Abstract
Cyclin-dependent kinase (Cdk) both promotes mitotic entry (spindle assembly and anaphase) and inhibits mitotic exit (spindle disassembly and cytokinesis), leading to an elegant quantitative hypothesis that a single cyclin oscillation can function as a ratchet to order these events. This ratchet is at the core of a published ODE model for the yeast cell cycle. However, the ratchet model requires appropriate cyclin dose–response thresholds. Here, we test the inhibition of mitotic exit in budding yeast using graded levels of stable mitotic cyclin (Clb2). In opposition to the ratchet model, stable levels of Clb2 introduced dose-dependent delays, rather than hard thresholds, that varied by mitotic exit event. The ensuing cell cycle was highly abnormal, suggesting a novel reason for cyclin degradation. Cdc14 phosphatase antagonizes Clb2–Cdk, and Cdc14 is released from inhibitory nucleolar sequestration independently of stable Clb2. Thus, Cdc14/Clb2 balance may be the appropriate variable for mitotic regulation. Although our results are inconsistent with the aforementioned ODE model, revision of the model to allow Cdc14/Clb2 balance to control mitotic exit corrects these discrepancies, providing theoretical support for our conclusions.
Keywords: cell cycle, cyclin, mathematical model, mitosis, oscillator
Introduction
Chromosome transmission to daughter cells occurs in discrete steps. In S. cerevisiae, this sequence begins with Start, followed by budding, DNA replication and SPB duplication, mitotic spindle assembly, anaphase, spindle disassembly and cytokinesis. An orderly execution of these steps produces two daughter cells with hereditary material identical to the parent cell. Execution of these steps out-of-order can produce aneuploid or inviable progeny. Two general mechanisms could define the order of the cell cycle: dependency between steps (each step requiring completion of the previous step), or an independent central timer that activates each step in the proper order (Hartwell et al, 1974; Murray and Kirschner, 1989). Under either mechanism, accuracy and fidelity could be greatly increased by extrinsic surveillance mechanisms (checkpoints) that prevent initiation of one step until the preceding step is complete (Hartwell and Weinert, 1989).
Cyclins and cyclin-dependent kinase (Cdk) provide excellent candidates for a central controlling timer. In all eukaryotes, mitotic cyclin–Cdk activity rises and falls once per division. Mitotic cyclins (CLB1, 2, 3, and 4 in S. cerevisiae) are required for mitotic entry (spindle assembly and anaphase). However, overexpression of mitotic cyclin prevents mitotic exit (spindle disassembly and cytokinesis), resulting in telophase arrest (Surana et al, 1993). If high cyclin levels induce mitotic entry, and mitotic exit is restrained until cyclin level drops below a lower threshold (Murray and Kirschner, 1989; King et al, 1994; Stern and Nurse, 1996; Zachariae and Nasmyth, 1999; Morgan and Roberts, 2002; Morgan, 2007; Figure 1A), then oscillation of mitotic cyclin would obligatorily order sequential events of mitotic entry and exit. The result is ‘ratchet'-like control of mitosis: a single rise and fall of cyclin–Cdk activity toggles activation and inhibition of both mitotic entry and exit, producing one and only one sequential execution of each step. Although elegant and supported by experimental data, this hypothesis depends critically on quantitative values of cyclin response thresholds; the model fails if mitotic entry can be driven by cyclin levels too low to inhibit mitotic exit, or exit can be inhibited only by levels not achieved in a normal cell cycle and thus, necessarily above any entry threshold.
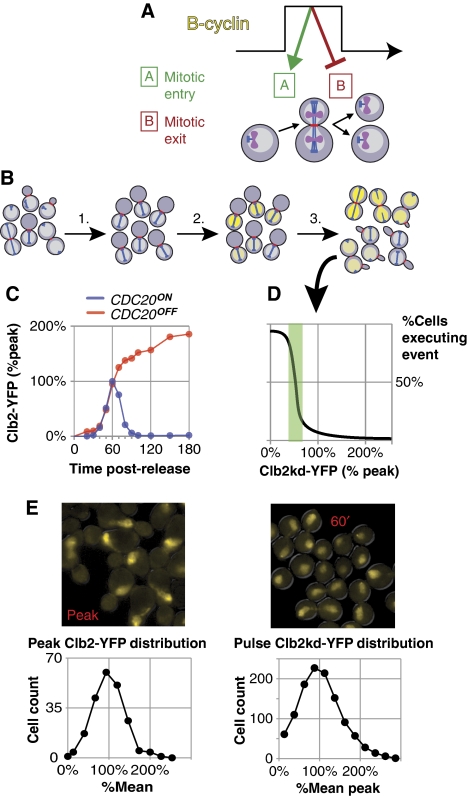
Figure 1.
Measurement of the post-anaphase response to undegradable Clb2p in single cells. (A) Hypothetical control of mitotic order: high B-cyclin promotes mitotic entry and inhibits mitotic exit, whereas low B-cyclin permits exit but not entry. In this model cyclin oscillation dictates ordering. (B) Method schematic using PGAL1 → CLB2kd-YFP GAL4-rMR PMET3 → CDC20 cells, with phenotypic markers CFP–TUB1 (blue) and MYO1–mCherry (red). (1) Metaphase block; (2) Clb2kd–YFP pulse by brief incubation with deoxycorticosterone to activate Gal4–rMR; (3) Synchronous release to anaphase by Cdc20 induction; variation in Clb2kd–YFP dosage affects mitotic exit; Clb2kd concentration and exit phenotypes can be correlated. (C) Measurement of endogenous Clb2–YFP reference levels. PMET3 → CDC20 bar1Δ CLB2-YFP clb1,3,4Δ (BD141-52D) cultures were synchronized with α-factor and released to either a metaphase block or a second α-factor block. [Clb2–YFP] level was measured every 10 min. (D) Peak [Clb2–YFP] and steady state [Clb2–YFP] levels at metaphase arrest, as measured in (C), define the relevant concentration ranges for the hypothetical plot, [Clb2kd–YFP] versus process execution, generated from the cells described in (B). The green box illustrates a hypothetical threshold (steepest decrease, passing through 50% completion) in units of peak [Clb2–YFP]. (E) Single cell distribution of Clb2–YFP at peak concentration (left, BD128-37A) and of a typical Clb2kd–YFP pulse at 60 min post-release from metaphase arrest (right, BD127-11B).
In S. cerevisiae, oscillation of cyclin–Cdk activity is largely a product of periodic cyclin transcription and proteolysis. The anaphase-promoting complex (APC), bound to one of two activators, Cdc20 or Cdh1 (Visintin et al, 1997; Yeong et al, 2000), mediates Clb proteolysis (Irniger et al, 1995). APCCdc20 also mediates proteolysis of the anaphase inhibitor Pds1, promoting anaphase (Cohen-Fix et al, 1996). Subsequently, APCCdh1 completes Clb2 degradation (Schwab et al, 1997; Yeong et al, 2000). Absence of Cdc20 causes metaphase arrest and stabilizes Clb2 (Hartwell and Smith, 1985; Sethi et al, 1991; Lim et al, 1998; Irniger, 2002; Wäsch and Cross, 2002). Mutant Clb2, lacking Cdc20 and Cdh1 recognition domains (Clb2kd), is stable and causes a post-anaphase block when expressed from the endogenous locus (Wäsch and Cross, 2002). The latter observation supports the ratchet model, as endogenous expression of undegradable Clb2 drives cells into mitosis, but cells then fail to exit because this cyclin cannot be degraded.
The stoichiometric inhibitor, Sic1, can reduce cyclin–Cdk activity independently of cyclin proteolysis. Sic1 is normally expressed during mitotic exit (Schwob et al, 1994). Its overexpression can rescue normally lethal genetic backgrounds in which mitotic cyclin degradation is blocked (Wäsch and Cross, 2002; Archambault et al, 2003; Cross, 2003; Thornton and Toczyski, 2003). In such rescued strains, alternating accumulation and proteolysis of Sic1 may substitute for oscillating cyclin levels, periodically inhibiting cyclin–Cdk activity and providing the rise and fall of cyclin–Cdk activity (Thornton et al, 2004) required for viability according to ‘ratchet' models. Sic1 levels are periodic both because of cyclin–Cdk-mediated proteolysis (Schwob et al, 1994; Verma et al, 1997) and periodic transcription (Knapp et al, 1996).
The Cdc14 phosphatase is required for mitotic exit in budding yeast (Visintin et al, 1998). From G1 phase until mitosis, Net1 anchors Cdc14p in the nucleolus; Cdc14 is released into the rest of the cell only during mitosis (Shou et al, 1999). It is then thought to promote SIC1 transcription by dephosphorylating Swi5, the major SIC1 transcription factor, and to promote Sic1 stability by dephosphorylating Sic1 itself (Visintin et al, 1998). Cdc14 also dephosphorylates Cdh1, promoting its association with the APC and driving the degradation of many proteins including mitotic cyclins (Visintin et al, 1998). Cdc14 release from the nucleolus depends on the FEAR pathway in early anaphase, and later on the mitotic exit network (Shou et al, 1999; Stegmeier et al, 2002). The integration of Cdc14 release with the cyclin–Cdk cycle is still incompletely understood.
The complete cell-cycle control system has been modeled mathematically using systems of ordinary differential equations (Chen et al, 2004), and the efficacy of this model in accounting for many mutant backgrounds has been established (Chen et al, 2004; Cross et al, 2005). Two subsequent models concentrating solely on mitotic exit have also been proposed: one to account for the regulation of Cdc14 release from the nucleolus (Queralt et al, 2006) and another for describing reciprocal negative regulation of Clb2 and Sic1, and its possible consequences for mitotic irreversibility (Lopez-Aviles et al, 2009). These latter two models do not attempt to provide a comprehensive view of cell cycle control, but demonstrate the plausibility of dynamic molecular mechanisms that may regulate mitotic exit.
Here we devise a method to determine quantitative relationships between mitotic cyclin levels and the ability to carry out multiple steps in mitotic exit. The results challenge the cyclin-based ratchet model but can be reconciled by incorporating Cdc14 as a general antagonist of Clb2-Cdk, directly removing Clb2–Cdk-dependent phosphorylations that inhibit mitotic exit. Similarly, the quantitative cell cycle model (Chen et al, 2004) that incorporates a cyclin-based ratchet as the central determinant of mitotic entry and exit, predicts our results poorly, but editing the model to include Cdc14 as a direct Clb2 antagonist rescues the model's ability to match our results.
Results
Development of a method to load pre-anaphase cells with titrated, physiological levels of undegradable mitotic cyclin
Any ratchet model for mitotic control by cyclin–Cdk complex must predict that high but physiological levels of mitotic cyclin-dependent kinase activity will inhibit mitotic exit. We developed a method to introduce a fixed level of a stable version of the major mitotic cyclin, Clb2, before anaphase (Figure 1B). In the absence of degradation, Clb2 expressed from its endogenous promoter accumulates above its normal peak level attained in cycling cells (Figure 1C); this is also true of Clb2kd expressed from its endogenous promoter (data not shown). Therefore, completely blocking Clb2 degradation results in a super-physiological level of cyclin. To study the ability of undegradable Clb2 at physiological levels (i.e. at or around normal peak levels), we induced a pulse of fluorescent, undegradable Clb2 (Clb2kd–YFP) (Wäsch and Cross, 2002) expression from the GAL1 promoter using a transient pulse of deoxycorticosterone in cells containing a hormone-responsive Gal4–rMR fusion (Picard, 2000; Supplementary methods; Supplementary Figures 1–8). The resulting Clb2kd pulse remained stable throughout subsequent release from a metaphase cdc20 block (Supplementary Figure 1C). CLB2–YFP at the endogenous locus was fully functional (Supplementary Figure 2), indicating that the YFP tag did not significantly affect Clb2 function.
We measured Clb2kd concentration in ‘peak-equivalent' units, where one peak-equivalent is the average peak Clb2 level observed in a synchronous cell cycle (Figure 1C). In this study, we have not calibrated peak Clb2 level in absolute terms; previous results suggest a value around 3000 molecules per cell (Cross et al, 2002). Clb2–YFP levels measured by immunoblotting in α-factor-synchronized cultures were identical with or without Cdc20 expression up to the time of peak accumulation (60 min post-release); thereafter these levels diverge (Figure 1C; Supplementary Figures 3 and 4). As the peak level and timing are probably dependent on activation of Clb2p degradation by APCCdc20 (Yeong et al, 2000), this result indicates that the 60-min peak closely approximated the average single-cell peak (Supplementary Figure 3). We used cdc20-depleted cells released for 60 min, followed by a 45-min incubation in cycloheximide, to obtain a distribution of Clb2–YFP fluorescence in cells at the time of peak expression. (The incubation in cycloheximide was required to allow maturation of Clb2–YFP fluorescence; cdc20 depletion was required to prevent Clb2–YFP degradation during this interval; Figure 1E). Detailed analysis of quantitative immunoblotting measurements and averaged single-cell fluorescence measurements for Clb2–YFP and Clb2kd–YFP levels show a close agreement between the two methods, indicating that we can accurately estimate Clb2kd–YFP levels in peak-equivalent units in single cells (Supplementary Figure 1D).
Timing and magnitude of peak Clb2–YFP concentration in this procedure were the same in the presence or absence of the other mitotic cyclins CLB 1, 3, or 4 (Supplementary Figure 2). This leads to the important conclusion that ‘peak-equivalent' Clb2 is a physiologically meaningful level; it provides sufficient mitotic cyclin for timely induction of mitotic entry. These measurements are consistent with the original characterization of clb1, 3, and 4Δ cells that remain fully viable with near-normal cell cycle kinetics (Fitch et al, 1992). Our protocol generated a unimodal distribution of single-cell Clb2kd–YFP levels in cdc20-blocked cells that was similar to the distribution observed for peak wild-type Clb2 (Figure 1E).
Single-cell measurements of inhibition of mitotic exit events by graded levels of undegradable mitotic cyclin
On release from the metaphase block, endogenous Clb2 was completely degraded, regardless of Clb2kd level (Supplementary Figure 9), suggesting that mitotic cyclins Clb1, 3, and 4 are also degraded (Baumer et al, 2000). Furthermore, simultaneous deletion of CLB1, 3, and 4 did not affect the Clb2kd dose–response curves (Supplementary Figure 10). Thus, it is unlikely that endogenous mitotic cyclins are confounding our analysis.
We were surprised at the efficiency of degradation of endogenous Clb2 under these conditions, as this degradation probably requires Cdh1. Cdh1 is inactivated by cyclin–Cdk complex (Zachariae et al, 1998), and so might be expected to be inactive in the presence of high Clb2kd. It is possible that Clb2–Cdk complex is inefficient at inhibiting Cdh1, compared with Clb5 or G1 cyclin–Cdk kinases (Yeong et al, 2001).
We correlated Clb2kd–YFP levels in individual cells to mitotic exit events, including spindle disassembly, initiation of bud ring contraction, completion of cytokinesis, and new bud formation (Figure 2A and B). These data allowed us to generate inhibitory concentration curves for each event.
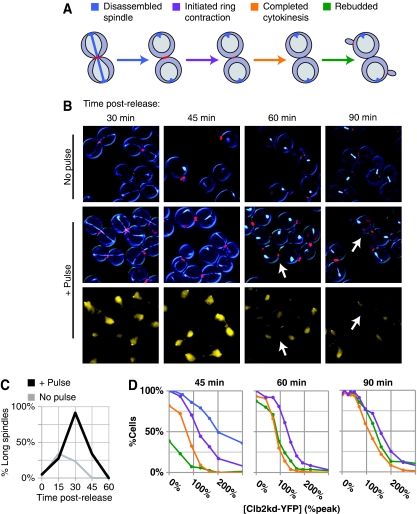
Figure 2.
Peak-equivalent undegradable Clb2 permits mitotic exit. Cells (BD127-11B) pulsed with Clb2kd–YFP were collected at 15-min intervals after release from metaphase block. (A) CFP–TUB1 and MYO1–mCherry allow monitoring of anaphase spindle disassembly, initiation and completion of actomyosin ring contraction, and new bud formation. (B) Representative images with or without the Clb2kd–YFP pulse. For pulsed cells, merged Myo1–mCherry and CFP–Tub1 images are shown above (image processing for readability as described in Supplementary methods), Clb2kd–YFP below. White arrows indicate cells that rebud before completing cytokinesis. (C) Anaphase spindles versus time post-release plotted for pulsed and unpulsed cultures. Average [Clb2kd–YFP] level dilutes from 150 to 100% peak after 60 min (Supplementary Figure 1c) because of increase in cell mass. (D) Mitotic exit events (A) versus [Clb2kd–YFP] level (as schematized in Figure 1C), at 15-min intervals post-release. Unpulsed cells have completed cytokinesis and rebudded by 60-min post-release. As spindle disassembly is completed in nearly all pulsed cells by 60 min post-release (C), spindle disassembly versus [Clb2kd–YFP] level was only measured at 45 min. Color scheme for plots is as in Figure 2A.
Although strong overexpression of stable Clb2 was reported to block spindle disassembly (Surana et al, 1993), a culture pulsed with an average of peak-equivalent Clb2kd only delayed spindle disassembly for ∼15 min (Figure 2B and C). In single cells, persistence of long spindles at 45 min post-release required about two peak-equivalents of Clb2kd (Figure 2D), and by 60 min, even cells with very high Clb2kd levels had disassembled spindles (Figure 2C).
High-dose Clb2kd (>2 peak-equivalents) stably blocked cytokinesis and rebudding. In contrast, one peak-equivalent of Clb2kd delayed but did not block these events; cells slowly constricted their Myo1 rings and frequently formed new buds before completing cytokinesis (budding almost never occurs before completion of cytokinesis in unpulsed cells; Figure 2B). This aberrant phenotype correlates well with the following observations. First, levels of Clb2kd that inhibited cytokinesis and rebudding were similar (1–2 peak-equivalents). Second, these inhibitory levels increased over time at different rates (Figure 2D); by 60 min post-release, rebudding could occur at a Clb2kd level that still inhibited completion of cytokinesis. Strongly delayed cytokinesis was directly observable by time-lapse microscopy of Clb2kd-pulsed cells, as was the occurrence of bud formation before completion of cytokinesis (Supplementary Movies 1 and 2).
These results suggest that the maximal Clb2 concentration attained during a normal cell cycle cannot stably block mitotic exit. Furthermore, Clb2 inhibition of mitotic exit does not reflect a discrete threshold, but rather dose-dependent delays that are process-specific. Spindle disassembly is least sensitive to Clb2kd level, followed by onset of cytokinesis, rebudding, and completion of cytokinesis.
The high level of Clb2kd needed to inhibit rebudding suggests that cells can commit to a new division cycle without reducing Clb2 concentration below peak or without completing cytokinesis. Consistent with this idea, inhibition of SPB duplication after anaphase required at least 1–2 peak-equivalents of Clb2kd, as suggested by comparison with cytokinesis in Clb2kd–YFP-pulsed cells (Figure 3A and B; Supplementary Figure 11). The high requirements for Clb2 inhibition of mitotic exit events compared to normal peak levels suggest that, in a normal cell cycle, Clb oscillation may not suffice to explain why these processes occur in sequence and without repetition. A stable block to mitotic exit requires a level of Clb2 that is rarely, if ever, attained in a normal cell cycle.
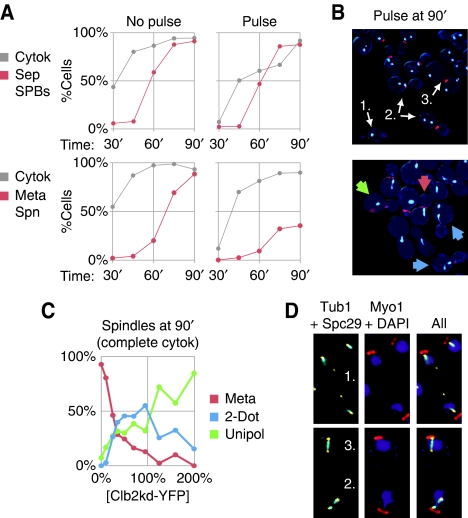
Figure 3.
Persistent Clb2 after mitotic exit causes aberrant spindle assembly in the next cell cycle. (A) SPB duplication and separation (red, top, BD144c-8B) or metaphase spindle formation (red, bottom, BD127-11B) are plotted with completion of cytokinesis (gray) versus time post-release from cdc20 arrest for an unpulsed control and a low Clb2kd YFP pulse (70% peak at the beginning of the experiment, diluting to 40% peak by the end). (B) To the right of each graph are fluorescence microscopy images of cells 90 min post-release: Myo1–mCherry merged with Spc29–CFP (top) or CFP–Tub1 (bottom). For Spc29–CFP, cells with one, two, or three SPBs are indicated (arrows). For CFP–Tub1, arrows label cells with bar-shaped metaphase spindles (red), unipolar spindles (green) or short discontinuous ‘2-dot' spindles (blue). (C) Spindle morphology versus Clb2kd–YFP concentration among cells that have completed cytokinesis at 90 min post-release. Phenotypes charted are described in (B, bottom). (D) Cells bearing CFP–Tub1 and Spc29–YFP (YL178-1-12-2) were pulsed with untagged Clb2kd and released for 90 min from cdc20 arrest. Three spindle types are shown: (1) One dim SPB distant from DAPI staining that does not colocalize with tubulin (top), (2) two SPBs with discontinuous tubulin arrays (bottom), or (3) a normal bipolar spindle (bottom).
Abnormalities in the second cycle after mitotic exit in the presence of undegradable Clb2
Intriguingly, in cells with moderately high Clb2kd level (⩾one-half peak-equivalent), the duplicated SPBs did not nucleate a normal metaphase spindle in the succeeding cell cycle (Figure 3A and B; Supplementary Figure 12)—a spindle-like tubulin structure formed that was attached to only one of the two SPBs (Figure 3C and D; Supplementary Figure 13). In addition, similarly low Clb2kd concentrations abrogated mating factor arrest. Control-unpulsed cells released into mating factor exited mitosis and then arrested without buds or separated SPBs (Figure 4A), whereas cells pulsed with low levels of Clb2kd rebudded (Figure 4B; Supplementary Figure 14), activated the CLN2 G1 cyclin promoter (Figure 4C), and duplicated and separated SPBs, despite the presence of mating factor (Supplementary Figure 15). These findings suggest a previously unsuspected rationale for mitotic cyclin degradation: even if mitotic cyclin levels are not high enough to block mitotic exit, cells that do exit exhibit multiple defects in the succeeding cell cycle.
Figure 4.
Persistent Clb2 after mitotic exit eliminates G1 mating factor sensitivity. A bar1Δ strain (BD127b-9D) was released from cdc20 arrest into α-factor-containing medium (A) without or with (B) a Clb2kd–YFP pulse. Merged YFP, mCherry, and DIC channels are shown. Unbudded cells (black), complete (red) or partial (blue) cytokinesis, and rebudding (green) are plotted versus time post-release for unpulsed cells (A) or versus Clb2kd–YFP concentration 60-min post-release for pulsed cells (B). (C) Cells bearing Myo1–mCherry and GFP fused to the Cln2 degron under the control of the CLN2 promoter (BD143a-21C) were released from cdc20 arrest into α-factor-containing medium with or without a pulse of untagged Clb2kd. Merged GFP, mCherry and DIC channels are shown at 0 and 75 min post-release.
Pulsed Clb2kd is associated with constant histone H1 kinase activity through mitotic exit, and does not display significant regulation by Swe1 or Sic1
Thus far, we have measured the effect of mitotic cyclin concentration on mitotic exit. As cyclins affect cellular processes by activating Cdk, it was important to determine whether our Clb2kd pulses generate an equivalent level of Cdk activity. Therefore, we tested the effects of known Cdk inhibitors, Swe1 and Sic1 (Booher et al, 1993; Schwob et al, 1994), on the Clb2kd dose–response relationships described above, and also directly measured the in vitro Clb2–Cdk kinase activity generated throughout our protocol.
Swe1, the homolog of the Wee1 Cdk-inhibitory kinase, has the potential to downregulate Clb2kd–Cdk activity. However, deletion of Swe1 did not significantly alter the Clb2kd dose–response curves (Supplementary Figure 16). For this reason, we assume that Swe1 did not limit Clb2kd–Cdk activity under the conditions of our assay. This assumption is consistent with the finding that Swe1 is degraded during mitosis and does not reaccumulate until bud emergence in the subsequent cycle (Sia et al, 1998).
Sic1 is another prominent candidate for limiting Clb2kd–Cdk activity (see Introduction section). SIC1 deletion resulted in many aberrant/inviable cells even without a Clb2kd pulse (Nugroho and Mendenhall, 1994; data not shown), preventing the use of sic1Δ cells for reliable assignment of phenotypes to undegradable cyclin. Therefore, we evaluated the role of SIC1 in limiting Clb2kd–Cdk activity by examining the effects of increases in SIC1 gene dosage. Interpretation of these experiments relies on known characteristics of Sic1–Clb2 interaction. Sic1 stoichiometrically binds Clb2–Cdk complexes, removing them from the active pool. We reasoned that if Clb2kd thresholds for inhibiting mitotic exit were set by exceeding a Sic1 blockade, then doubling Sic1 expression levels should sharply increase these thresholds.
In contrast, there are two possible reasons why doubling Sic1 expression might have little or no effect on the thresholds. First, Sic1 levels may not rise high enough to significantly inhibit near-peak Clb2—the relative levels are directly relevant since Sic1 is a stoichiometric inhibitor. Peak Sic1 levels in cycling cells were previously estimated to be less than peak Clb2 levels (Cross et al, 2002). Second, Clb2–Cdk complex phosphorylates Sic1, promoting its degradation, and Clb2 can also phosphorylate and inactivate Swi5, the main SIC1 transcription factor. Therefore, persistent Clb2kd–Cdk activity may prevent significant Sic1 accumulation, in which case doubling Sicl expression may have little effect on Clb2kd dose–response thresholds.
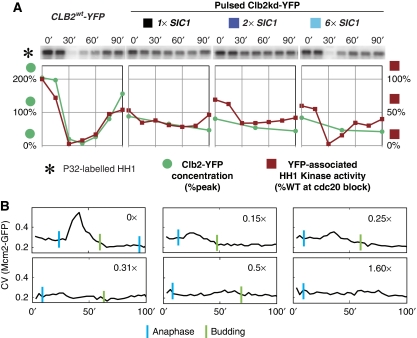
We made 2 × SIC1 and 6 × SIC1 strains by ectopic integration of SIC1. SIC1 transcription increased in proportion to copy number (Figure 5A). 2 × SIC1 did not decrease sensitivity of cytokinesis to Clb2kd dosage. Indeed, sensitivity was only moderately decreased by 6 × SIC1 (Figure 5B).
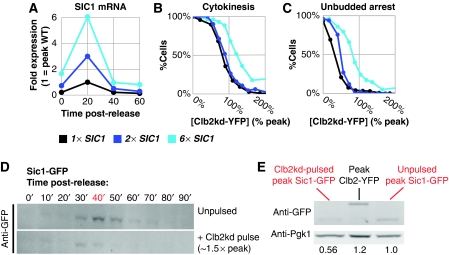
Figure 5.
SIC1 does not set the Clb2kd inhibitory threshold for mitotic exit. (A) SIC1 expression for two multi-copy SIC1 alleles relative to wild type (strains listed below), measured by RT–qPCR performed on samples released from metaphase block into α-factor. SIC1 RNA was standardized to ACT1 RNA, and is expressed as a ratio to peak SIC1 expression in the wild-type background. (B) Cytokinesis completion versus [Clb2kd-YFP] level at 60 min post-release in SIC1WT (BD127b-9D), 2 × SIC1 (BD138a-5), and 6 × SIC1 (BD138a-1) cells pulsed with Clb2kd–YFP. (C) Mating factor arrest versus [Clb2kd–YFP] level at 60 min post-release in SIC1WT (BD127b-9D), 2 × SIC1 (BD138a-5), and 6 × SIC1 (BD138a-1) cells pulsed with Clb2kd–YFP. (D, E) Peak SIC1–GFP is less than peak CLB2–YFP and this is decreased further in pulsed cells. Cells bearing Sic1–GFP (YL205-7-5-4) were released from metaphase arrest (MET-CDC20) with or without an untagged Clb2kd pulse, and Sic1–GFP concentration was monitored by western blot (using anti-GFP antibody). (D) SIC1–GFP levels after release from metaphase in unpulsed versus pulsed cells. (E) Direct comparison of the peaks from (D), as well as peak Clb2–YFP. Quantification (below) was performed by standardizing to Pgk1 loading control and normalizing to wild-type peak SIC1–GFP.
When we examined second-cycle responses to Clb2kd, which occurred at sub-peak Clb2kd levels, such as failure of SPB duplication or failure of mating-factor arrest, (see above), the 2 × SIC1 cassette clearly decreased sensitivity to Clb2kd (Figure 5C). This result confirmed that the increased SIC1 gene dosage could yield increased Sic1 protein with the capability of inhibiting Clb2kd in our protocol. This increased Sic1 production was presumably overwhelmed by titration and/or degradation with higher (near-peak) Clb2kd levels, accounting for the identical inhibitory dose–response of cytokinesis to Clb2kd in 1 × SIC1 and 2 × SIC1 backgrounds (Figure 5B).
To provide a direct biochemical correlate to these genetic experiments, we examined Sic1 accumulation in our protocol with or without a Clb2kd pulse. Sic1–GFP accumulation was transient with or without the pulse, but pulsed cells accumulated an approximately two-fold lower level of Sic1 (Figure 5D and E). As noted above, this could be due to Clb2kd-mediated Sic1 degradation or due to Clb2kd-mediated inhibition of SIC1 transcription, or both. In either case, this finding could explain why increasing SIC1 gene dosage does not affect the thresholds. Importantly, this result is observed with only a single endogenous copy of tagged SIC1.
Parallel immunoblotting using anti-GFP antibody against peak Sic1–GFP and peak Clb2–YFP (wild-type Clb2 from a synchronous time course, as in Figure 1C) showed that peak Clb2–YFP is probably higher than peak Sic1–GFP when standardized to total cell protein (Figure 5E). This quantitative comparison is preliminary; comparing the abundance of proteins of different molecular weight is not trivial, and careful titration of the quantification by serial dilution of all samples and standards has not been carried out. Nevertheless, it suggests that peak Sic1 cannot completely titrate peak Clb2, leaving APC activators, Cdc20 and Cdh1, as primary regulators Clb2–CDK activity. Both conclusions corroborate previous analyses using different methods (Cross et al, 2002; Wäsch and Cross, 2002). As the level of Sic1–GFP in Clb2kd-pulsed cells was even lower, it is unlikely that sufficient Sic1 can accumulate to inhibit peak-equivalent Clb2kd–CDK activity.
To confirm directly that neither Swe1 nor Sic1, nor any other inhibitor, significantly inhibited Clb2–Cdk kinase activity in our experiments, we measured Clb2-associated protein kinase activity in vitro. In control unpulsed 1 × SIC1 (i.e. wild-type) cells, wild-type Clb2 protein and associated in vitro histone H1 kinase activity were completely eliminated 30 min after release from the cdc20 block. In contrast, in Clb2kd-pulsed 1 × SIC1 and 2 × SIC1 cells, kinase activity of a sub-peak-equivalent level of Clb2kd remained constant throughout the time-course (Figure 6A). 6 × SIC1 caused brief inhibition of Clb2kd-associated kinase 30 min after release, correlating with the modest effect of 6 × SIC1 on cytokinesis. Clb2-associated kinase activity per unit protein was similar for Clb2kd and wild-type Clb2 (Figure 6A). Thus, peak-equivalent Clb2kd–Cdk activity is constant throughout our protocol.
Figure 6.
Peak-equivalent undegradable Clb2 yields fully active Clb2–Cdk. (A) Strains from Figure 5A and B, MET-CDC20 CLB2WT-YFP (BD128-41A) control were pulsed with deoxycorticosterone, released from metaphase block, and sampled for protein concentration and YFP-associated in-vitro HH1 kinase activity. [Clb2WT–YFP] and [Clb2kd–YFP] levels measured in units of peak [Clb2–YFP]. Kinase activity is standardized to the metaphase-arrested CLB2WT-YFP level. (B) Mcm2–GFP nuclear localization as an in vivo Clb2–CDK activity marker. Cells bearing Mcm2–GFP and Htb2–mCherry (BD142a-13C) were released from metaphase arrest (MET-CDC20) with or without a Clb2kd–YFP pulse. Coefficient of variation (CV) of Mcm2–GFP signal in individual cells was used as a proxy for Mcm2 nuclear localization (higher values represent higher nuclear concentration, see Materials and methods section). Representative traces from cells with indicated Clb2kd–YFP concentration (numbers in the upper right corner) are shown (n=35). Blue bars=nuclear separation (anaphase); green bars=budding.
Mcm2 nuclear transport as a biosensor for in vivo Clb2kd kinase activity
The MCM complex, required for pre-replicative complex formation at DNA replication origins, is excluded from the nucleus by Clb-dependent phosphorylation (Labib et al, 1999; Nguyen et al, 2000). Using time-lapse microscopy to detect Mcm2–GFP fusion expressed from the endogenous locus, we observed sharp nuclear accumulation of Mcm2 shortly after the release of the cdc20 block in cells not pulsed with Clb2kd (Figure 6B). Pulsed Clb2kd delayed or completely blocked Mcm2 nuclear accumulation; the dose–response data suggested that even sub-peak Clb2kd levels were able to substantially limit Mcm2 nuclear accumulation. This in vivo finding confirms the conclusions reached above that pulsed Clb2kd-associated kinase activity is constitutive in our protocol.
Cdc14 is released from the nucleolus on schedule independently of Clb2kd levels, providing a possible mechanism to allow mitotic exit in the presence of high levels of undegradable Clb2
Cdc14, a phosphatase that reverses Cdk-mediated phosphorylation, is required for mitotic exit in budding yeast (Visintin et al, 1998). Although the results above indicate that the known functions of Cdc14 in activating Sic1 or Cdh1 do not contribute to mitotic exit in Clb2kd-expressing cells, Cdc14 may reverse many other Cdk-mediated phosphorylations (Irniger, 2002; Wäsch and Cross, 2002; Sullivan and Morgan, 2007). Thus Cdc14 may directly antagonize Clb2 inhibition of mitotic exit, without affecting Clb2-associated kinase activity.
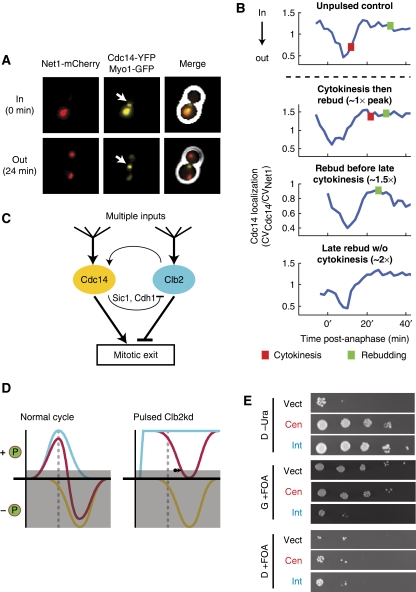
From G1 until mitosis, Net1 anchors Cdc14p in the nucleolus; Cdc14 is released into the rest of the cell only during mitosis (Shou et al, 1999). Using a quantitative assay for Cdc14 release based on co-localization of Cdc14–YFP with Net1–mCherry (Figure 7A; Lu and Cross, 2009), we observed efficient Cdc14 release in all pulsed cells, even in cells expected to contain super-peak-equivalent levels of Clb2kd based on blocks of cytokinesis and rebudding (Figure 7B). Quantification of the kinetics and amplitude of this Cdc14 release event revealed no relationship with Clb2kd levels (data not shown; Y Lu and FR Cross, in preparation). Thus, the balance between Clb2–Cdk activity and released Cdc14 phosphatase may be the relevant variable in ordering mitotic events (Figures 7C and D). In agreement with this model, extra copies of CDC14 rescued the viability of heterozygous CLB2kd/CLB2 diploids (CLB2kd and CDC14 under endogenous promoters) (Figure 7E).
Figure 7.
Mitotic exit may be regulated by kinase/phosphatase balance. (A, B) Cdc14 release from the nucleolus was observed by time-lapse fluorescence microscopy in all cells pulsed with undegradable Clb2. (A) Composite phase contrast, Cdc14–YFP, Net1–mCherry, and Myo1–GFP images of a metaphase-blocked cell (t=0 min) and a cell undergoing release of Cdc14 from the nucleolus (t=24 min). Arrows indicate Myo1–GFP at the bud neck. (B) Cdc14 localization traces at the nucleolus after release from metaphase block for four phenotypes generated by pulsed Clb2kd (YL176-3C). Cdc14 localization was quantified as the ratio of the coefficients of variation (CV=s.d./mean) of pixel intensities for Cdc14–YFP and Net1–mCherry. [Clb2kd] level was estimated by phenotypic correlation with [Clb2kd–YFP] level (Figure 2). (C) Model for co-regulation of mitotic exit by Cdc14 and Clb2–Cdk. (D) Left: normal cells; Clb–Cdk kinase activity (blue) falls after anaphase (dashed line), decreasing phosphorylation of targets. Simultaneous Cdc14 phosphatase release (yellow) further decreases substrate phosphorylation. Mitotic exit occurs when net phosphorylation (red) drops below a threshold (gray box). Right: in the presence of stable Clb2–Cdk activity, Cdc14 can still drive substrate dephosphorylation to allow mitotic exit with a delay (arrow). (E) Additional copies of CDC14 rescue CLB2kd lethality. A diploid strain (ALG876: cdc14/CDC14-YFP CLB2,kd/CLB2 GAL-SIC1(2 × ) ura3/ura3), was transformed with one of three URA3-containing plasmids: vector control (pRS416), centromeric (pRS316-CDC14), or integrating (pRS406-CDC14). Transfomants were grown in galactose and plated in serial dilutions (10-fold) onto glucose (D) or galactose (G) that either lacked uracil (-Ura) or contained 5-FOA. All strains were fully viable on G-Ura (Supplementary Figure 17).
The timing of partial Mcm2–GFP re-accumulation in the nucleus in cells containing low levels of Clb2kd (Figure 6D) was approximately coincident with the timing of Cdc14 release, consistent with the idea that localization of the Mcm complex is controlled by a balance between Clb2-dependent phosphorylation and Cdc14-dependent dephosphorylation. However, we do not have direct biochemical evidence that Cdc14 dephosphorylates Mcm proteins, and the relationship between Cdc14 activity and Mcm relocalization may not be straightforward (Braun and Breeden, 2007).
We noted that eventual mitotic exit in Clb2kd-pulsed cells was frequently significantly delayed after the initial Cdc14 release event, whereas mitotic exit in unpulsed controls is nearly coincident with Cdc14 release. This delay could reflect dilution of Clb2kd concentration as cells grow; we have also observed additional cycles of Cdc14 release under these conditions, which could contribute to ultimate mitotic exit in these experiments (Y Lu and FR Cross, in preparation).
A previous computational model accounts poorly for the quantitative dose–response relationship between Clb2kd and mitotic exit
Chen et al. have presented a model that formalized the cell cycle control network as a set of ordinary differential equations, integrating many experimental results. In this model, the criterion for cell division (mitotic exit) is the reduction in the active Clb2 level below an inhibitory threshold KEZ.
This model was not accurate in simulation of our results (Figure 8A and B), whether we substituted initiation or completion of cytokinesis from our experimental results for model-predicted cell division. The model predicted significantly greater Clb2kd-induced delays than the measured values. Most importantly, modeling a doubling of SIC1 dosage strikingly reduced the simulated delay per unit Clb2kd, whereas it had little effect on the actual delay (Figures 5B and 8B); doubling SIC1 dosage was also predicted to result in a striking decrease in Clb2kd-associated kinase activity on mitotic exit, in contrast to our experimental results (Figures 6C and 8D).
Figure 8.
Computational modeling of mitotic exit in the presence of undegradable Clb2p with or without direct Cdc14 antagonism. (A) Measured intervals from spindle elongation to initiation (blue) or completion (red) of cytokinesis. At 15-min intervals after release, the Clb2kd concentration, that yielded 50% event execution, is plotted against the time from 50% spindle elongation in the culture (this is the same experiment as presented in Figure 2C and D, with the data extracted in this different manner to allow direct comparison with the computational model). Initial Clb2kd concentrations were extrapolated from ∼33% decay over 90 min (Supplementary Figure 1c). (B–D) Simulations of the cdc20-block Clb2kd-pulse protocol (Figure 1B) using either the standard ODE model published by , or a revised model in which Cdc14 directly opposes Clb2 to promote mitotic exit (the criterion for mitotic exit in the original model, Clb2 – KEZ=0, with KEZ=0.3, was altered to Clb2 – (1.5 × Cdc14) – KEZ′=0, with KEZ=0.6). SIC1 expression was also reduced four-fold by decreasing basal and regulated SIC1 synthesis parameters (details of the simulation and revised model in Supplementary Information). The threshold for initiation of DNA replication was increased from 1 to 5 for reasons detailed in Supplementary Information. The model was implemented in MATLAB code revised from that described in Chen et al (2004) to explicitly take account of the cdc20 block-release protocol. For comparison with experimental data, ‘spindle elongation' was arbitrarily taken to occur 10 min after reactivation of CDC20 synthesis. In all other respects, the original and revised models are identical. Code can be made available on request. (B) Initial Clb2kd concentration versus simulated intervals from ‘spindle elongation' to mitotic exit (compare with Figure 6A), using both the standard (left) and revised (right) models. 1 × , 2 × , and 6 × SIC1 simulations are plotted (blue, green, and red, respectively); these were generated by multiplying the SIC1 synthesis parameters by 1, 2 or 6. (C, D) Clb2 protein (green) and kinase activity (red) in peak-equivalent units, simulated with the standard (top) and revised (bottom) models. Compare with experimental data in Figure 4C. (C) Clb2WT protein without the Clb2kd pulse. (D) Peak Clb2kd pulse simulations in 1 × , 2 × , and 6 × SIC1 backgrounds.
For simulated exit to occur in the model, Clb2 activity must be reduced below a hard threshold; activity of undegradable Clb2 can only decrease in the model due to Sic1 inhibition. Thus, this model is structurally unable to account for our results, because at a near-inhibitory Clb2 level, doubling SIC1 expression will necessarily sharply increase the threshold for inhibition, in contrast to the observation. The error in prediction of the Clb2kd-associated kinase activity comes from the same source.
Simple revision of the computational model to allow direct Cdc14–Clb2 antagonism greatly increases concordance of the model with quantitative characterization of Clb2kd mitotic exit inhibition
In the model, Cdc14 inactivates Clb2 only indirectly, through activation of the inhibitors Sic1 and Cdh1 (and also Cdc6 as a minor inhibitor). We hypothesized above that Clb2 kinase–Cdc14 phosphatase balance controls mitotic exit (Figure 7C and D). To conservatively reflect this idea in the model, we altered the ‘exit criterion'. Instead of requiring active Clb2 to drop below a threshold KEZ, we altered the model such that mitotic exit would occur when the quantity (Clb2–(k × Cdc14)) dropped below a threshold KEZ, where k is a parameterization of the strength of Cdc14 relative to Clb2.
Independent evidence suggests that the original model may overexpress SIC1 (Supplementary Information); we suggest that a four-fold reduction in SIC1 expression is reasonably justified on biochemical and genetic grounds (Supplementary Figure 18), and we incorporate this change in our model revision.
Finally, we systematically varied the strength parameter k and the exit threshold KEZ to obtain an improved fit between the revised model and our experimental data relating Clb2kd levels and delay in mitotic exit (Figure 8B). The revised model has a strength parameter k of 1.5 (putting Cdc14 on the same order of activity as Clb2) and an increased KEZ (from 0.3 to 0.6; necessary to rescale because of reduction of overall activity due to the inclusion of Cdc14). This model fits the data acceptably at wild-type SIC1 levels, and importantly, shows essentially no sensitivity to doubling of SIC1 gene dosage, and only a moderate response to 6 × SIC1, as observed experimentally (Figure 5B), and in contrast to the original model (Figure 8B and D).
The revised model has essentially no effect on the timing of accumulation of cell cycle regulators and cell cycle events with otherwise ‘wild-type' parameters, compared with the original model (Figure 8C; Supplementary Figure 19; Supplementary Information), and the revision significantly improved accuracy with respect to a number of genetic results not available at the time of the original model's generation (Supplementary Figures 18 and 20; Supplementary Information).
We could also use the original and revised models to reconstruct predicted kinase activity curves through our protocol in cells pulsed with 1 × Clb2kd (empirical data in Figure 6A). We observed major errors in the original model due to excessive Sic1 inhibition of the modeled kinase activity; in contrast, we observed a striking correlation between the kinase activity profile simulated by the revised model and the empirical results (Figure 8D). We emphasize that the latter finding was an ‘unselected' consequence of the model revision.
Wäsch and Cross (2002) observed that mitotic exit could occur efficiently in the absence of Cdh1 and Sic1 (Wäsch and Cross, 2002). In those experiments, Clb2-associated kinase was essentially constant at a moderate level, throughout mitotic exit. The model of Chen et al (2004) predicts a sharp approximately eight-fold downregulation of Clb2-associated kinase in cdh1 sic1 cells, mediated by hypothetical Cdc6 inhibition; the revised model gives a much more accurate prediction of at most a 40% reduction in Clb2-associated kinase level in cdh1 sic1 cells undergoing mitotic exit (data not shown). This contrast is especially pertinent in the present context, as mitotic exit in cdh1 sic1 cells was absolutely dependent on Cdc14 release (Wäsch and Cross, 2002).
We emphasize that this model revision is only a sketch of a solution, as we did not attempt to evaluate the revised model with respect to all the constraining genetic and biochemical information used to construct the original model; indeed, we have already observed that the revised model is less effective in some such cases than the original (Supplementary Information). However, the revisions we suggest are simple, empirically justified on independent grounds, and provide an excellent fit to our highly quantitative data set. For these reasons, we think that our modeling results provide theoretical support for our conclusions, and could guide further model evolution.
Discussion
Here we have quantitatively tested a hypothesis regarding the control of the cell cycle by cyclin–Cdk complexes (Murray and Kirschner, 1989; King et al, 1994; Stern and Nurse, 1996; Zachariae and Nasmyth, 1999; Morgan and Roberts, 2002; Morgan, 2007), which suggests that oscillation of cyclin locks mitotic entry and exit events into a specific order. We found that stable Clb2 exerts process-specific, dose-dependent delays on mitotic exit (Figure 2D), rather than a single all-or-none threshold; importantly, significant delays in any process may require non-physiological Clb2 levels. Our measurements suggest that additional regulation, beyond Clb–Cdk inhibition of mitotic exit, is necessary to fully explain the order of steps in the cell cycle. Cdc14 release could provide a major counterbalance, even to high Clb–Cdk activity, by direct dephosphorylation of Clb–Cdk substrates. Introducing this modification to a previous theoretical treatment of control of the budding yeast cell cycle may strongly improve the model's ability to simulate our quantitative results.
The counterbalance between Cdc14 and CIp–Cdk could be substrate-specific, based on differential affinities. This could explain different Clb2kd dose-responsiveness for different mitotic exit events. Mcm2–GFP nuclear re-accumulation, for example, seems highly sensitive to Clb2kd levels, compared with spindle disassembly, cytokinesis and rebudding. Too few relevant phosphorylation/dephosphorylation substrates have been identified to begin testing the biochemical basis for these differences.
Subcellular localization of key regulators could also be important. Clb2 and released Cdc14 are concentrated in the nucleus, spindle pole bodies and bud neck (Figures 2B and 7A). If local levels of Clb2 and Cdc14 deviate over time from their global oscillation, then measurements of whole-cell activity could be misleading. Oscillation of the Cdk/Cdc14 balance near their relevant substrates may coordinate mitotic entry and exit events (Sullivan and Morgan, 2007). Incorporating such spatial considerations into a quantitative model will be a major challenge for the future studies.
Other regulatory mechanisms could also contribute to order. Functional dependency (Hartwell et al, 1974) and intrinsic process duration (e.g. the time it takes to build a new spindle pole body) could provide effective cell cycle ordering, especially when reinforced by checkpoints for occasional process failure (Hartwell and Weinert, 1989). It has recently been shown that transcriptional oscillation (Orlando et al, 2008) and cyclical centrosome duplication (McCleland and O'Farrell, 2008) can occur independently of cyclin–Cdk oscillation.
These repeated Cdk-independent oscillations could be integrally involved in cell cycle timing, provided the intrinsic frequency of these independent oscillators is close to the cyclin–Cdk oscillator frequency, and assuming some mechanism(s) for occasional modulation of the phase of these oscillators by cyclin–Cdk activity. In any case, cyclin–Cdk-independent oscillation of cell cycle events suggests the possibility of intrinsic ‘momentum' of cell cycle events, in the face of fixed cyclin–Cdk levels, consistent with the results reported here.
Materials and methods
Loading undegradable mitotic cyclin into pre-anaphase cells
CLB2∷GAL1-CLB2Δdb,ken1,2-YFP ADH1pr-GAL4-rMR cdc20∷MET3-HA3-CDC20 cells, grown in raffinose medium lacking methionine, were incubated for 150 min in 2 mM methionine to turn off MET3-CDC20 and induce a metaphase block. Clb2kd–YFP was induced for 30 min in 10 μM deoxycorticosterone; subsequently, 45 min in glucose+methionine allowed maturation of Clb2–YFP fluorescence. Methionine removal turned on MET3-CDC20 and released the metaphase block. Re-addition of methionine after 45 min collected cells at a second metaphase block.
Quantitative fluorescence microscopy
Cells were lightly fixed in 4% paraformaldehyde. YFP fluorescence was quantified from single unenhanced exposures, after single-cell masking and background subtraction. For analysis of spindles, cytokinesis rings and buds, 3–5 0.3-μm, contrast-enhanced optical sections were combined. These procedures yielded single-cell correlations between Clb2kd–YFP levels and mitotic exit phenotypes. Cdc14–Net1 colocalization was measured essentially as described by Lu and Cross (2009), using the coefficient of variation (CV; s.d. divided by mean) of Cdc14–YFP signal over the cell, divided by the CV of Net1–mCherry. CV decreases as signal spreads through the cell. The use of the CV ratio standardizes apparent dispersal of Cdc14 to that of Net1, controlling for changes in focal plane or Net1/nucleolar morphological variations.
Population Clb2kd–YFP measurement by immunoblot
Clb2–YFP 60-min peak and Clb2kd–YFP pulse samples were serially diluted two-fold into clb2Δ extract for calibration. Blots were probed with anti-GFP (Roche), anti-Clb2, and anti-Pgk1 (Santa Cruz Biotechnology) (loading control). ECL signal was imaged using a Fujifilm DarkBox+CCD camera and quantified using MultiGauge (Fujifilm) software (linear detection range). Multiple comparisons were performed for each time point.
Clb2kd inhibitory threshold calculation
Inhibitory thresholds were measured at different time points after event completion in the unpulsed control. The percentage of Clb2kd peak-equivalence (%peak) was determined by:
where Fcell is the YFP fluorescence of an individual pulsed cell; Wpulse the quantified anti-YFP immunoblot signal from the pulsed population; Wpeak the quantified anti-YFP immunoblot signal from peak synchronized wild-type samples.
Cells were sorted into Clb2kd-YFP bins containing >60 cells, and phenotypes of cells in these bins yielded inhibitory Clb2kd concentrations (Figure 1C).
ODE modeling
An implementation of the Chen et al. (2004) model in MatLab was modified to model precisely the cdc20 block-release protocol used in these experiments; this model was used for testing the previous and variant models for modeled biological and biochemical responses. The code can be made available on request (fcross@mail.rockefeller.edu).
Additional information on methods and strain list can be found in Supplementary materials.
Supplementary Material
Supplementary methods, Model discussion, Strain list, Supplementary figures S1–20
Acknowledgments
We thank L Schroeder for experimental assistance; A Amon, G Charvin, K Nasmyth, D Picard, E Schiebel, and F Yeong for providing strains and plasmids; V Archambault, N Buchler, S Di Talia, S Haase, J Novatt, C Oikonomou, J Robbins, ED Siggia and J Skotheim for discussions and critical readings of the paper; L Bai for quantitative PCR assistance; N Buchler for advice on GAL1 promoter induction and reversible promoters; M Niepel and C Strambio-de-Castillia for cell fixation and microscopy techniques; MP Rout for discussions and for use of reagents and laboratory facilities; and ED Siggia and BT Chait for guidance. This study was supported by a grant from the National Institutes of Health to FRC.
Author contributions: Experimental work performed by BJD, YL, and ALP. YL developed the Cdc14 localization assay. BJD developed the Clb2kd dose–response measurement method, with assistance from BLT. MatLab mask and contrast-enhancement routines were written by BLT and FRC, respectively. The paper was written by BJD and FRC, and edited by all co-authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Archambault V, Li CX, Tackett AJ, Wäsch R, Chait BT, Rout MP, Cross FR (2003) Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. Mol Biol Cell 14: 4592–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer M, Braus GH, Irniger S (2000) Two different modes of cyclin clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett 468: 142–148 [DOI] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW (1993) Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J 12: 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun KA, Breeden LL (2007) Nascent transcription of MCM2-7 is important for nuclear localization of the minichromosome maintenance complex in G1. Mol Biol Cell 18: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ (2004) Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell 15: 3841–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10: 3081–3093 [DOI] [PubMed] [Google Scholar]
- Cross FR (2003) Two redundant oscillatory mechanisms in the yeast cell cycle. Dev Cell 4: 741–752 [DOI] [PubMed] [Google Scholar]
- Cross FR, Archambault V, Miller M, Klovstad M (2002) Testing a mathematical model of the yeast cell cycle. Mol Biol Cell 13: 52–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Schroeder L, Kruse M, Chen KC (2005) Quantitative characterization of a mitotic cyclin threshold regulating exit from mitosis. Mol Biol Cell 16: 2129–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B (1992) Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell 3: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D (1985) Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ (1974) Genetic control of the cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K (1995) Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81: 269–278 [DOI] [PubMed] [Google Scholar]
- Irniger S (2002) Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett 532: 7–11 [DOI] [PubMed] [Google Scholar]
- King RW, Jackson PK, Kirschner MW (1994) Mitosis in transition. Cell 79: 563–571 [DOI] [PubMed] [Google Scholar]
- Knapp D, Bhoite L, Stillman DJ, Nasmyth K (1996) The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol 16: 5701–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Diffley JF, Kearsey SE (1999) G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol 1: 415–422 [DOI] [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U (1998) Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol 8: 231–234 [DOI] [PubMed] [Google Scholar]
- Lopez-Aviles S, Kapuy O, Novak B, Uhlmann F (2009) Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature 459: 592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cross FR (2009) Mitotic exit in the absence of separase activity. Mol Biol Cell 20: 1576–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, O'Farrell PH (2008) RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr Biol 18: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO, Roberts JM (2002) Oscillation sensation. Nature 418: 495–496 [DOI] [PubMed] [Google Scholar]
- Morgan DO (2007) The Cell Cycle: Principles of Control. Published by New Science Press in association with Oxford University Press; Distributed inside North America by Sinauer Associates, Publishers, London, Sunderland, MA [Google Scholar]
- Murray AW, Kirschner MW (1989) Dominoes and clocks: the union of two views of the cell cycle. Science 246: 614–621 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ (2000) Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr Biol 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Nugroho TT, Mendenhall MD (1994) An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol Cell Biol 14: 3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB (2008) Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D (2000) Posttranslational regulation of proteins by fusions to steroid-binding domains. Methods Enzymol 327: 385–401 [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125: 719–732 [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693 [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Sethi N, Monteagudo MC, Koshland D, Hogan E, Burke DJ (1991) The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol 11: 5592–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244 [DOI] [PubMed] [Google Scholar]
- Sia RA, Bardes ES, Lew DJ (1998) Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J 17: 6678–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108: 207–220 [DOI] [PubMed] [Google Scholar]
- Stern B, Nurse P (1996) A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet 12: 345–350 [PubMed] [Google Scholar]
- Sullivan M, Morgan DO (2007) Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol 8: 894–903 [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J 12: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP (2003) Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol 5: 1090–1094 [DOI] [PubMed] [Google Scholar]
- Thornton BR, Chen KC, Cross FR, Tyson JJ, Toczyski DP (2004) Cycling without the cyclosome: modeling a yeast strain lacking the APC. Cell Cycle 3: 629–633 [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460 [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- Wäsch R, Cross FR (2002) APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418: 556–562 [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U (2000) Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28–Clb2 mitotic kinase and the role of Cdc20. Mol Cell 5: 501–511 [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Wang Y, Surana U (2001) Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol Cell Biol 21: 5071–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W (1998) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev 13: 2039–2058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, Model discussion, Strain list, Supplementary figures S1–20