are thus difficult to translate to clinical settings.13 Fluorescence-based imaging techniques tend to be more suitable for human applications but have high background noise to overcome and exhibit poor deep tissue penetration except at wave-lengths greater than 600 nm.26 Nuclear imaging techniques, such as gamma camera imaging, SPECT, PET, or CT are the most widely implemented in clinical use and clinical trials for gene therapy vectors27–29 but are often not practical or economical for use in the in vitro setting. These techniques often use short-lived radioactive tracers and are less sensitive than the best light-emitting technologies.30,31
As a result of our ultimate goal to implement our new construct clinically in the context of vector localization, nuclear technology and fluorescent imaging clearly lend themselves best to these endeavors. To that end, we desired to build on constructs previously shown to be promising for clinical use in a manner that may yield a surrogate end point for future clinical investigation of CRAd therapy in the context of treatment of human ovarian cancer. In light of previous genetic modifications, such as the chimeric 5/3 fiber, which was shown to enhance Ad infectivity and replication in ovarian cancer cells,32–34 and the 24 bp deletion in E1, cancer targeting of the Ad replication and the cytolytic potential of the virus,16,34,35 it was important to ensure that additional genetic modifications did not abolish the above improvements in the viral biologic properties.
Owing to the replicative nature of CRAds, for any reporter to be a dynamic reflection of viral propagation and lateralization, the reporter must be genetically engineered into the Ad genome, as opposed to any “one-time” chemical labeling or staining of the virus components, unable to be “inherited” by the progeny particles. We chose the capsid protein pIX gene locale, as opposed to expression of the corresponding transgenes in the E3 or other Ad genomic regions, commonly used for transgene expression. A theoretical advantage of capsid labeling is that any viral particle-associated reporter moiety is functional inside or outside the cell and can be directly visualized shortly after virus administration. These labeled viral particles do not depend on imaging transgene expression or viral replication for visualization. Expression of the imaging transgene, amplified with each viral DNA replication cycle, augments the signal of the virus-associated reporters and depends on the productive infection of the virus. However, after the signal amplification, the capsid-associated signal is likely to be overwhelmed and no longer capable of providing any significant advantage over the transgene expression.
Furthermore, not all types of reporters can function outside the cell owing to the nature of the biochemical mechanism underlying their function and thus provide all of the advantages of genetic capsid labeling. Although the functions of imaging reporters, such as mRFP1 or HSV-tk, do not directly depend on the cell metabolism, only fluorescent reporters can effectively function while being outside the living cell. Despite the fact that the HSV-tk enzyme associated with viral particles can efficiently phosphorylate its substrates even in a test tube, as was demonstrated by this and previous studies,13 it is unlikely to provide a clinically significant imaging benefit outside the cell. The reason is that for the signal to be visualized, the converted product needs to accumulate within the cell to generate a detectable imaging signal. On the other hand, theoretically, association with internalized viral particles could enable the HSV-tk reporter to generate some imaging signal prior to the onset of gene expression and viral replication while coordinated expression with pIX could allow imaging of Ad replication via expression of the transgene in infected cells.
Several studies have used dual- or triple-reporter fusions as imaging tags to enable multimodality imaging applications. In one recent study, a tk-green fluorescent protein (GFP) was expressed in the E3 region of an infectivity-enhanced “delta 24” CRAd (Ad5/3-Δ24-TK-GFP). This CRAd demonstrated effective killing of ovarian cancer cells in vitro, which correlated with GFP expression. Delivery of ganciclovir (GCV) immediately after infection abrogated viral replication and enhanced killing of some cell lines via a bystander effect. The CRAd demonstrated improved antitumor potency over the respective replication-deficient virus with GCV in mouse models of ovarian cancer. However, GCV did not further enhance the efficacy of Ad5/3-Δ24-TK-GFP in vivo, suggesting that the cytotoxic (antitumor) function of HSV-tk may not be compatible with the CRAd oncolytic function.36 The imaging benefit of HSV-tk, however, was not analyzed in that study. Tumor imaging by GFP fluorescence allowed tracking of viral replication; however, the ability of GFP fluorescence to penetrate in mammalian tissues is limited and can be used primarily for superficial tumors.
The advantages of multimodality imaging, including nuclear imaging capabilities of HSV-tk, have been demonstrated.29,37 Ponomarev and colleagues demonstrated the feasibility of noninvasive multimodality fluorescence, bioluminescence, and nuclear imaging of a triple-fusion reporter gene, designed in the context of a retrovirus vector, to simplify and establish proportional coexpression of the three imaging reporter subunits.37 In addition to this triple fusion, the functionality of HSV-tk in the context of fusion proteins has also been demonstrated for HSV-tk/GFP,38 GFP/HSV-tk,39 and HSV-tk/Renilla luciferase40 configurations, suggesting that HSV-tk retains functional activity regardless of its N- or C-terminal position in a fusion.
Our results are consistent with this observation and demonstrate activity of HSV-tk as an internal subunit (component) of a trifusion including N-terminally fused pIX protein and C-terminally fused mRFP1. In the aggregate, we were able to document construction, rescue, propagation, purification, and validation of a genomically stable virus, incorporating both mRFP1 and HSV-tk in the form of a large fusion protein with pIX. Furthermore, modification of the pIX protein with tk-mRFP1 fusion was noted to have a minimal effect on viral specificity, efficiency of replication or oncolysis. The tk moiety of the fusion protein was active and easily imaged in culture, and its activity was undiminished compared with the single-modality controls. These gamma imaging results should be expected to translate to PET/CT activity, given selection of an appropriate radioactive tracer.35 We documented the dynamic in vitro imaging capability of mRFP1 within the fusion protein and subjectively demonstrated proportional fluorescence associated with cytopathic evidence of CRAd replication in multiple ovarian cancer cell lines while sparing normal human cells.
Ultimately, we have created the first reported infectivity-enhanced, ovarian cancer–targeted CRAd with a genetically engineered capsid protein pIX that carries a C-terminal dual-imaging reporter fusion protein composed of both nuclear and red fluorescent imaging modalities. On the basis of the presented data, in vivo validation of the new agent in murine ovarian cancer models using both spectral imaging and SPECT/CT or micro-PET should be performed. Implementation of clinical trials using efficient CRAds with imaging reporters may allow researchers to further understand the mechanisms of viral clearance or visualize clinically relevant nontarget areas of virus sequestration as well as tissue types, facilitating CRAd amplification. Given the lack of virotherapy clinical trials using CRAds with localizing reporters, these data offer a stepping-stone toward a clinical trial that gives hope of gaining incremental inroads into the unknown barriers that prevent meaningful CRAd efficacy in ovarian cancer.
Acknowledgments
Many thanks to Minghui Wang for prompt qPCR analysis.
Financial disclosure of authors: This work was supported by National Institutes of Health grants RO1CA121187 and 5R01CA108585 and University of Alabama at Birmingham's Small Animal Imaging shared facility (P30CA013148). Dr. Joanne T. Douglas holds equity in Vectorlogics, Inc..
Footnotes
Financial disclosure of reviewers: None reported.
References
- 1.American Cancer Society . Cancer facts and figures 2009. American Cancer Society; Atlanta: 2009. [Google Scholar]
- 2.Alemany R, Balague C, Curiel DT. Replicative adenoviruses for cancer therapy. Nat Biotechnol. 2000;18:723–7. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- 3.Kimball KJ, Numnum TM, Rocconi RP, Alvarez RD. Gene therapy for ovarian cancer. Curr Oncol Rep. 2006;8:441–7. doi: 10.1007/s11912-006-0073-x. [DOI] [PubMed] [Google Scholar]
- 4.Kanerva A, Zinn ZR, Peng PW, et al. Noninvasive dual modality in vivo monitoring of the persistence and potency of a tumor targeted conditionally replicating adenovirus. Gene Ther. 2005;12:87–94. doi: 10.1038/sj.gt.3302387. [DOI] [PubMed] [Google Scholar]
- 5.Rivera AA, Davydova J, Schierer S, et al. Combining high selectivity of replication with fiber chimerism for effective adenoviral oncolysis of CAR-negative melanoma cells. Gene Ther. 2004;11:1694–702. doi: 10.1038/sj.gt.3302346. [DOI] [PubMed] [Google Scholar]
- 6.Ono HA, Le LP, Davydova JG, et al. Noninvasive visulaization of adenovirus replication with a fluorescent reporter in the E3 region. Cancer Res. 2005;22:10154–8. doi: 10.1158/0008-5472.CAN-05-1871. [DOI] [PubMed] [Google Scholar]
- 7.Hakkarainen T, Hemminki A, Curiel DT, Wahlfors J. A conditionally replicative adenovirus that codes for a TK-GFP fusion protein (Ad5Delta24TK-GFP) for evaluation of the potency of oncolytic virotherapy combined with molecular chemotherapy. Int J Mol Med. 2006;18:751–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Black ME, Kokoris MS, Sabo P. Herpes simplex virus-1 thymidine kinase mutants created by semi-random sequence mutagenesis improve prodrug-mediated tumor cell killing. Cancer Res. 2001;61:3022–6. [PubMed] [Google Scholar]
- 9.Tang Y, Le LP, Matthews QL, et al. Derivation of a triple mosaic adenovirus based on modification of the minor capsid protein IX. Virology. 2008;377:391–400. doi: 10.1016/j.virol.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Vellinga J, de Vrij J, Myhre S, et al. Efficient incorporation of a functional hyper-stable single-chain antibody fragment protein-IX fusion in the adenovirus capsid. Gene Ther. 2007;14:664–70. doi: 10.1038/sj.gt.3302908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le LP, Le HN, Dmitriev IP, et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98:203–14. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Le L, Sibley DA, et al. Genetic incorporation of HSV-1 thymidine kinase into the adenovirus protein IX for functional display on the virion. Virology. 2005;338:247–58. doi: 10.1016/j.virol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Matthews QL, Sibley DA, Wu H, et al. Genetic incorporation of a herpes simplex virus type 1 thymidine kinase and firefly luciferase fusion into the adenovirus protein IX for functional display on the virion. Mol Imaging. 2006;5:510–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Le LP, Everts M, Dmitriev IP, et al. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging. 2004;3:105–16. doi: 10.1162/15353500200404100. [DOI] [PubMed] [Google Scholar]
- 15.Kanerva A, Zinn KR, Chaudhuri TR, et al. Enhanced therapeutic efficacy for ovarian cancer with a serotype 3 receptor-targeted oncolytic adenovirus. Mol Ther. 2003;8:449–58. doi: 10.1016/s1525-0016(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhu ZB, Lu B, Park M, et al. Development of an optimized conditionally replicative adenoviral agent for ovarian cancer. Int J Oncol. 2008;32:1179–88. doi: 10.3892/ijo_32_6_1179. [DOI] [PubMed] [Google Scholar]
- 17.Fallaux FJ, Kranenburg O, Cramer SJ, et al. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–22. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 18.He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Martín C, Glasgow JN, Borovjagin A, et al. Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. J Mol Biol. 2008;383:923–34. doi: 10.1016/j.jmb.2008.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heise C, Hermiston T, Johnson L, et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6:1134–9. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 21.Dmitriev IP, Kashentseva EA, Curiel DT. Engineering of adenovirus vector containing heterologous peptide sequences in the C terminus of capsid protein IX. J Virol. 2002;76:6893–9. doi: 10.1128/JVI.76.14.6893-6899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nettelbeck DM, Rivera AA, Balagué C, et al. Novel oncolytic adenoviruses targeted to melanoma: specific viral replication and cytolysis by expression of E1A mutants from the tyrosinase enhancer/promoter. Cancer Res. 2002;62:4663–70. [PubMed] [Google Scholar]
- 23.Gerber S, Folkers G. A new method for quantitiative determination of tritium-labeled nucleoside kinase products adsorbed on DEAE-cellulose. Biochem Biophys Res Commun. 1996;225:263–7. doi: 10.1006/bbrc.1996.1164. [DOI] [PubMed] [Google Scholar]
- 24.Rivera AA, Davydova J, Schierer S, et al. Combining high selectivity of replication with fiber chimerism for effective adenoviral oncolysis of CAR-negative melanoma cells. Gene Ther. 2004;11:1694–702. doi: 10.1038/sj.gt.3302346. [DOI] [PubMed] [Google Scholar]
- 25.Sarkioja M, Kanerva A, Salo J, et al. Noninvasive imaging for evaluation of the systemic delivery of capsid-modified adenoviruses in an orthotopic model of advanced lung cancer. Cancer. 2006;107:1578–88. doi: 10.1002/cncr.22209. [DOI] [PubMed] [Google Scholar]
- 26.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs A, Voges J, Reszka R, et al. Positron-emission tomography of vector-mediated gene expression in gene therapy for gliomas. Lancet. 2001;358:727–9. doi: 10.1016/s0140-6736(01)05904-9. [DOI] [PubMed] [Google Scholar]
- 28.Yaghoubi S, Barrio JR, Dahlbom M, et al. Human pharmacokinetic and dosimetry studies of [(18)F]FHBG: a reporter probe for imaging herpes simplex virus type-1 thymidine kinase reporter gene expression. J Nucl Med. 2001;42:1225–34. [PubMed] [Google Scholar]
- 29.Blasberg RG, Tjuvajev JG. Herpes simplex virus thymidine kinase as a marker/reporter gene for PET imaging of gene therapy. Q J Nucl Med. 1999;43:163–9. [PubMed] [Google Scholar]
- 30.Wetterwald A, van der Pluijm G, Que I, et al. Optical imaging of cancer metastasis to bone marrow: a mouse model of minimal residual disease. Am J Pathol. 2002;160:1143–53. doi: 10.1016/S0002-9440(10)64934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De A, Lewis XZ, Gambhir SS. Noninvasive imaging of lentiviral-mediated reporter gene expression in living mice. Mol Ther. 2003;7:681–91. doi: 10.1016/s1525-0016(03)00070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami Y, Li H, Lam JT, et al. Substitution of the adenovirus serotype 5 knob with a serotype 3 knob enhances multiple steps in virus replication. Cancer Res. 2003;63:1262–9. [PubMed] [Google Scholar]
- 33.Kanerva A, Mikheeva GV, Krasnykh V, et al. Targeting adenovirus to the serotype 3 receptor increases gene transfer efficiency to ovarian cancer cells. Clin Cancer Res. 2002;8:275–80. [PubMed] [Google Scholar]
- 34.Bauerschmitz GJ, Lam JT, Kanerva A, et al. Treatment of ovarian cancer with a tropism modified oncolytic adenovirus. Cancer Res. 2002;62:1266–70. [PubMed] [Google Scholar]
- 35.Curiel DT. The development of conditionally replicative adenoviruses for cancer therapy. Clin Cancer Res. 2000;6:3395–9. [PubMed] [Google Scholar]
- 36.Raki M, Hakkarainen T, Bauerschmitz GJ, et al. Utility of TK/GCV in the context of highly effective oncolysis mediated by a serotype 3 receptor targeted oncolytic adenovirus. Gene Ther. 2007;14:1380–8. doi: 10.1038/sj.gt.3302992. [DOI] [PubMed] [Google Scholar]
- 37.Ponomarev V, Doubrovin M, Serganova I, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–51. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs A, Dubrovin M, Hewett J, et al. Functional coexpression of HSV-1 thymidine kinase and green fluorescent protein: implications for noninvasive imaging of transgene expression. Neoplasia. 1999;1:154–61. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paquin A, Jaalouk DE, Galipeau J. Retrovector encoding a green fluorescent protein-herpes simplex virus thymidine kinase fusion protein serves as a versatile suicide/reporter for cell and gene therapy applications. Hum Gene Ther. 2001;12:13–23. doi: 10.1089/104303401450924. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Yu YA, Shabahang S, et al. Renilla luciferase--Aequorea GFP (Ruc-GFP) fusion protein, a novel dual reporter for real-time imaging of gene expression in cell cultures and in live animals. Mol Genet Genomics. 2002;268:160–8. doi: 10.1007/s00438-002-0751-9. [DOI] [PubMed] [Google Scholar]