Abstract
We show that salinixanthin, the light-harvesting carotenoid antenna of xanthorhodopsin, can be reconstituted into the retinal protein from Gloeobacter violaceus expressed in E. coli. Reconstitution of gloeobacter rhodopsin with the carotenoid is accompanied by characteristic absorption changes and the appearance of CD bands similar to those observed for xanthorhodopsin that indicate immobilization and twist of the carotenoid in the binding site. As in xanthorhodopsin, the carotenoid functions as a light-harvesting antenna. The excitation spectrum for retinal fluorescence emission shows that ca. 36% of the energy absorbed by the carotenoid is transferred to the retinal. From excitation anisotropy, we calculate the angle between the two chromophores as ca. 50°, similar to that in xanthorhodopsin. The results indicate that gloeobacter rhodopsin binds salinixanthin in a similar way as xanthorhodopsin, and suggest that it might bind a carotenoid also in vivo. In the crystallographic structure of xanthorhodopsin, the conjugated chain of the carotenoid lies on the surface of helices E and F, and the 4-keto-ring is immersed in the protein at van der Waals distance from the ionone ring of the retinal. The 4-keto-ring is in the space occupied by a tryptophan in bacteriorhodopsin, which is replaced by the smaller glycine in xanthorhodopsin and gloeobacter rhodopsin. Specific binding of the carotenoid and its light-harvesting function are eliminated by a single mutation of the gloeobacter protein that replaces this glycine with a tryptophan. This indicates that the 4-keto-ring is critically involved in carotenoid binding, and suggests that a number of other recently identified retinal proteins, from a diverse group of organisms, could also contain carotenoid antenna since they carry the homologous glycine near the retinal.
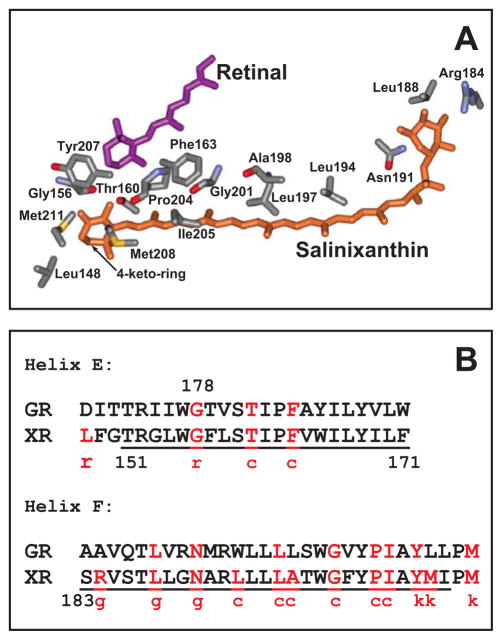
Carotenoids play a major role in light-harvesting in the blue-green region of the spectrum, and in photoprotection, in the complex chlorophyll based photosynthetic apparatus (1–5). Their presence in a retinal protein as a light-harvesting component was established only recently, xanthorhodopsin of Salinibacter ruber being the first example of such a complex (6). The retinal-based light-driven proton pump of the archaea, bacteriorhodopsin (7), does not contain carotenoids, although the carotenoid bacterioruberin binds to another such pump, the archaerhodopsin of the archaeon Halorubrum (8, 9), without a light-harvesting function (9, 10). Salinixanthin (11), a component of the light-driven proton pump xanthorhodopsin, is the major carotenoid of the extremely halophilic eubacterium Salinibacter ruber (12). This C40 carotenoid has an acyl glycoside at one end, a chain with 11 double bonds and a conjugated ring with a 4-oxo (keto) group at the other. When bound to xanthorhodopsin (6), it serves as a light-harvesting antenna (6, 10). It transfers 40–45% of the absorbed quanta to retinal (6, 13) in a fast, femtosecond process (14) that involves the short lived S2 state of the carotenoid and the S1 state of the retinal (13, 14). In carotenoids and polyenes with more than three conjugated double bonds, the lowest excited state S1 (21Ag−- like) is symmetry forbidden (3, 15–17). The intense absorption bands in the blue-green region are from the transition to the S2 (11Bu+-like) state. The carotenoid S1 state is populated as a result of fast internal conversion from the S2. The energy level of the carotenoid S1 is below the S1 of the retinal (3, 13), so it cannot serve as an energy donor for the retinal chromophore. This is different from carotenoid-bacteriochlorophyll pairs in light harvesting complexes, where both S2 and S1 states serve as energy donors (3, 18)). In retinal proteins with protonated Schiff bases (as bacteriorhodopsin and xanthorhodopsin), the lowest excited state is a strongly-allowed 11Bu-like state (19), which is responsible for the broad and intense absorption band in the visible range. This state is characterized by large oscillator strength and large transition dipole moment (19), as the S2 state of the carotenoid. The short distance between the salinixanthin and retinal chromophores in xanthorhodopsin and their favorable mutual orientation (13, 20) result in significant electronic coupling (21) between the two conjugated chains of the chromophores (of ca. 160–210 cm−1 (14)) and efficient excitation energy transfer from the carotenoid to the retinal chromophore (13). This simple single-chromophore carotenoid antenna doubles the light-harvesting capability of the retinal protein, and confers additional optical cross-section, particularly in the blue-green region where absorption of the retinal chromophore is low. Binding of the carotenoid is accompanied by sharpening of the vibronic bands in its absorption spectrum and the appearance of optical activity that indicates immobilization of the 4-keto-ring and the conjugated chain in an asymmetric conformation (22). This conformation is controlled by the retinal. Removal of the latter by hydrolysis eliminates the characteristic spectral features of the bound carotenoid (6, 22, 23). The crystallographic structure of xanthorhodopsin identified the residues that constitute the binding site for the carotenoid (20), see Figure 1A and Supporting Information, Section I. It confirmed the asymmetric conformation of the carotenoid deduced from spectroscopic properties, with its 4-keto-ring turned 82 degrees out of plane and immobilized near the β-ionone ring of retinal. Most of the polyene chain is at the lipid-protein interface, and interacts with a number of residues, as shown in Figure 1A and B, while the 4-keto-ring is immersed in the protein in a site close to the β-ionone ring of retinal. To accommodate it, the bulky tryptophan-138 which occupies this site in bacteriorhodopsin (24), is replaced by the smaller glycine in xanthorhodopsin (20), Figure 2.
Figure 1.
A. Conformation of salinixanthin and residues within 4 Å of salinixanthin in xanthorhodopsin, based on the 3DLL coordinate set (20). The acyl tail of the carotenoid is in contact with lipids, and is not shown. The distances between the side chains and carotenoid are presented in Supporting Information, section I. B. Conservation of residues that interact with the carotenoid in xanthorhodopsin in gloeobacter rhodopsin. Letters c, g, k, and r stand for the chain, glucoside, keto-group, and ring, respectively, and indicate part of the carotenoid in the vicinity of a residue.
Figure 2.
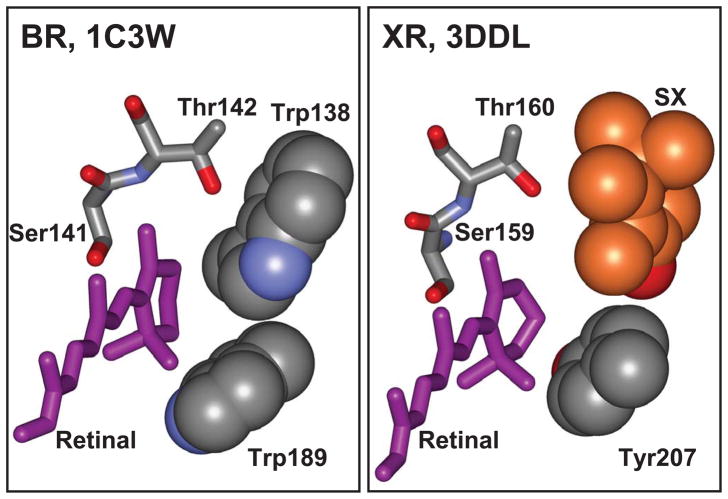
Binding site for the retinal β-ionone ring. A. Bacteriorhodopsin, 1C3W (24). B. Xanthorhodopsin, 3DLL (20); the 4-keto-ring of salinixanthin is shown in orange color.
The question arises whether xanthorhodopsin is unique as a light-harvesting retinal protein/carotenoid complex. Early observations of the multi band action spectra of phototaxis of Haematococcus pluvialis (25) implied that carotenoids or flavins might be involved in the photoreception in this organism as antennae also (26). The development of genomic sequencing in the last decade resulted in the discovery of numerous genes of diverse but related (27–33) retinal proteins in proteobacteria (the proteorhodopsins) and other marine bacteria (34–37), freshwater bacteria (38), cyanobacteria (39, 40), fungi (30, 41) and algae (42), some of which were characterized by heterologous expression in E. coli and other organisms and a few in their native hosts. Several genes of these retinal proteins, from a variety of organisms exhibit high homology to xanthorhodopsin and form a clade (43–45). Some of them might bind a carotenoid, as judged by the fact that the bulky tryptophan residue near the retinal ionone ring of bacteriorhodopsin is replaced with glycine, as in xanthorhodopsin, (for a representative list of organisms and protein sequences see Supporting Information, Section II).
The genome of cyanobacterium Gloeobacter violaceus (46) contains a gene for a retinal protein (accession number NP_923144), highly homologous to the xanthorhodopsin gene (43) with ca. 50% identical amino acid residues (see Supporting Information, Section III). Eleven out of sixteen residues which are in the vicinity of the carotenoid in xanthorhodopsin (Figure 1B), are conserved in gloeobacter rhodopsin, including the glycine (residue 156 in xanthorhodopsin and 178 in gloeobacter rhodopsin sequence) that replaces tryptophan in the binding site, see Supporting Information, Section 2. Gloeobacter rhodopsin has been expressed heterologously in E. coli, and reconstituted with retinal (30). It shows features peculiar to proton pumps (40), and its photocycle is similar to those of proteorhodopsin (47) and xanthorhodopsin (6, 48). It constitutes a good test system for the determinants of carotenoid binding in xanthorhodopsin-like proteins.
Cells of Gloeobacter violaceus contain three carotenoids: β-carotene, oscillol diglycoside, and echinenone (49). The last is the 4-oxo derivative of β-carotene and has a 4-keto-ring similar to salinixanthin (50, 51). To test the possibility that gloeobacter rhodopsin might form a complex with carotenoids and use it as a light-harvesting antenna, we attempted to reconstitute the protein expressed in E. coli with salinixanthin, easily extractable from S. ruber where it accounts to > 96% of the total carotenoids (11). Earlier it was shown that carotenoids can be reconstituted into light-harvesting complexes (52, 53), and we were able to remove and reconstitute salinixanthin in xanthorhodopsin also (unpublished results).
We report here that addition of salinixanthin to gloeobacter rhodopsin produces a functional complex with spectroscopic features similar to those found in xanthorhodopsin. This suggests that the gloeobacter protein may bind a similar carotenoid also in vivo. In order to investigate the role of the 4-keto-ring of the carotenoid in its binding, a mutant was constructed in which the glycine in the common binding site of the rings of the two chromophores was replaced by a tryptophan, as in bacteriorhodopsin. The latter would create steric hindrance for the 4-keto-ring in its binding site. This single mutation eliminated specific binding of the carotenoid and energy transfer from it to retinal, indicating that ability of the protein to make a complex with the carotenoid strongly depends on accommodation of the 4-keto-ring.
Materials and Methods
Gloeobacter rhodopsin was expressed in E. coli, solubilized in 0.02% n-dodecyl-β-D maltopyranoside (DDM1), and purified using its 6x His-tag as described in (40). The G178W mutant was produced using same procedures as for other mutants of this protein (40). Salinixanthin was extracted with acetone/methanol (7:3) from cell membranes of Salinibacter ruber and lipids were removed by precipitation with cold acetone (11). It was stored as ethanol solution at −20°C. For mixing with the protein, the ethanol was evaporated and the carotenoid solubilized in 0.15 % DDM. The extinction coefficient for gloeobacter rhodopsin at the 541 nm maximum was estimated to be ca. 50,000 M−1cm−1 from spectral changes after hydrolysis of the Schiff base with hydroxylamine. The extinction for salinixanthin was assumed to be ca. 140,000 M−1cm−1 (13). The absorption spectra were recorded on a Shimadzu 1701 spectrophotometer in 4×10 mm cells (4 mm pathway). The fluorescence and fluorescence excitation spectra corrected for the light intensity (N) were recorded on an SLM-Aminco spectrofluorimeter, as described recently (13).
Results and Discussion
Gloeobacter rhodopsin binds salinixanthin
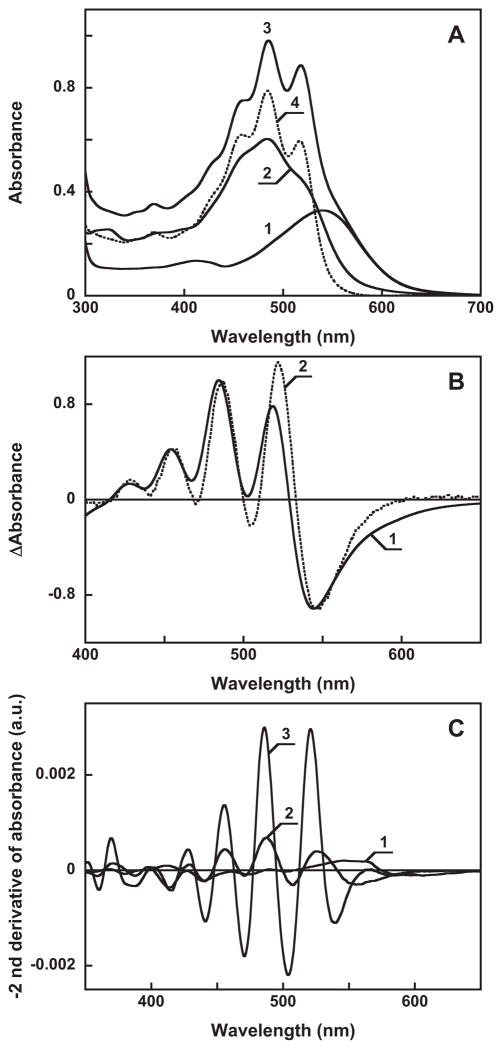
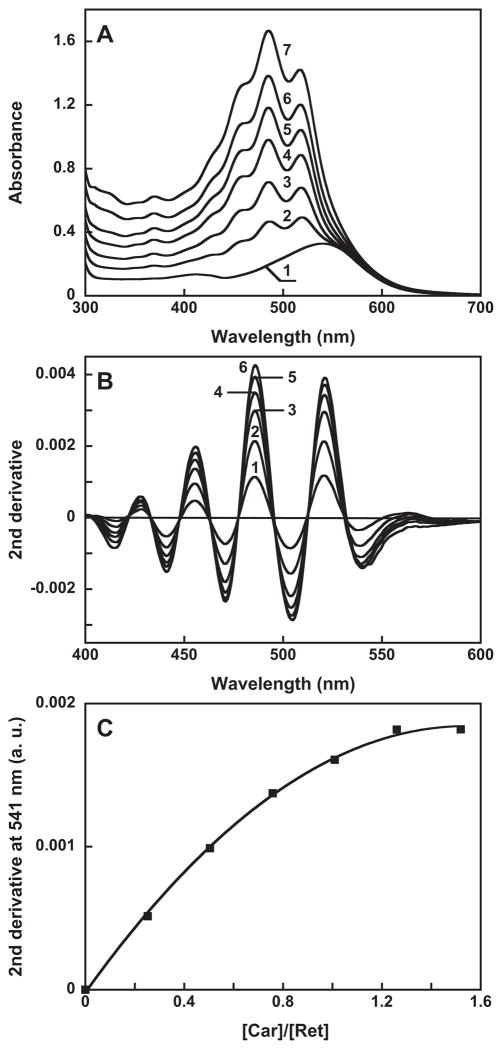
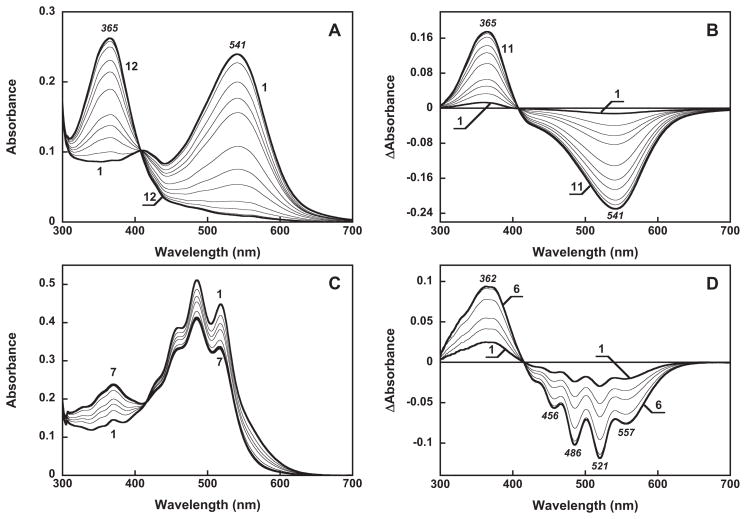
Figure 3 presents evidence for specific binding of salinixanthin by gloeobacter rhodopsin. Gloeobacter rhodopsin solubilized in detergent has a single absorption maximum at 541 nm at pH 7.2 (Figure 3A, spectrum 1). Salinixanthin in solution exhibits a spectrum with poorly resolved vibronic maxima (spectrum 2). Upon addition of salinixanthin to the protein, the spectrum of the product shows characteristic narrowing of the carotenoid vibronic bands that indicates specific binding (Figure 3A, spectra 3 and 4). In the difference spectrum, “spectrum 4 minus spectrum 2”, the sharp positive bands at 521 and 486 nm are from increase of extinction at the maximum of the carotenoid, whereas the negative band at 532 nm is from narrowing of the band (Figure 3B, spectrum 1). These features are similar to those observed for binding of salinixanthin to xantho-opsin upon addition of retinol (23) (Figure 3B, spectrum 2), and upon reconstitution of carotenoid-free xanthorhodopsin with salinixanthin (our unpublished result). Incorporation of the carotenoid occurs with a time-constant of ca. 5 min (data not shown). The amount of bound carotenoid can be estimated from the amplitude of the second derivative of the absorption spectra (Figure 3C). The narrowing of absorption bands upon binding results in increase in the amplitude of their second derivative (which is inversely proportional to the square of the half-width), as evident from comparison of the second derivative of untreated gloeobacter rhodopsin, non-bound salinixanthin, and gloeobacter rhodopsin reconstituted with salinixanthin (spectra 1–3, respectively, in Figure 3C). Subsequent additions of the carotenoid result in additional binding (Figure 4A), until the available binding sites are saturated at a ca. 1:1 carotenoid/retinal stoichiometry. The amount of bound carotenoid was estimated from the second derivative of absorption spectra (Figure 4B) by plotting the amplitude at 541 nm, a wavelength at which its value for the free carotenoid is zero. Such a titration curve is shown in Figure 4C.
Figure 3.
A. Absorption spectra of: 1, gloeobacter rhodopsin, 16 μ in 0.02% DDM, pH 7.2, 100 mM NaCl, 4 mm pathlength; 2, 8 μ salinixanthin in the same buffer; 3, mixture after adding 8 μ of salinixanthin to 16 μ to the gloeobacter rhodopsin; 4, spectrum 3 minus spectrum 1. B. Absorption changes of salinixanthin upon binding to: 1, gloeobacter rhodopsin (difference between spectrum 4 and 2 in panel A); 2, xanthorhodopsin (taken from (23), Figure 6A, curve 4). C. Second derivatives of absorption spectra 1 through 3 in Figure 3A (multiplied by minus 1).
Figure 4.
Titration of gloeobacter rhodopsin with salinixanthin. A. Absorption spectra of: 1, untreated retinal protein, 16 μ 2 through 7, after addition of 4, 8,11,13,16 and 19 μ of salinixanthin. B. Second derivatives of spectra 2 through 7 in panel A (multiplied by minus 1). C. Titration curve, with the Y axis proportional to the amount of bound salinixanthin (in arbitrary units), estimated from the amplitude of second derivative of the difference spectra vs. the amount of added carotenoid (after correction for dilution).
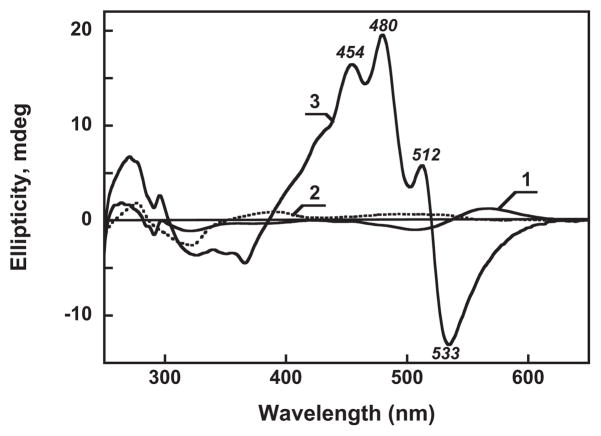
The binding of the carotenoid is accompanied by changes in the circular dichroism spectra as well (Figure 5). Gloeobacter rhodopsin exhibits a weak CD signal (spectrum 1) with a positive band at 567 nm and a negative band at 507 nm. Free carotenoid in detergent shows a signal with even fewer features (spectrum 2) with a very weak band at ca. 490 nm. Upon binding, a characteristic structured, intense CD spectrum appears, with maxima at 512, 480 and 454 nm and a minimum at 533 nm (spectrum 3), similar to those seen in xanthorhodopsin (22). It indicates that when bound, the carotenoid, otherwise non-chiral, acquires a conformation similar to that in xanthorhodopsin.
Figure 5.
Circular dichroism spectra of: 1, untreated gloeobacter rhodopsin in 0.02% DDM, pH 7.2, 100 mM NaCl; 2, salinixanthin in the same buffer; 3, after reconstitution. Concentration of gloeobacter rhodopsin and salinixanthin, ca. 16 μM. Absorption spectra of the samples are shown in Figure 3A, spectra 1, 2 and Figure 4A curve 6, respectively.
Binding of salinixanthin is controlled by the retinal
Hydrolysis of the retinal Schiff base of gloeobacter rhodopsin with hydroxylamine results in formation of retinal oxime, with a strong blue-shift of the maximum, as expected for any rhodopsin (Figures 6A, B). The same reaction of the protein after its reconstitution with salinixanthin causes changes in the carotenoid absorption bands as well (Figures 6C and D). The carotenoid loses the resolution of the vibronic bands upon hydrolysis, as in xanthorhodopsin, but to a somewhat lesser extent. The results indicate that conformation of the carotenoid is controlled by the retinal, apparently from direct interaction of their rings as in xanthorhodopsin (20, 23). Unlike in xanthorhodopsin, however, the carotenoid retains a somewhat structured spectrum even after complete hydrolysis of the retinal Schiff base. There might be two reasons for this: either a fraction of retinal oxime remains in the binding site (23), or some degree of immobilization of the bound carotenoid occurs in the absence of retinal.
Figure 6.
Absorption changes accompanying cleavage of the Schiff base bond by 0.2 M hydroxylamine in gloeobacter rhodopsin before (A and B) and after reconstitution with salinixanthin (C, D) at pH 7.2 in the dark. A and C, absolute spectra; B and D, difference spectra. The spectra shown in A were taken at 0, 5, 10 15, 30, 45, 60, 90 120, 180 and 240 min after addition of hydroxylamine. The spectra in B obtain by subtraction of initial spectrum (at zero time) from the rest. The spectra shown in C were taken at 0, 5, 10, 15, 30, 60 and 90 min.
Salinixanthin acts as a light-harvesting antenna to gloeobacter rhodopsin
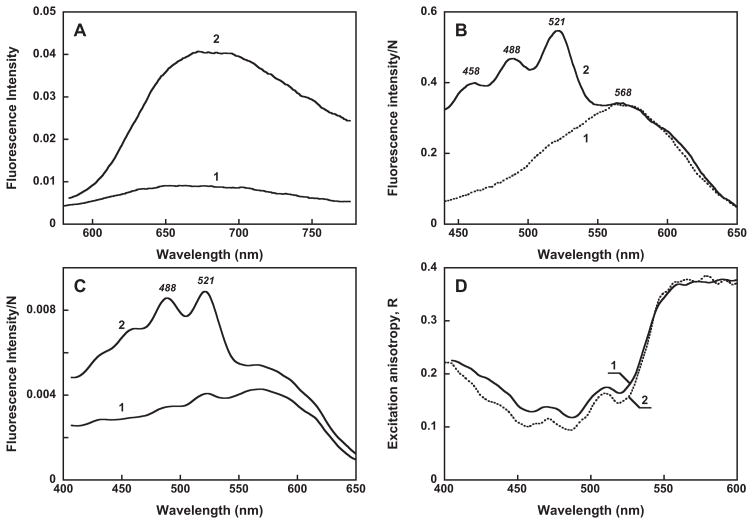
Evidence for excitation-energy transfer from salinixanthin to retinal was obtained by measuring the excitation spectrum for retinal chromophore fluorescence before and after reconstitution with salinixanthin. The fluorescence of rhodopsins exhibits complex behavior. The retinal chromophore of gloeobacter rhodopsin shows a broad fluorescence emission band in the far-red region, with a maximum at ca. 670 nm at pH 7.2. It undergoes a red-shift to 680 nm and ca. 4 fold increase in intensity when the pH is lowered to 4.5, from protonation of the Schiff base counter-ion (Figure 7A), similar to 2.5 fold increase that we had found for xanthorhodopsin (13). In bacteriorhodopsin the protonation of the counter-ion is accompanied by a larger increase in the quantum yield of retinal emission, by more than an order of magnitude (54, 55), from a longer lifetime of the excited state (56), and a larger shift in the absorption spectrum. The latter is about 40 nm in bacteriorhodopsin (57, 58), compared to 3–5 nm in xanthorhodopsin (48) and in gloeobacter rhodopsin. The maximum of the excitation spectrum for retinal emission of gloeobacter rhodopsin, sampled at 720 nm, is at ca. 568 nm at pH 4.5 (Figure 7B, spectrum 1). It is thus substantially red-shifted from the absorption maximum of the retinal chromophore at 545 nm at this pH. This deviation of the excitation spectrum from the absorption spectrum suggests that there is a species with protonated counter-ion and a high fluorescence quantum yield that absorbs at ca. 568 nm, and the occupancy of this species at pH 4.5 is only partial (we estimate at ca. 30%). The fraction of this fluorescent species is partial even at low pH since the absorption maximum remains at 548 nm when the pH is lowered from 4 to 2 (not shown).
Figure 7.
Fluorescence, excitation and anisotropy spectra. A. Fluorescence spectra of gloeobacter rhodopsin at pH, 7.2 (spectrum 1) and 4.5 (spectrum 2). B. Excitation spectra for emission at 720 nm corrected for the light intensity N, pH 4.5: 1, untreated gloeobacter rhodopsin; 2, after reconstitution with salinixanthin. C. Excitation spectra of gloeobacter rhodopsin reconstituted with salinixanthin: 1, under parallel (both vertical) and 2, perpendicular (vertical excitation/horizontal emission) orientation of polarizers. Absorbance of the sample at the maximum was 0.32. D. Excitation anisotropy: 1, gloeobacter rhodopsin reconstituted with salinixanthin; 2, xanthorhodopsin, from (13)
Most important is that the excitation spectrum after reconstitution with the carotenoid exhibits additional maxima at 521, 488 and 458 nm (Figure 7B, spectrum 2) from carotenoid absorption, which clearly indicates energy transfer from the carotenoid to retinal. The fluorescence spectra obtained upon excitation at the retinal band (at 545 nm) and at the carotenoid 520 nm band have similar shape (data not shown), indicating that fluorescence of the carotenoid itself does not contribute substantially at 720 nm. From the comparison of the relative amplitudes of the carotenoid and retinal bands in the xanthorhodopsin (13) and the gloeobacter rhodopsin excitation spectra, the efficiency of energy transfer in the latter can be estimated of being at least 80% of that in xanthorhodopsin (i.e., > 36 ± 4%). The excitation spectra obtained under parallel (all vertical, VV) and perpendicular (vertical/horizontal, VH) polarizations are very different; the latter exhibits a much larger contribution from the carotenoid component (Figure 7C), as was observed also for xanthorhodopsin (13). From the similar shape of the excitation anisotropy in the two pigments (Figure 7D), it follows that the salinixanthin and retinal chromophores in gloeobacter rhodopsin are also not parallel but make an angle calculated to be ca. 50±5°. In xanthorhodopsin this value was 56±3° (13).
The results show that salinixanthin, the light harvesting antenna of xanthorhodopsin, binds to gloeobacter rhodopsin in a conformation similar to that in xanthorhodopsin, and apparently to a similar binding site. Eleven of the 16 side chains of residues that contact salinixanthin in xanthorhodopsin are conserved in the gloeobacter protein (homologues of Gly156, Thr160, Phe163, Asn191, Leu188, Leu197, Gly201, Pro204, Ile205, Tyr207, and Met211 in the xanthorhodopsin sequence). Several of these are in the common binding site that immobilizes the 4-keto-ring of the carotenoid and the ionone ring of retinal (Gly156, Tyr207 and Met 211). Leu197 and Ile205 interact with the carotenoid side chain in a slot along helix F, thus providing immobilization of the polyene chain and determining the overall angle between the two chromophores (20). The conformation of salinixanthin in gloeobacter rhodopsin results in energy transfer almost as efficient as in xanthorhodopsin. There are two implications. The first is that gloeobacter rhodopsin can serve as a good model for getting more insights into carotenoid binding and the mechanism of energy transfer, utilizing the already developed high-yield expression system (30, 40) that will enable exploration of the role of protein residues in binding and energy transfer. The roles of the conserved Gly156 and Tyr207 are of particular interest. Second, gloeobacter rhodopsin probably binds a carotenoid with a similar structure also in vivo. Future experiments dealing with reconstitution of this retinal protein with the carotenoids found in Gloeobacter violaceus (major components are 3-carotene and oscillol diglycoside; minor component echinenone (49)) will test this.
Effect of the substitution of Gly in the ring binding site for a Trp on carotenoid binding
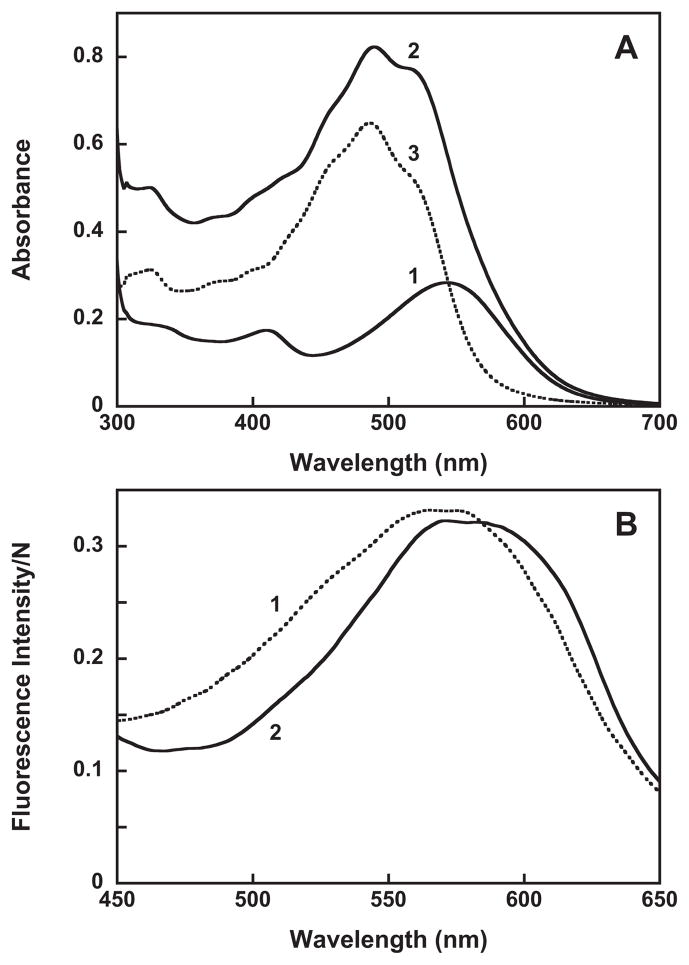
An intriguing question is the importance of the 4-keto-ring in carotenoid binding. Comparison of the structures of bacteriorhodopsin and xanthorhodopsin at the ionone ring of the retinal (Figure 2) indicates that the carotenoid 4-keto-ring in xanthorhodopsin occupies the space of Trp138 in bacteriorhodopsin. If same is true for gloeobacter rhodopsin, then replacing Gly178 (homologous to Gly156 in xanthorhodopsin) by the much bulkier tryptophan will create steric hindrance to the 4-keto-ring, and that might hinder carotenoid binding and energy transfer. The results show that this is indeed the case. As shown in Figure 8A, addition of salinixanthin to the G178W mutant does not cause significant changes in the carotenoid absorption spectrum. It does not cause any chirality changes either (data not shown), and there is no evidence for excited-state energy transfer, as the carotenoid bands are missing from the excitation spectrum (compare spectra 2 in Figure 8B and Figure 7B). Thus, a single mutation that prevents the entry of the 4-keto-ring into the binding site eliminates specific binding of the carotenoid conjugated chain, and most importantly energy transfer to retinal. This result emphasizes the role of the 4-keto-ring in the overall binding and function of the salinixanthin as antenna, and the role of the glycine in providing space for the carotenoid binding. Many members of xanthorhodopsin clade with high homology (harbored by Thermus aquaticus, Roseiflexus sp. RS-1, Alpha proteobacterium, Actinobacteria and others) and even proteins with smaller homology (30%) contain glycine near the retinal ring (see Supporting Information, Section II) and might bind carotenoid antennas, whereas some highly homologous proteins as in Methylophilales bacterium HTCC2181 (44) contain Trp, as in bacteriorhodopsin, and would not be able to accommodate a carotenoid ring similar to that in salinixanthin near the retinal. This does not exclude the possibility that they might bind different carotenoids in some other way.
Figure 8.
A. Absorption spectra of: 1, G178W mutant of gloeobacter rhodopsin, ca. 16 μM in 0.02% DDM, 20 mM MOPS, pH 7.2, 100 mM NaCl; 2, after addition of 12 μM salinixanthin; and 3, spectrum 2 minus spectrum 1, indicating no binding of salinixanthin to the G178W mutant. B. Excitation spectra of retinal emission sampled at 720 nm before (spectrum 1) and after addition of 16 μM salinixanthin (spectrum 2).
In conclusion, the results indicate that gloeobacter rhodopsin, the representative of a group of homologous retinal-proteins with potential carotenoid binding sites, binds salinixanthin, the C40 carotenoid antenna of xanthorhodopsin from S. ruber. The carotenoid acquires a conformation similar to that in xanthorhodopsin and functions as a light-harvesting antenna. The single mutation G178W creates steric hindrance for the entry of the 4-keto-ring of the carotenoid into its binding site, and eliminates carotenoid binding and energy transfer. The results imply that gloeobacter rhodopsin and the other similar rhodopsins bind carotenoid as an accessory light-harvesting antenna, and that the conserved glycine in the primary structure might indicate the presence of a carotenoid antenna.
Supplementary Material
Footnotes
Abbreviations: DDM, n-dodecyl-β-D maltopyranoside; MOPS, 3-[N-morpholino]propanesulfonic acid.
Supporting Information Available
Distances between salinixanthin and side chain residues in xanthorhodopsin, list of retinal proteins with high homology to xanthorhodopsin, and alignment of xanthorhodopsin and gloeobacter rhodopsin. This material is available free of charge via the Internet at http://pubs.acs.org/
This work was supported in part by grants from the National Institutes of Health (GM29498), the Department of Energy (DEFG03-86ER13525) to J.K.L and the U.S. Army Research Office (W911NF-09-1-0243) to S.P.B, and by a Korea Research Foundation Grant (KRF2004-042-C00113) to K.H.J., the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science & Technology, Korea to K.H.J., the second stage of Brain Korea 21 graduate Fellowship Program for A.R.C.
References
- 1.Frank HA, Cogdell RJ. Carotenoids in photosynthesis. Photochem Photobiol. 1996;63:257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 2.Cogdell RJ, Fyfe PK, Howard TD, Fraser N, Isaacs NW, Freer AA, McKluskey K, Prince S. The Structure and Function of the LH2 Complex from Rhodopseudomonas acidophila Strain 10050, with Special Reference to the Bound Carotenoids. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The Photochemistry of Carotenoids. Kluwer Academic Publishers; Dordrecht/Boston/London: 1999. pp. 71–80. [Google Scholar]
- 3.Polivka T, Sundström V. Ultrafast dynamics of carotenoid excited states - from solution to natural and artificial systems. Chem Rev. 2004;104:2021–2071. doi: 10.1021/cr020674n. [DOI] [PubMed] [Google Scholar]
- 4.Wilson A, Punginelli C, Gall A, Bonetti C, Alexandre M, Routaboul JM, Kerfeld CA, van Grondelle R, Robert B, Kennis JTM, Kirilovsky D. A photoactive carotenoid protein acting as light intensity sensor. Proc Natl Acad Sci USA. 2008;105:12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berera R, van Stokkum IHM, d’Haene S, Kennis JTM, van Grondelle R, Dekker JP. A mechanism of energy dissipation in Cyanobacteria. Biophys J. 2009;96:2261–2267. doi: 10.1016/j.bpj.2008.12.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oesterhelt D, Stoeckenius W. Functions of a new photoreceptor membrane. Proc Natl Acad Sci USA. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura K, Kouyama T. Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J Mol Biol. 2008;375:1267–1281. doi: 10.1016/j.jmb.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Mukohata Y, Ihara K, Uegaki K, Miyashita Y, Sugiyama Y. Australian Halobacteria and their retinal-protein ion pumps. Photochem Photobiol. 1991;54:1039–1045. doi: 10.1111/j.1751-1097.1991.tb02127.x. [DOI] [PubMed] [Google Scholar]
- 10.Boichenko VA, Wang JM, Antón J, Lanyi JK, Balashov SP. Functions of carotenoids in xanthorhodopsin and archaerhodopsin, from action spectra of photoinhibition of cell respiration. Biochim Biophys Acta. 2006;1757:1649–1656. doi: 10.1016/j.bbabio.2006.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutnaes BF, Oren A, Liaaen-Jensen S. New C40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J Nat Prod. 2002;65:1340–1343. doi: 10.1021/np020125c. [DOI] [PubMed] [Google Scholar]
- 12.Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Micrbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- 13.Balashov SP, Imasheva ES, Wang JM, Lanyi JK. Excitation energy-transfer and the relative orientation of retinal and carotenoid in xanthorhodopsin. Biophys J. 2008;95:2402–2414. doi: 10.1529/biophysj.108.132175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polívka T, Balashov SP, Chábera P, Imasheva ES, Yartsev A, Sundström V, Lanyi JK. Femtosecond carotenoid to retinal energy transfer in xanthorhodopsin. Biophys J. 2009;96:2268–2277. doi: 10.1016/j.bpj.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson BS, Kohler BE. A low-lying weak transition in the polyene αω-diphenyloctatetraene. Chem Phys Lett. 1972;14:299–304. [Google Scholar]
- 16.Schulten K, Karplus M. On the origin of a low-lying forbidden transition in polyenes and related molecules. Chem Phys Lett. 1972;14:305–309. [Google Scholar]
- 17.Christensen RL. The electronic states of carotenoids. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The Photochemistry of Carotenoids. Kluwer Academic Publishers; Dordrecht/Boston/London: 1999. pp. 137–157. [Google Scholar]
- 18.Cong H, Niedzwiedzki DM, Gibson GN, LaFountain AM, Kelsh RM, Gardiner AT, Cogdell RJ, Frank HA. Ultrafast time-resolved carotenoid to-bacteriochlorophyll energy transfer in LH2 complexes from photosynthetic bacteria. J Phys Chem B. 2008;112:10689–10703. doi: 10.1021/jp711946w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birge RR, Zhang CF. Two-proton double resonance spectroscopy of bacteriorhodopsin. Assignment of the electronic and dipolar properties of the low-lying 1Ag*-like and 1Bu*+-like π, π* states. J Chem Phys. 1990;92:7178–7195. [Google Scholar]
- 20.Luecke H, Schobert B, Stagno J, Imasheva ES, Wang JM, Balashov SP, Lanyi JK. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc Natl Acad Sci USA. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholes GD. Long-range resonance energy transfer in molecular systems. Ann Rev Phys Chem. 2003;54:57–87. doi: 10.1146/annurev.physchem.54.011002.103746. [DOI] [PubMed] [Google Scholar]
- 22.Balashov SP, Imasheva ES, Lanyi JK. Induced chirality of the light-harvesting carotenoid salinixanthin and its interaction with the retinal of xanthorhodopsin. Biochemistry. 2006;45:10998–11004. doi: 10.1021/bi061098i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imasheva ES, Balashov SP, Wang JM, Smolensky E, Sheves M, Lanyi JK. Chromophore interaction in xanthorhodopsin - retinal dependence of salinixanthin binding. Photochem Photobiol. 2008;84:977–984. doi: 10.1111/j.1751-1097.2008.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 25.Litvin FF, Sineshchekov OA, Sineshchekov VA. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 26.Sineshchekov OA, Litvin FF. Molecular Mechanisms of Biological Effects of Optical Radiation (in Russian) Nauka; Moscow: 1988. Mechanisms of Phototaxis of Microorganisms; pp. 212–227. [Google Scholar]
- 27.Ruiz-Gonzales MX, Marin I. New insights into the evolution history of type 1 rhodopsins. J Mol Evol. 2004;58:348–358. doi: 10.1007/s00239-003-2557-8. [DOI] [PubMed] [Google Scholar]
- 28.Spudich JL, Jung K-H. Microbial Rhodopsins: Phylogenetic and Functional Diversity. In: Briggs WR, Spudich JL, editors. Handbook of photosensory receptors. Wiley-VCH; Darmstadt: 2005. pp. 1–23. [Google Scholar]
- 29.Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 30.Brown LS, Jung KH. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem Photobiol Sci. 2006;5:538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- 31.Sharma AK, Spudich JL, Doolitle WF. Microbial rhodopsins: functional versatility and genetic mobility. Trends in Microbiology. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Adamian L, Ouyang Z, Tseng YY, Liang J. Evolutionary patterns of retinal-binding pockets of type I rhodopsins and their functions. Photochem Photobiol. 2006;82:1426–1435. doi: 10.1562/2006-02-14-RA-802. [DOI] [PubMed] [Google Scholar]
- 33.Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles? Nature Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 34.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 35.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 36.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R, Pedros-Alio C, Pinhassi J. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 38.Sharma AK, Sommerfeld K, Bullerjahn GS, Matteson AR, Wilhelm SW, Jezbera J, Brandt U, Doolittle WF, Hahn MW. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 2009;3:726–737. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- 39.Vogeley L, Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL, Luecke H. Anabaena sensory rhodopsin: A photochromic color sensor at 2.0 Å. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miranda MRM, Choi AR, Shi L, Bezerra AG, Jung KH, Brown LS. The photocycle and proton translocation pathway in a cyanobacterial ion-pumping rhodopsin. Biophys J. 2009;96:1471–1481. doi: 10.1016/j.bpj.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waschuk SA, Bezerra JAG, Shi L, Brown LS. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma AK, Nesbø CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC, Oh HM, Kitner JB, Vergin KL, Rappé MS. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 45.Sharma AK, Zhaxybayeva O, Papke RT, Doolitle WF. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environental Microbiology. 2008;10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura Y, Kaneko T, Sato S, Mimuro M, Miyashita H, Tsuchiya T, Sasamoto S, Watanabe A, Kawashima K, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Nakazaki N, Shimpo S, Takeuchi C, Yamada M, Tabata S. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Research. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 47.Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 48.Imasheva ES, Balashov SP, Wang JM, Lanyi JK. pH-dependent transitions in xanthorhodopsin. Photochem Photobiol. 2006;82:1406–1413. doi: 10.1562/2006-01-15-RA-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takaichi S, Mochimaru M. Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci. 2007;64:2607–2619. doi: 10.1007/s00018-007-7190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Britton G, Liaaen-Jensen S, Pfander H, editors. Carotenoids Handbook. Birkhauser Verlag; Basel, Boston, Berlin: 2004. [Google Scholar]
- 51.Goodwin TW. Animals. II. Chapman and Hall; London, New-York: 1984. The Biochemistry of the Carotenoids. [Google Scholar]
- 52.Davis CM, Bustamante PL, Loach PA. Reconstitution of bacterial core light-harvesting complexes of Rhodobacter sphaeroides and Rhodospirillum rubrum with isolated α- and β-polypeptides, bacteriochlorophyll a and carotenoid. J Biol Chem. 1995;270:5793–5804. doi: 10.1074/jbc.270.11.5793. [DOI] [PubMed] [Google Scholar]
- 53.Frank HA. Incorporation of carotenoids into reaction center and light-harvesting pigment-protein complexes. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The Photochemistry of Carotenoids. Kluwer Academic Publishers; Dordrecht/Boston/London: 1999. pp. 223–234. [Google Scholar]
- 54.Kouyama T, Kinosita K, Ikegami A. Excited-state dynamics of bacteriorhodosin. Biophys J. 1985;47:43–54. doi: 10.1016/S0006-3495(85)83875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balashov SP, Litvin FF, Sineshchekov VA. Photochemical processes of light energy transformation in bacteriorhodopsin. In: Skulachev VP, editor. Physicochemical Biology Reviews. Harwood Academic Publishers GmbH; UK: 1988. pp. 1–61. [Google Scholar]
- 56.Song L, El-Sayed MA, Lanyi JK. Protein catalysis of the retinal subpicosecond photoisomerization in the primary process of bacterial photosynthesis. Science. 1993;261:891–894. doi: 10.1126/science.261.5123.891. [DOI] [PubMed] [Google Scholar]
- 57.Mowery PC, Lozier RH, Chae Q, Tseng Y-W, Taylor M, Stoeckenius W. Effect of acid pH on the absorption spectra and photoreactions of bacteriorhodopsin. Biochemistry. 1979;18:4100–4107. doi: 10.1021/bi00586a007. [DOI] [PubMed] [Google Scholar]
- 58.Fischer U, Oesterhelt D. Chromophore equilibria in bacteriorhodopsin. Biophys J. 1979;28:211–230. doi: 10.1016/S0006-3495(79)85172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.