Abstract
Strong positive Darwinian selection acts on two sperm fertilization proteins, lysin and 18-kDa protein, from abalone (Haliotis). To understand the phylogenetic context for this dramatic molecular evolution, we obtained sequences of mitochondrial cytochrome c oxidase subunit I (mtCOI), and genomic sequences of lysin, 18-kDa, and a G protein subunit. Based on mtDNA differentiation, four north Pacific abalone species diverged within the past 2 million years (Myr), and remaining north Pacific species diverged over a period of 4–20 Myr. Between-species nonsynonymous differences in lysin and 18-kDa exons exceed nucleotide differences in introns by 3.5- to 24-fold. Remarkably, in some comparisons nonsynonymous substitutions in lysin and 18-kDa genes exceed synonymous substitutions in mtCOI. Lysin and 18-kDa intron/exon segments were sequenced from multiple red abalone individuals collected over a 1,200-km range. Only two nucleotide changes and two sites of slippage variation were detected in a total of >29,000 nucleotides surveyed. However, polymorphism in mtCOI and a G protein intron was found in this species. This finding suggests that positive selection swept one lysin allele and one 18-kDa allele to fixation. Similarities between mtCOI and lysin gene trees indicate that rapid adaptive evolution of lysin has occurred consistently through the history of the group. Comparisons with mtCOI molecular clock calibrations suggest that nonsynonymous substitutions accumulate 2–50 times faster in lysin and 18-kDa genes than in rapidly evolving mammalian genes.
Keywords: lysin/abalone/Haliotis/fertilization/intron/positive selection/adaptive sweep/protein evolution
Cell surface recognition proteins, which provide the molecular interface between individuals interacting in competition, reproduction, or pathogenicity are subjected to strong selection. For example, conflicting interests of pathogens and hosts may cause antigen and immune proteins to evolve rapidly in a race for survival (1). Molecular signals of adaptive evolution have been found in the excess of nonsynonymous (Dn) over synonymous (Ds) nucleotide substitution in a variety of intercellular recognition genes (2, 3). Some of the highest Dn/Ds ratios are found in comparisons of cDNA sequences of sperm protein genes from abalone (Haliotis) that interact species-specifically with egg surface proteins during fertilization (4–8). Positive selection on sperm proteins mediating species-specific fertilization suggests that adaptive evolution might be involved in establishing reproductive isolation between species. A variety of explanations for strong positive selection on fertilization proteins have been proposed (8, 9). Recently, studies of genes encoding the abalone egg receptor for lysin have revealed a molecular mechanism for adaptive co-evolution of lysin (10).
Abalone lysin is released by the sperm acrosome reaction and nonenzymatically creates a hole in the egg vitelline envelope (8) by binding to a high molecular weight glycoprotein (7). The 18-kDa protein, also released from the sperm acrosome, coats the acrosomal process and may mediate membrane fusion between sperm and egg (11). Despite the high sequence divergence between these two sperm proteins, congruent patterns of secondary structure prediction indicate they arose by gene duplication (6, 8). Between-species comparisons of cDNAs revealed excess nonsynonymous nucleotide substitution over synonymous substitution; Dn/Ds ratios ranged from 0.95–4.10 for lysin (4, 5) and 0.76–4.67 for 18 kDa (6).
Information on phylogenetic relationships and divergence times of abalone species could illuminate patterns of molecular evolution in these genes and contribute to knowledge of the evolution of species-specific fertilization. Many of the present California abalone species are represented as Pliocene and Pleistocene fossils (12, 13), and some of these have been matched to fossils from the Miocene (14). However, abalone fossils are relatively rare, and it has not been possible to use them to infer relationships nor to date divergence times among species. Phylogenetic relationships based on lysin cDNA sequences (15) are the most complete yet published for Haliotis. However, nonneutral evolution of lysin genes might affect phylogenetic reconstruction, particularly if selection differs with lineage.
Here we report a phylogenetic reconstruction of partial mitochondrial cytochrome c oxidase subunit I (mtCOI) sequences of abalone. We also compare intron sequences within and between species for lysin and 18-kDa genes. For positive selection to act, genetic variation must be present, and some gamete recognition proteins are highly polymorphic (9). However, a preliminary survey of cDNA sequences revealed little evidence of within species polymorphism in abalone lysins (4, 5). We searched further for variation in red abalone by surveying mtCOI sequences, and introns of lysin, 18-kDa, and G protein loci.
MATERIALS AND METHODS
Abalone Samples and Template DNA.
Red abalone (Haliotis rufescens) were collected in California at La Jolla, Santa Barbara, San Miguel Island, and Mendocino. Animals were of both sexes with shell lengths ranging from 9 to 22 cm. Haliotis corrugata and Haliotis fulgens were collected at La Jolla. A peripheral epipodial tentacle was clipped from each living abalone, frozen on dry ice, and stored at −80°C. Samples of Haliotis sorenseni, Haliotis kamtschatkana, Haliotis walallensis, Haliotis cracherodii, Haliotis discus hannai, Haliotis cyclobates, Haliotis rubra, Haliotis midae, and Haliotis iris were collected at localities previously described (4, 5) and shipped in 50% ethanol.
To prepare template DNA, a single abalone tentacle, or piece of ethanol-preserved testis, was washed in 20 mM Tris (pH 7.6) and 20 mM EDTA and macerated in 200–500 μl of the same solution containing 10% (vol/vol) chelating resin (Sigma) in a 1.5-ml tube. The tube was boiled 5 min, vortex mixed, centrifuged, and stored at −20°C.
PCR Amplification of mtDNA Sequences.
A segment of the mtCOI gene was amplified from purified H. rufescens genomic DNA by using universal primers (16). Using the resulting sequence, abalone-specific mtCOI primers F1 (TGATCCGGCTTAGTCGGAACTGC) and R1 (GATGTGTTGAAATTACGGTCGGT) were designed. These primers correspond to nucleotide positions 64–86 and 622–644 in the complete mtDNA sequence of a chiton (17). mtCOI PCRs (50–100 μl) contained each primer at 0.4 μM, Taq polymerase (Promega) at 10 units/ml, Taq polymerase buffer (Promega), 1.5 mM MgCl2, 0.2 mM of each dNTP, and 1–2 μl of template DNA. Thermal cycle settings were 94°C for 30 sec, 53°C for 30 sec, and 72°C for 60 sec for 35 cycles.
PCR Amplification of Genomic DNA Sequences.
Species-specific primers were designed from lysin and 18-kDa coding sequences of H. rufescens, H. corrugata, and H. fulgens (4, 6) and used in amplifications with tentacle or testis DNA templates. These exon primers produced PCR products that span introns. The following forward (f) and reverse (r) primers are identified by nucleotide positions in the H. rufescens lysin and 18-kDa cDNA sequences (GenBank accession nos. M34388 and L36552). The lysin primers were “16-Af”: 13-32; “16-Cr”: 252–230; “16-4f”: 330–348; “16-12r”: 462–439. The 18-kDa primers were “18-10f”: 49–68; “18-3r”: 279–259; “18-4f”: 350–371; “18-12r”: 501–481. Based on previous alignments (4, 6), similarly positioned primers were also made for H. fulgens lysin and 18-kDa cDNAs (GenBank accession nos. M59972 and L36589). Primers s1 and s2 (18) span an intron in the abalone G protein α subunit gene.
PCRs (50–100 μl) contained each primer at 0.4 μM, Taq polymerase (Promega) at 10 units/ml, Taq Extender (Stratagene) at 10 units/ml and Taq Extender buffer, 0.2 mM of each dNTP, and 1–2 μl of template DNA. Reactions were incubated at 94°C for 1 min prior to starting the thermal cycling. Thermal cycle settings for genomic amplifications were 94°C for 30 sec, 55°C for 30 sec, and 72°C for 60–300 sec for 35–38 cycles.
Sequencing PCR Products.
PCR products from complete reactions or excised agarose gel bands were purified by using QiaQuick spin columns (Qiagen, Chatsworth, CA). Cycle sequencing reactions were performed by using the Prism fs Kit (Applied Biosystems) and included 60–90 ng of PCR product. mtCOI PCR products were sequenced by using the two PCR primers. Lysin, 18-kDa and G protein PCR products were sequenced by using the PCR primers and internal primers. Sequences were obtained by using an automated sequencer (Applied Biosystems).
Because sequences were obtained directly from PCR products, nucleotide differences between the two alleles of nuclear genes might be masked; this study detected alleles that predominated in amplification. In the survey of intraspecific polymorphism, all red abalone individuals supported amplification with the lysin and 18-kDa primers. A few sequence positions had overlapping peaks on chromatograms. However, in most cases such double peaks showed the same pair of nucleotides in all sequences, which probably reflects sequencing artefacts rather than allelic polymorphism.
Sequence Analysis.
Sequences were aligned by inspection using sequencher 3.0 (Gene Codes). Sequences have been deposited in GenBank under accession numbers AF060835–AF060854 for mtCOI, AF070955–AF070960 for G protein intron, and AF076819–AF076837 for lysin and 18-kDa introns. mega (19) was used to obtain transition/transversion ratios, genetic distances, Dn and Ds distances, differences in deduced amino acid sequences, and bootstrapped neighbor joining trees. The GCG program rnafold (20) was used to search for secondary structure in introns. Parsimony trees were obtained by using paup (21). Stepmatrices appended to the data file were used to introduce transversion weighting. Maximum likelihood and neighbor joining trees were obtained by using phylip (22).
RESULTS
mtDNA Phylogeny.
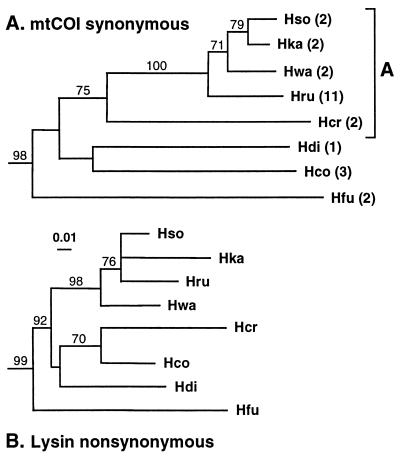
mtCOI sequences were obtained from 12 abalone species. Aligned nucleotide sequences of 528 bases included 168 variable sites and 138 phylogenetically informative sites. Eight north Pacific species form a clade distinct from four outgroup species from Australia (H. cyclobates and H. rubra), New Zealand (H. iris), and South Africa (H. midae) (Fig. 1A). Seven species inhabit the North American coast and overlap in California; the remaining species is from Japan (H. discus hannai). Nucleotide differences among the eight species were all synonymous. Four California species (H. rufescens, H. sorenseni, H. kamtschatkana, and H. walallensis) group together (Fig. 1A) and are about 3% or less divergent in mtCOI (Table 1). These species are 7.5–8.4% divergent from H. cracherodii. Together, these five California species are referred to as “group A.” Sequence differences in the remaining California species comparisons range from 9.7 to 12.9% (Table 1).
Figure 1.
(A) North Pacific clade from a neighbor joining tree based on synonymous distances (mega; ref. 19) in 528 bp of mtCOI from 12 abalone species. Percent support (70% or higher) in 500 bootstrap searches is shown on branches. (Bar = 1%.) Unless otherwise indicated, samples were collected in California. Hru: H. rufescens; Hso: H. sorenseni; Hwa: H. walallensis; Hka: H. kamtschatkana; Hcr; H. cracherodii; Hco: H. corrugata, Hdi: H. discus hannai (Japan); Hfu: H. fulgens. The first five of these species are designated group A. The number of individuals sequenced is shown in parentheses. Bootstrap values on branches defining each species are >95%. The tree included four additional species (data not shown): H. cyclobates (Australia), H. rubra (Australia), H. midae (South Africa), H. iris (New Zealand), and was midpoint rooted. For additional sample collection details, see ref. 5. (B) North Pacific clade from a neighbor-joining tree based on Dn distances in lysin coding sequences from the same 12 abalone species (4, 5).
Table 1.
mtCOI distances used to estimate lysin and 18-kDa nonsynonymous rate in California abalone
| Species* | K2P%† | COI Div.,‡ Mya | Third-Tv%§ | Tv Div.,¶ Mya | COI Ds%‖ | Lysin Dn%** | Lysin Rate,‡‡ ×10−9 | 18-kDa Dn%†† | 18-kDa Rate,§§ ×10−9 |
|---|---|---|---|---|---|---|---|---|---|
| Hru-Hso | 2.5 | 1.3 | <1 | <1 | 9.9 | 5.8 | 22 | 8.1 | 36 |
| Hru-Hka | 3.1 | 1.6 | <1 | <1 | 12.3 | 10.7 | 33 | 18.5 | 58 |
| Hru-Hwa | 2.7 | 1.4 | 0 | <1 | 10.8 | 10.1 | 36 | ||
| Hso-Hka | 1.0 | 0.5 | 0 | <1 | 3.7 | 8.4 | 84 | 14.5 | 145 |
| Hso-Hwa | 1.5 | 0.8 | <1 | <1 | 6.0 | 6.4 | 40 | ||
| Hka-Hwa | 2.1 | 1.1 | <1 | <1 | 8.3 | 13.6 | 62 | ||
| Hru-Hcr | 8.4 | 4.2 | 1.7 | 3.4 | 37.3 | 21.8 | 27 | ||
| Hso-Hcr | 7.7 | 3.9 | 2.3 | 4.6 | 34.0 | 20.8 | 26 | ||
| Hka-Hcr | 7.5 | 3.8 | 2.3 | 4.6 | 32.7 | 26.1 | 33 | ||
| Hwa-Hcr | 7.5 | 3.8 | 1.7 | 3.4 | 33.1 | 22.7 | 28 | ||
| Hru-Hco | 9.9 | 5.0 | 4.1 | 8.2 | 43.6 | 14.6 | 9 | 82.6 | 50 |
| Hru-Hfu | 12.5 | 6.3 | 4.8 | 9.6 | 61.3 | 24.9 | 13 | 81.1 | 42 |
| Hso-Hco | 10.4 | 5.2 | 3.5 | 7.0 | 46.4 | 13.7 | 10 | 75.6 | 54 |
| Hso-Hfu | 12.9 | 6.5 | 5.4 | 10.8 | 64.9 | 25.4 | 12 | 76.9 | 36 |
| Hka-Hco | 10.2 | 5.1 | 3.5 | 7.0 | 44.9 | 18.1 | 13 | 82.3 | 59 |
| Hka-Hfu | 11.7 | 5.9 | 5.4 | 10.8 | 56.7 | 24.5 | 11 | 85.5 | 40 |
| Hwa-Hco | 9.7 | 4.9 | 4.1 | 8.2 | 42.9 | 15.9 | 10 | ||
| Hwa-Hfu | 12.5 | 6.3 | 4.8 | 9.6 | 62.2 | 23.8 | 12 | ||
| Hcr-Hco | 11.2 | 5.6 | 6.0 | 12.0 | 55.2 | 13.1 | 5 | ||
| Hcr-Hfu | 11.2 | 5.6 | 6.7 | 13.4 | 54.3 | 27.8 | 10 | ||
| Hco-Hfu | 12.3 | 6.2 | 9.3 | 18.6 | 62.4 | 24.0 | 6 | 86.9 | 23 |
Species designations as in Fig. 1. First 10 comparisons are within group A.
Kimura two-parameter pairwise percent distances (mtCOI all sites).
Divergence time (Myr) calculated by using COI calibration: 2% pairwise divergence per Myr.
Percent transversion differences at third base positions of codons.
Divergence time (Myr) calculated using cytochrome b third base transversion calibration: 0.5% pairwise divergence per Myr (24).
Synonymous substitutions (Ds) per 100 synonymous sites in mtCOI.
Nonsynonymous substitutions (Dn) per 100 nonsynonymous sites in mature lysin coding sequences (4, 5).
Lysin Dn/(2× divergence time) = estimated nonsynonymous substitutions/site/year × 10−9.
Nonsynonymous substitutions (Dn) per 100 nonsynonymous sites in mature 18 kDa coding sequences (6). Note: 18 kDa cDNA sequence reported for H. assimilis, a subspecies of H. kamtschatkana.
The 18-kDa Dn/(2× divergence time) = estimated nonsynonymous substitutions/site/year × 10−9. Note: Nonsynonymous rate estimates within group A were calculated by using all sites (2% per Myr); rate estimates for more distant comparisons were calculated by using third position transversions (0.5% per Myr).
Abalone mtCOI third codon positions showed an average of 66% A+T content (range, 62–72% in different species). Such bias increases susceptibility to saturation of nucleotide changes, which impacts phylogenetic reconstructions and molecular clock calibrations (23, 24). Reflecting these effects, the ratio of transitions (ts) to more slowly accumulating transversions (tv) decreases with greater divergence in comparisons of abalone mtCOI sequences. Between the five group A species the average ts/tv ratio is 10.5 (range, 7–15). For more distant California comparisons the average ts/tv ratio is 5.5 (range, 2.9–7.3). In comparisons between California species and southern hemisphere outgroup species the ratios decline to 1–2, indicating saturation. The ts/tv ratios in group A are similar to those found in mtDNA of other closely related species of the temperate marine gastropods Nucella (24) and Tegula (25). Based on these values, transversion weighting was applied to phylogenetic reconstructions. Fig. 1A shows the neighbor joining tree based on synonymous distances and equal weighting. The same topology between species was also obtained by using all nucleotide sites in both paup and maximum likelihood reconstructions using 1:1, 2:1, and 4:1 transversion weighting. With 8:1, 10:1, and 12:1 transversion weighting, the branches for H. fulgens and H. corrugata exchanged positions, but otherwise topologies remained the same.
Rate of Molecular Evolution of Lysin and 18-kDa Genes.
Comparison of the mtCOI tree (Fig. 1A), which reflects synonymous differentiation, with the corresponding lysin tree (Fig. 1B) based on Dn distances shows that accumulation of these different kinds of nucleotide changes occurs on a similar scale. Dn values between species in both lysin and 18 kDa are similar to, or greater than, mtCOI Ds distances (Table 1). This result is most apparent in closely related species (group A) where saturation of mtCOI (and lysin) is minimal. In Fig. 2, the number of amino acid differences in pairwise comparisons of lysins or 18-kDa proteins is plotted against the corresponding mtCOI divergence for all 12 abalone species.
Figure 2.
Number of amino acid differences between lysins or 18-kDa proteins plotted against corresponding mtCOI Kimura two parameter corrected percent difference. All pairwise comparisons of mature lysin from 12 species (4, 5), or mature 18 kDa from 5 species (6), are shown. Multiple changes per site are likely to be substantial in comparisons with >8–10% mtCOI difference.
Estimates of the rate of mtCOI evolution range from 1.6 to 2.6% per million years (Myr) in different marine invertebrate phyla (26–28). North Pacific species of Tegula, which probably diverged in the early Pliocene, show mtCOI divergence rates in this range (25). Based on these rate estimates, we used a calibration of 2% mtCOI divergence per Myr to estimate abalone divergence times (Table 1). The four most closely related species, which have mtCOI distances 3% or less, probably diverged <2 million years ago (Mya). Distances between these species and H. cracherodii indicate divergence about 4 Mya.
Distant comparisons of abalone mtCOI with multiple changes per site may underestimate divergence times. For such comparisons, the relatively slow accumulation of transversions has been used to improve divergence estimates (23, 24). Studies of this kind have included two gastropod genera, Nucella (24) and Littorina (29), which have North Pacific species. These studies examined transversions at third base positions of codons in sequences from the mitochondrial cytochrome b gene, and estimated divergence rates to be 0.4–0.7% per Myr. This calibration (0.5% per Myr) was applied to give approximate divergence times for abalone species in which transitions are saturating. The results suggest California species may have diverged up to 18.6 Mya (Table 1). Divergence calculated between north Pacific species and the southern hemisphere outgroup species ranges from 40 to 70 Mya (data not shown). Transversion substitution rates depend on the availability of 4-fold sites; the greater the proportion of such sites per codon, the greater the expected rate (23). The ratio of 4-fold to 2-fold sites at third base positions in cytochrome b sequences is near 1:1, but in the abalone mtCOI sequences this ratio is 1.6:1. Hence the cytochrome b transversion calibration may underestimate divergence rates and overestimate divergence times when applied to mtCOI sequences. This provides a useful comparison with the 2% per Myr mtCOI calibration, which may underestimate divergence times due to saturation (Table 1).
Based on the divergence time estimates and Dn distances between species for lysin (4, 5) and 18 kDa (6), rates of nonsynonymous substitution per site per year were estimated. For closely related species of group A, divergence times used for these calculations were based on the 2% per Myr mtCOI calibration. Remaining estimates used the 0.5% per Myr third base transversion calibration. The range of the resulting estimates is 5–84 × 10−9 substitutions/nonsynonymous site/year for lysin and 23–145 × 10−9 substitutions/nonsynonymous site/year for 18 kDa (Table 1).
Between–Species Comparisons of Lysin and 18-kDa Introns and Exons.
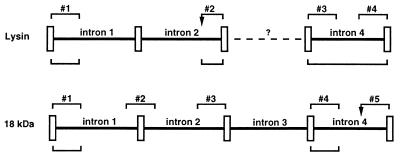
Abalone cDNA sequences for lysin and 18 kDa (4–6) were used to design primers for genomic DNA amplifications. Comparisons between genomic and cDNA sequences indicated intron positions (Fig. 3). Three pairs of introns are in identical positions in the two genes. Each intron pair also shares the same phase in the interrupted codon. Concordance of intron position and phase in lysin and 18-kDa genes confirms their origin by gene duplication (6). Sequence similarity was not found in comparisons of corresponding homologous introns between the two genes, indicating substantial divergence since duplication. The position of an intron in lysin corresponding to the third intron in 18 kDa could not be determined (30).
Figure 3.
Diagram of exons and introns of lysin and 18-kDa genes (not to scale), showing segments sequenced. Rectangles denote the five exons. Sequences compared between H. rufescens individuals are shown by numbered brackets above the gene (Table 3). Comparisons between species (H. rufescens, H. corrugata, and H. fulgens) are shown by brackets below (Table 2). Arrows in segment 2 of lysin and segment 5 of 18 kDa indicate positions of a poly-G region and CA microsatellite.
Complete sequences of lysin intron 4 and partial sequences of lysin intron 1 were obtained for H. rufescens, H. corrugata, and H. fulgens. Partial sequences of 18-kDa introns 1 and 4 were obtained from H. rufescens and H. fulgens (Fig. 3). Comparisons between species showed low levels of intron sequence divergence, ranging from 2.7 to 6.9% (Table 2). Lysin intron segments compared between species had 3.2- to 6.7-fold fewer nucleotide differences than lysin exons (Table 2). In the most extreme case, 18-kDa exons of H. rufescens and H. fulgens differ by 81.3%, yet introns differ by only 3.4–6.8%.
Table 2.
| Gene region‡ | Hru-Hco | Hru-Hfu | Hco-Hfu |
|---|---|---|---|
| Lysin§ | |||
| Exons (420 bp) | 13.7 ± 2.0 | 24.1 ± 2.8 | 22.1 ± 2.6 |
| Exons-Dn | 14.6 ± 2.3 | 24.9 ± 3.2 | 24.0 ± 3.2 |
| Exons-Ds | 10.6 ± 3.5 | 21.3 ± 5.3 | 16.0 ± 4.4 |
| Intron 1 (410 bp) | 2.9 ± 0.8 | 3.6 ± 0.9 | 4.6 ± 1.1 |
| Intron 2 (220 bp) | 2.7 ± 1.4 | 5.5 ± 2.0 | 6.9 ± 2.2 |
| Intron 4 (1557 bp) | 3.5 ± 0.6 | 5.4 ± 0.7 | 6.1 ± 0.9 |
| 18 kDa¶ | |||
| Exons (447 bp) | 83.6 ± 7.7 | 81.3 ± 7.5 | 92.8 ± 8.4 |
| Exons-Dn | 82.6 ± 8.6 | 81.1 ± 8.5 | 86.9 ± 8.9 |
| Exons-Ds | 87.0 ± 17.5 | 78.2 ± 15.4 | 114.2 ± 23.8 |
| Intron 1 (400 bp) | ND | 3.4 ± 0.9 | ND |
| Intron 4 (345 bp) | ND | 6.8 ± 1.5 | ND |
| G protein | |||
| Intron (640 bp) | 7.8 ± 1.6 | 6.2 ± 1.4 | 8.1 ± 1.2 |
Introns also differ between species in several indels and direct repeats (data not shown). For the completely sequenced lysin intron 4, sliding windows along the aligned sequences confirmed that differentiation between species is low throughout the intron. Searches for base complementarity within this intron revealed little potential for RNA secondary structure. For comparison with lysin and 18-kDa introns, a G protein intron was sequenced from the same three species. Divergence of the G protein intron between species is 6.2–8.1%, which is similar to the values found for lysin and 18-kDa introns (Table 2).
Lysin intron differentiation between species is 2.3–5.9 times lower than Ds calculated for lysin exons. Likewise, 18-kDa intron differentiation is 12–23 times lower than Ds in 18-kDa exons (Table 2). To examine high Ds in exons, silent differences were counted in 4-fold codons. In lysin, 23 4-fold positions are shared in seven California species; there are only two differences (both in H. fulgens). In 18 kDa, four 4-fold positions are shared among five species; only one position differs (in H. fulgens). Similarly, among the four most closely related species there are no differences in 44 shared 4-fold positions of lysin and two differences in 38 shared 4-fold positions of 18 kDa. Silent differentiation in exons may actually be similar to the low levels of differentiation observed in introns. When Dn is high, calculated values of Ds might be inflated (31, 32).
Genetic Variation in Red Abalone.
Red abalone (H. rufescens) were sampled from La Jolla, Santa Barbara, San Miguel Island, and Mendocino, representing a range of ≈1,200 km. To survey within species variation in lysin and 18-kDa genes, exon/intron sequence segments (Fig. 3) were obtained. Most of these exon/intron segments were obtained from 6 to 19 individuals (Table 3), including at least 3 individuals from both range extremes. Over 29,000 nucleotides were surveyed; only 2 nucleotide differences were found. Sequence chromatograms went out of register at a CA microsatellite in 18-kDa intron 4 and at a poly-G region in lysin intron 2 (Fig. 3, Table 3), revealing length variation between alleles at these two sites.
Table 3.
Nucleotide sequence variation in red abalone
| Region* | Intron | n† | Length‡ | Differences§ |
|---|---|---|---|---|
| Lysin | ||||
| 1 | 1 | 6 | 410 | 1 ts |
| 2 | 2 | 6 | 280 | poly-G |
| 3 | 4 | 6 | 455 | 1 ts |
| 4 | 4 | 6 | 410 | 0 |
| 18 kDa | ||||
| 1 | 1 | 12 | 480 | 0 |
| 2 | 1, 2 | 19 | 460 | 0 |
| 3 | 2 | 8 | 480 | 0 |
| 4 | 4 | 2 | 430 | 0 |
| 5 | 4 | 2 | 400 | CA-repeat |
| G protein | 1 | 6 | 750 | 1.4% |
| mtCOI | — | 11 | 528 | 0.4% |
Lysin and 18-kDa exon/intron sequence segments as shown in Fig. 2.
Number of individuals sequenced.
Bases of sequence compared.
Nucleotide differences in each comparison; ts: transition; poly-G region and CA repeat show length variation; average percent difference shown for mtCOI and G protein intron.
G protein introns sequenced from six red abalone (three from La Jolla and three from Mendocino) ranged from 0 to 2.2% different (average, 1.4%) between individuals (Table 3), showing that genetic variation exists in other introns of this species. mtCOI sequences from 11 individuals include 7 variable positions with 5 transition and 2 transversion differences. Percent sequence differences among all pairwise comparisons range from 0 to 1.0% (average, 0.4%). Phylogenetic reconstructions based on mtCOI from 12 abalone species (Fig. 1A), which included all 11 red abalone sequences, consistently showed them to be monophyletic with 98–100% bootstrap support. No evidence of geographic structure in red abalone mtDNA haplotypes was found.
DISCUSSION
Lysin and 18-kDa Introns.
Introns from lysin and 18-kDa reveal two unusual patterns of molecular evolution: intron divergence between species is substantially lower than exon divergence, and intron nucleotide polymorphism within species is low. Generally, introns have little functional constraint and accumulate neutral mutations including indels. As a result, intron sequences compared between species typically show several times more differentiation than nonsynonymous sites in exons (32). However, in comparisons among three abalone species, nonsynonymous differences in lysin and 18-kDa genes are several times greater than the corresponding intron differences (Table 2). High divergence of exons relative to introns has also been reported for snake venom phospholipase A2 loci (33, 34). These loci show high Dn/Ds ratios and are subjected to positive selection, presumably because prey can evolve resistance to venoms. Snake venom introns might evolve slowly because of functional constraints (34, 35); alternatively, positive selection may cause rapid divergence of exons, leaving introns relatively unchanged (33). In genes of the major histocompatibility complex, exons are highly polymorphic and evolve by positive selection, yet introns are relatively conserved, probably as a result of the homogenizing effects of recombination (36).
Lysin and 18-kDa introns might be subject to unknown functional constraints. However, in comparisons between species, these introns show indels, low differentiation along all sequences, and differentiation similar to the G protein intron (Table 2). These results suggest that molecular evolution at neutral, or nearly neutral, regions of the abalone genome is generally low when compared with coding regions of the two fertilization genes.
Introns and silent sites in adjacent exons are expected to accumulate differences at similar rates (32). However, Ds values calculated in lysin and 18-kDa exons are substantially greater than differences in introns (Table 2). A similar discrepancy was found in snake venom loci (33). Lysin and 18-kDa comparisons between the most closely related abalone species show mostly nonsynonymous differences and high Dn/Ds ratios, but in comparisons of more divergent species, Dn and Ds values converge (5, 6, 8). For divergent comparisons of these genes, accumulation of a large number of changes, most of which are nonsynonymous, might lead to overestimation of synonymous differences.
Low Polymorphism and Adaptive Sweeps.
High interspecific divergence and low polymorphism have been found in genes mediating sexual recognition in green algae and other phyla (37). A previous survey of lysin cDNA sequences from seven individual red abalone revealed only one transition in the 3′ untranslated region of one individual (5). The present survey revealed little variation in exon/intron segments of both lysin and 18-kDa genes. Low levels of genetic variation within species may reflect a variety of processes, including gene conversion, strong purifying selection, population bottlenecking, selective sweeps resulting from adaptive evolution, and selection on linked loci in regions of low recombination (38, 39).
Low levels of polymorphism in sperm protein genes are not paralleled by similar low levels elsewhere in the nuclear and mitochondrial genomes of red abalone. Furthermore, a study of allozyme variation in this species revealed variation at four polymorphic loci (40). In the present study, polymorphism was found in both G protein introns and mtCOI. mtDNA is particularly susceptible to loss of variation by drift in small populations (41); hence low variation at sperm protein loci was not caused directly by population bottlenecking. The strikingly high Dn/Ds ratios characteristic of these abalone fertilization genes indicate that lack of polymorphism is probably the signature of adaptive sweeps resulting from strong positive selection. The pattern is seen in both loci, suggesting independent sweeps of lysin and 18-kDa genes unless they are closely linked.
Lysin and 18-kDa Evolution Versus mtCOI.
North Pacific abalone species group together, distinct from southern hemisphere species, in phylogenetic reconstructions based on synonymous distances in mtCOI and nonsynonymous distances in lysin (Fig. 1). Both kinds of molecular evolution involve a rapid accumulation of nucleotide differences. In mtCOI this is largely caused by a regular fixation of silent mutations. However, in the sperm protein loci, positive selection appears to create a similar pattern for nonsynonymous substitution. This pattern contrasts with that seen in phylogenetic reconstructions of protein evolution in which rapid evolution is highly episodic (42, 43). Lysin evolves by positive selection, whereas mtCOI evolves primarily neutrally, yet there are striking similarities between the two trees (Fig. 1).
Nonsynonymous distances between species in lysin and 18 kDa exons rival or exceed synonymous distances in mtCOI (Table 1, Fig. 1). This result is remarkable considering the higher rates of mutation and fixation found in mtDNA relative to nuclear DNA (41). The relationship between amino acid differences in lysin versus nucleotide differences in mtCOI (Fig. 2) suggests that roughly 20 amino acid differences accumulate in lysins during the time required for 3% divergence in mtCOI. By using the mtCOI rate calibration of 2% per Myr and an abalone generation time of 3 years (44) allows us to estimate that, on average, along lineages leading to the closely related abalone species, one amino acid change was fixed in lysin per 105 years, or per 3 × 104 generations. Fixation of new variants of 18 kDa may have occurred twice as rapidly (Fig. 2).
Among the most rapidly evolving mammalian proteins are relaxin and gamma-interferon, which have been estimated to accumulate 2.4–2.8 × 10−9 substitutions/nonsynonymous site/year in primate versus rodent comparisons (45). Rapidly evolving eosinophil defense proteins of the ribonuclease family also show up to 2.0 × 10−9 substitutions/nonsynonymous site/year in comparisons among primates (46). The range of nonsynonymous rates estimated for the two abalone sperm protein genes (Table 1) is ≈2- to 50-fold higher than the estimates for these rapidly evolving mammalian genes. The highest rates for the two sperm protein genes derive from the most closely related comparisons, in which both mtCOI and sperm protein genes are least likely to have accumulated multiple changes per site. By contrast, the mammalian protein rates were estimated from comparisons that encompass substantial divergence times: 80 Myr between primates and rodents (45) and 6–50 Myr within primates (46). However, abalone comparisons with divergence times in the 10–20 Myr range also give estimates for sperm proteins that rival the mammalian nonsynonymous rates (Table 1).
Strong selection for binding to a constantly changing egg receptor (10) may cause the adaptive diversification of abalone sperm proteins. The effects of this remarkable molecular evolution include the potential for rapid change of fertilization specificities in fragmented populations.
Acknowledgments
We thank W. Swanson, Y.-H. Lee, S. Anderson, R. McConnaughey and D. Leighton for abalone samples. We thank R. Burton, D. Geiger, M. Gould, M. Hellberg, R. Hudson, Y.-H. Lee, G. Moy, S. Palumbi, K. Roy, J. Stephano, W. Swanson, M. Tegner, L. Wodicka, and C. Wray for technical advice and discussions. We thank W. Swanson for comments on the manuscript. This work was supported by Fundación México-Estados Unidos para la Ciencia and National Institutes of Health Grant HD12986.
ABBREVIATIONS
- mtCOI
mitochondrial cytochrome c oxidase subunit I
- Dn
nonsynonymous
- Ds
synonymous
- Myr
million years
- Mya
million years ago
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF060835–AF060854, AF070955–AF070960, and AF076819–AF076837).
To whom reprint requests should be addressed. e-mail: vvacquier@ucsd.edu.
References
- 1.Hughes A L. In: Mechanisms of Molecular Evolution. Takahata N, Clark A G, editors. Tokyo, and Sinauer Associates, Sunderland, MA: Japan Scientific Societies; 1993. pp. 109–127. [Google Scholar]
- 2.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 3.Richman A D, Uyenoyama M K, Kohn J R. Science. 1996;273:1212–1216. doi: 10.1126/science.273.5279.1212. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y-H, Vacquier V D. Biol Bull. 1992;182:97–104. doi: 10.2307/1542183. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y-H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 6.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vacquier V D, Swanson W J, Lee Y-H. J Mol Evol. 1997;44:S15–S22. doi: 10.1007/pl00000049. [DOI] [PubMed] [Google Scholar]
- 9.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 10.Swanson W J, Vacquier V D. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- 11.Swanson W J, Vacquier V D. Biochemistry. 1995;34:14202–14208. doi: 10.1021/bi00043a026. [DOI] [PubMed] [Google Scholar]
- 12.Woodring W P. J Paleontol. 1931;5:34–39. [Google Scholar]
- 13.Lindberg D R. In: Abalone of the World. Shepherd S A, Tegner M J, Guzman del Proo S A, editors. Blackwell Scientific, Oxford: Fishing News Books; 1992. pp. 3–18. [Google Scholar]
- 14.Hertlein L G. Bull S Calif Acad Sci. 1937;36:93–97. [Google Scholar]
- 15.Lee Y-H, Vacquier V D. Mar Biol. 1995;124:267–278. [Google Scholar]
- 16.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 17.Boore J L, Brown W M. Genetics. 1994;138:423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wodicka L M, Morse D M. Biol Bull. 1991;180:318–327. doi: 10.2307/1542403. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park, PA: Pennsylvania State Univ.; 1993. , Version 1.02. [Google Scholar]
- 20.Genetics Computer Group. Genetics Computer Group Package. Madison, WI: Genetics Computer Group; 1991. [Google Scholar]
- 21.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Washington, DC: Smithsonian Institution; 1993. , Version 3.1.1. [Google Scholar]
- 22.Felsenstein J. phylip. Seattle: Univ. of Washington; 1995. , Version 3.54c. [Google Scholar]
- 23.Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 24.Collins T M, Frazer K, Palmer A R, Vermeij G J, Brown W M. Evolution. 1996;50:2287–2304. doi: 10.1111/j.1558-5646.1996.tb03617.x. [DOI] [PubMed] [Google Scholar]
- 25.Hellberg, M. E. (1998) Evolution in press.
- 26.Knowlton N, Weigt L A, Solorzano L A, Mills D K, Bermingham E. Science. 1993;260:1629–1632. doi: 10.1126/science.8503007. [DOI] [PubMed] [Google Scholar]
- 27.Palumbi S R. J Exp Mar Biol Ecol. 1996;203:75–92. [Google Scholar]
- 28.Schubart C D, Diesel R, Hedges S B. Nature (London) 1998;393:363–365. [Google Scholar]
- 29.Reid D G, Rumbak E, Thomas R H. Philos Trans R Soc London B. 1996;351:877–895. doi: 10.1098/rstb.1996.0082. [DOI] [PubMed] [Google Scholar]
- 30.Vacquier, V. D., Swanson, W. J., Metz, E. C. & Stout, C. D. (1998) Adv. Dev. Biochem. 5, in press.
- 31.Muse S V. Mol Biol Evol. 1996;13:105–114. doi: 10.1093/oxfordjournals.molbev.a025549. [DOI] [PubMed] [Google Scholar]
- 32.Hughes A L, Yeager M. J Mol Evol. 1997;45:125–130. doi: 10.1007/pl00006211. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima K-I, Ogawa T, Oda N, Hattori M, Sakaki Y, Kihara H, Ohno M. Proc Natl Acad Sci USA. 1993;90:5964–5968. doi: 10.1073/pnas.90.13.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John T R, Smith L A, Kaiser I I. Gene. 1994;139:229–234. doi: 10.1016/0378-1119(94)90761-7. [DOI] [PubMed] [Google Scholar]
- 35.Forsdyke D R. Mol Biol Evol. 1995;12:1157–1165. doi: 10.1093/oxfordjournals.molbev.a040273. [DOI] [PubMed] [Google Scholar]
- 36.Cereb N, Hughes A L, Yang S Y. Immunogenetics. 1997;47:30–36. doi: 10.1007/s002510050323. [DOI] [PubMed] [Google Scholar]
- 37.Ferris P J, Pavlovic C, Fabry S, Goodenough U W. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry A J, Ajioka J W, Kreitman M. Genetics. 1991;129:1111–1117. doi: 10.1093/genetics/129.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorit R L, Akashi H, Gilbert W. Science. 1995;268:1183–1185. doi: 10.1126/science.7761836. [DOI] [PubMed] [Google Scholar]
- 40.Gaffney P M, Rubin V P, Hedgecock D, Powers D A, Morris G, Hereford L. Aquaculture. 1996;143:257–266. [Google Scholar]
- 41.Avise J C. Molecular Markers, Natural History, and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 42.Wallis M. J Mol Evol. 1996;43:93–100. doi: 10.1007/BF02337353. [DOI] [PubMed] [Google Scholar]
- 43.Messier W, Stewart C-B. Nature (London) 1997;385:151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]
- 44.Parker D O, Haaker P L, Togstad H A. In: Abalone of the World. Shepherd S A, Tegner M J, Guzman del Proo S A, editors. Blackwell Scientific, Oxford: Fishing News Books; 1992. pp. 384–394. [Google Scholar]
- 45.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer Associates; 1991. [Google Scholar]
- 46.Rosenberg H F, Dyer K D, Tiffany H L, Gonzalez M. Nat Genet. 1995;10:219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]