Abstract
Modern genomics technologies generate huge data sets creating a demand for systems level, experimentally verified, analysis techniques. We examined the transcriptional response to DNA damage in a human T cell line (MOLT4) using microarrays. By measuring both mRNA accumulation and degradation over a short time course, we were able to construct a mechanistic model of the transcriptional response. The model predicted three dominant transcriptional activity profiles—an early response controlled by NFκB and c-Jun, a delayed response controlled by p53, and a late response related to cell cycle re-entry. The method also identified, with defined confidence limits, the transcriptional targets associated with each activity. Experimental inhibition of NFκB, c-Jun and p53 confirmed that target predictions were accurate. Model predictions directly explained 70% of the 200 most significantly upregulated genes in the DNA-damage response. Genome-wide transcriptional modelling (GWTM) requires no prior knowledge of either transcription factors or their targets. GWTM is an economical and effective method for identifying the main transcriptional activators in a complex response and confidently predicting their targets.
Keywords: modelling, transcription, microarrays, transcription factor activity
Introduction
Establishing major gene regulatory networks in a cell and modelling their interactions are key goals of systems biology (Camacho and Collins, 2009; Carter et al, 2009). Prevailing systems approaches usually aim to identify the components of a network from the literature, connect them in a consensus-based manner, and then map the intensities of interactions onto this framework (Mo and Palsson, 2009). This has advantage that the biological connections are experimentally verified, and, particularly in single-cell organisms like yeast, these approaches have led to excellent results (Liao et al, 2003; Workman et al, 2006). In multicellular organisms, however, modelling based primarily on documented network architecture is subject to certain limitations. Summarizing from the literature usually entails averaging evidence gathered from different cell types and in different experimental contexts. Molecules involved in multiple systems may be connected differently and may have different effects in each individual system. Network models built on this kind of data, although useful, risk being unrepresentative of specific situations in vivo.
An alternative approach is to identify connections within the whole-genome experimental data gathered in a system-specific manner using highly parallel genomics technologies such as gene expression microarrays. The resulting large data sets can then be analysed to reveal main expression patterns or else the main sources of variability in the data. The most commonly applied microarray analysis methods are generally derived from classical multivariate analysis. Typically, analysis of a data set will initially use statistical filtering (Welch t-test, ANOVA, Limma) to provide an effective summary of the main expression patterns in the transcriptome (Gentleman et al, 2004; Smyth, 2004). Differentially expressed genes can then be grouped into classes according to the similarity of their expression patterns using supervised or unsupervised clustering, or else the main sources of variability in the data can be detected and synthesized into principal components (PCA). These classifications then serve as the basis for identifying, often, large lists of genes with contrasting or predictive patterns of expression.
Although these approaches have been widely and successfully applied for traditional single gene pathway studies, high experimental costs and the lack of tools capable of extracting more than basic information has generally restricted interpretation of microarray data. Commonly, results are limited to the identification of the most differentially expressed genes in snapshots of an isolated, and often genetically modified biological system. To realize the full potential of microarrays and also of next-generation clonal sequencing technologies, it is necessary to develop flexible and efficient methods for using genome scale data to examine the transcriptome as a whole. Several groups have begun this task, applying Boolean and dynamic Bayesian network tools to predict the connectivity and behaviour of theoretical and specific biological systems (Husmeier, 2003; Zou and Conzen, 2005; Dojer et al, 2006). However, a widely applicable and experimentally verified method for genome wide transcriptional analysis is yet to be established. Our first aim was, therefore, to create a model capable of explaining as high a proportion of an induced transcriptional network as possible, solely on the basis of measurable data.
Lists of differentially expressed genes from microarray experiments are frequently extensive. Typically only low numbers of replicates (usually just three per condition) are run, and so the application of conventional statistics is seldom adequate to determine the extent to which these gene lists are interpretable. Our second goal, therefore, was to attribute a degree of confidence with which differential gene expression can be described as biologically meaningful.
We used the DNA-damage response in the MOLT4 human T-ALL cell line as a model system. The cells show a normal response to damage and die by apoptosis in a p53-dependent manner (Barenco et al, 2006). We have previously shown that by linking transcript levels measured in a time series after induction of damage, a dynamic picture of network activity can be created. We developed a mathematical modelling approach called Hidden Variable Dynamic Modelling (HVDM) (Barenco et al, 2006), available as an R package in Bioconductor (Barenco et al, 2009). HVDM incorporates RNA production and degradation terms and prior biological knowledge, namely genes known to be targets of a specific transcription factor, and deduces the activity of that transcription factor from microarray data.
HVDM predictions were often surprising. Many p53 targets were predicted accurately despite having quite different transcript time profiles. The most likely explanation for these findings was the effect of differential mRNA degradation rates. Transcripts with a high degradation rate, or short half-life, tend to track the shape of their activator more closely than those transcripts that degrade slowly. The influence of RNA degradation on transcript accumulation had received little attention in a systems context in which emphasis is typically put on mRNA production and activation. Recently, however, it has been shown that RNA turnover rates are central in shaping the response of yeast cells to different stimuli (Shalem et al, 2008).
Therefore, in this study we used microarrays to estimate the RNA degradation rates of all transcripts and used this information to generate a new approach we call genome-wide transcriptional modelling (GWTM). We applied GWTM to dissect the transcriptional response to DNA damage into its component activities, and identified, with confidence intervals, the targets of each activity.
Results
Model System—the response of human MOLT4 cells to ionizing radiation
Ionizing radiation causes double-strand DNA breaks and activates a complex transcriptional response in mammalian cells. Irradiation-activated genes are involved in a variety of cellular processes, including cell cycle arrest, apoptosis and cell survival. The network centres on the transcription factor and tumour suppressor, p53. We have previously modelled the p53 network using HVDM, generating a ranked list of p53 targets which we verified experimentally using siRNA (Barenco et al, 2006). This study also showed that many genes activated in the irradiation response network are independent of p53, or are co-regulated by another factor. We aimed to explain the behaviour of as high a proportion of the response as possible.
RNA degradation rates determine transcript time profiles
Conventional methods for identifying the number of activities involved in a complex transcriptional response are based solely on transcript levels. We have shown previously that clustering at this level is inefficient in identifying p53 targets in a complex response (Barenco et al, 2006). During the dynamic response to ionizing radiation, the rate of change of transcript concentration is a function of its production and degradation rates, and can be described by the following ordinary differential equation.
The first two terms on the right hand side of the equation are production terms. The rate at which a gene j is produced can be divided into a constant (or basal) transcription rate, Bj, and a rate that varies during the response as a consequence of transcription factor activity, f(t). As the activation profile of a gene is potentially gene specific, we will attach to f(t) an index, j. A transcription factor can act on multiple target genes, but its effect on each gene will depend on specific target gene sensitivity, Sj—for example, as a result of differing affinity for the promoter. The total production rate of the gene is, therefore, the sum of the constant term Bj and a time-dependent term Sjfj(t). The third term is a degradation term. mRNA molecules are degraded by nucleases at a rate, Dj. We assume the overall degradation rate to be proportional to transcript concentration Xj(t).
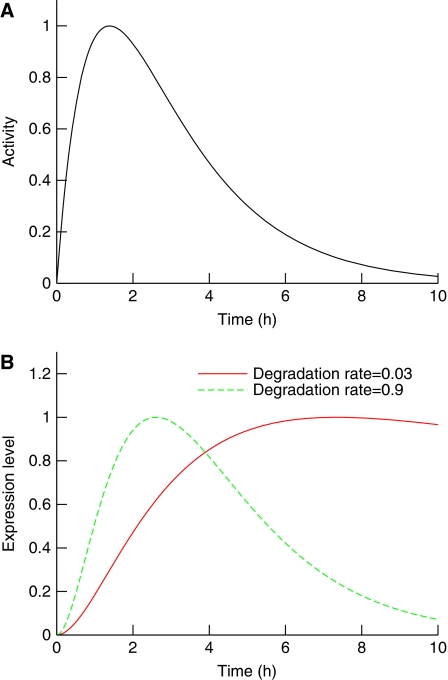
Exploration of the theoretical effects of RNA degradation on network modelling revealed that different transcript degradation rates could have a significant impact on the time profiles of gene expression data (Figure 1). The higher the degradation rate, the more the expression profile will tend to track the activator profile. In contrast, lower degradation rates are associated with comparatively slower responses.
Figure 1.
Transcript degradation rates determine the shape of the response. (A) The activity profile of a hypothetical transcription factor. (B) The corresponding normalized responses of two target genes with different degradation rates. At a higher degradation rate, the initial response and the subsequent decay are swifter, and the transcript profile corresponds better to the driving activity. In contrast, with a smaller degradation rate, the transcript response profile is delayed and peaks later.
Measuring transcript degradation rates allows modelling of transcript production
We then hypothesized that knowledge of global RNA degradation rates could be used to extract extensive hidden information about gene networks from microarray data.
To obtain degradation rates that are as specific as possible to our system, we measured them directly by activating the DNA-damage response for 4 h, blocking the transcription using actinomycin D, then running a microarray time course to measure transcript levels at intervals over the following 6 h (see Materials and methods and Supplementary information).
By measuring the degradation rate and transcript time profile on microarrays, we experimentally quantified the terms Dj and Xj(t) in our model equation above. From this information we can also infer the first derivative dXj(t)/dt. We then rearranged the model equation to collect these known terms on the LHS of the equation.
This creates an identifiable term, Gj, which is equivalent to a compound production term for a given transcript at a given time. Gj time profiles represent an affine transformation of the activation profiles of individual genes, that is, the profile may vary in amplitude or scale, but its shape remains the same. This shape invariance can be exploited to group genes together. Genes that share similar Gj profiles are likely to be directly controlled by the same regulatory activity. Often, but not exclusively, this could relate to active transcription factors.
Generation of discrete Gj profiles from microarray data
To generate Gj values for all changing transcripts, we used the same transcript time series values used in HVDM (Barenco et al, 2006). We assume that the damage response network is in equilibrium before irradiation, that is, the rate of change of its constituents is zero. Irradiating the cells disrupts the equilibrium and activates various transcription factors, including p53.
We defined Gj profiles above using a continuous framework, but experiments only provide measurements at specified time points. Therefore, we devised a method to obtain discrete Gj profiles. First, we calculated DjXj(t) by multiplying each transcript expression level at every time point by the corresponding transcript degradation rate measured independently. Approximation of the first derivative at the measurement points can be obtained using local Lagrange interpolation (see Mathematical methods section and (Barenco et al, 2006)). Using this method, individual slope estimates are linear combinations of the entries of Xj(t) and the first derivative vector can be written as A.Xj(t), where A is a square matrix containing the relevant coefficients. Therefore, individual, discrete Gj profiles can be calculated using
where I is the identity matrix. A entries only depend on the time vector (in our case 0, 2, 4, 6, 8, 10, and 12) and some of them are negative (see Materials and methods). A is not gene specific and can be computed just once and then applied to all gene time profiles. One profile of Gj values was generated for each experimental replicate time series (Gj,r; r=1, 2, 3).
Discovery of principal transcriptional activities using a graph representation
We proceeded to cluster the data using a Gj profile composed of three individual Gj,r profiles i.e. Gj=[Gj,1,Gj,2,Gj,3] with Pearson's correlation coefficient (which is unaffected by affine transformation of the underlying data). Genes that share a similar activation profile will have a correlation coefficient approaching 1. This should allow clustering of genes that share similar activation profiles. However, the real situation is more complicated because Gj is a composite production term. In the case of genes that are co-activated by two or more transcription factors, the Gj profile will represent a combination of the corresponding activation profiles. If this ‘blurring' is a common occurrence, then clearly separated clusters are unlikely to appear. In line with this, initial attempts to cluster Gj profiles using K-means clustering with a Pearson correlation coefficient did not identify sufficiently discrete activity profiles (see Supplementary information).
Experimental variability and measurement imprecision also confounded attempts at correlation. We observed that genes expressed just above detection limits tend to have a high level of noise, and that some degradation rates could not be calculated with sufficient precision (for example, because of very rapid degradation).
To solve these problems we applied a graph representation to locate regions where the density of Gj profiles is high. Graphical representation has previously been used in the context of gene expression profiles clustering (Sharan and Shamir, 2000). This has the advantage that it can overcome noise, imprecision and blurring by identifying groups of genes that are tightly associated with one another. We attached an edge to any pair of genes correlation of which was above a threshold, α (0.80). We then calculated the correlation coefficient between each replicate Gj profile and excluded genes with poorly correlated (β<0.45) replicates. We found that this effectively excluded genes hampered by a high level of noise.
Gj clustering identifies three global activities in the MOLT4 DNA-damage response
Using the effective Bron–Kerbosch algorithm (Bron and Kerbosch, 1973), we determined all maximal cliques in the graph defined above. Cliques are groups constituent genes of which are connected to every other gene in the clique. Maximal cliques are those cliques that are not a subset of a bigger clique. Maximal cliques having genes in common were then merged. We call these entities merged cliques and only considered those that comprised at least four genes. To check the robustness of the method and find the optimal values for the two parameters α and β, we systematically varied their respective values (see Materials and methods). By construction, the merged cliques constitute tight clusters and are, therefore, likely to correspond to the principal activities of the response.
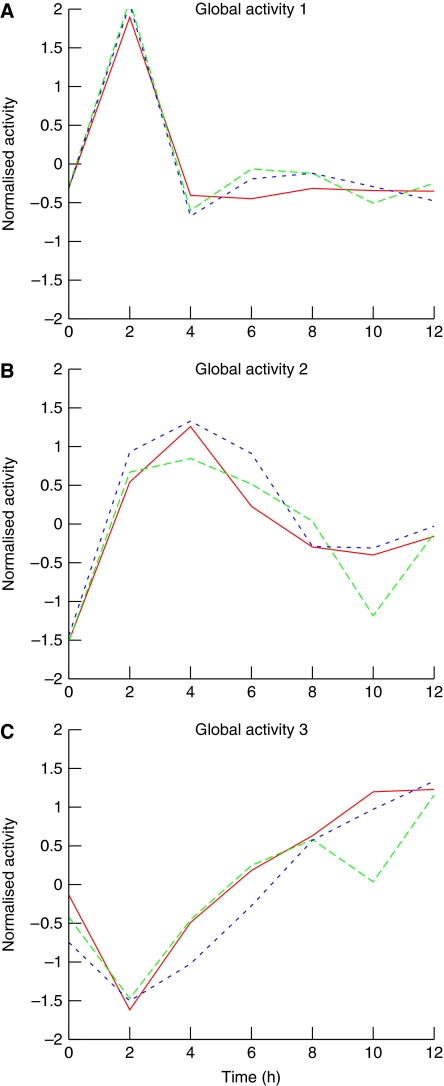
With a threshold value for α of 0.80 and β of 0.45, we found three merged cliques, indicating that there are at least three major global activity profiles in the DNA-damage response network (Figure 2). Activity profiles were then obtained by averaging the normalized profiles of its constituent genes. Merged clique 1 consisted of 51 transcripts, and the shape of the main activity corresponding to this first main clique is indicative of a strong early response, the activity profile peaks at 2 h and decays immediately afterwards (Figure 2A). The activity profile of merged clique 2 (15 transcripts) indicates a strong response that peaks 4 h after irradiation and then diminishes until reaching a plateau, at 8 h (Figure 2B). The eight transcripts in merged clique 3 have a very distinctive profile, which diminishes to a trough at 2 h and then increases at a constant rate reaching the starting value at about 5 h (Figure 2C). The three global activity profiles identified are robust, in that the merged cliques that lead to their discovery appear with a relatively wide range of values for these parameters (see Materials and methods). The constituent genes/transcripts of these cliques are provided in the Supplementary information (Supplementary Tables I–III).
Figure 2.
The three main activities in the DNA-damage response network. Individual activity profiles of the constituents of the merged cliques were normalized to an average of zero and an s.d. value of one. Global activity profiles were obtained by averaging these normalized profiles. Within each panel, individual curves represent a different replicate. (A) Global activity 1 corresponds to the activity of both NF-κB and c-Jun/AP-1, and has a rapid onset and decline. (B) Global activity 2 corresponds to the activity of p53, a transcription factor that is pivotal in the DNA-damage response network. (C) Global activity 3 likely corresponds to the transcription of genes in cells re-entering the cell cycle.
Identification of candidate transcription factors using bioinformatics methods
We adopted a bioinformatics approach to identify cis-regulatory domains, known transcription factor control and functional composition and connectivity of the genes in each merged clique (see Materials and methods).
Merged clique 1 (51 genes/transcripts)
We applied TRED (Jiang et al, 2007) to search for TF-binding motifs shared by constituents of MC1 (see Bioinformatic methods). This approach generated a clear enrichment of NF-κB-binding sites in the promoters of genes from merged clique 1 compared with a gene list derived from merged clique 2 (MC1 36% of screenable genes (n=30) had a NF-κB-binding site (matrix threshold >5) within 1000 bp (700 bp upstream and 300 bp downstream) of the transcription start site compared with 7% in MC2 (n=57). Although GeneTrail (Backes et al, 2007) failed to identify any known transcription factors significantly associated with the list, analysis of Gene Ontology categories using Ingenuity Pathway Analysis identified significant associations (P<0.05) with both NF-κB and the c-Jun-containing AP1 complex (see Bioinformatic methods). Included in these associations are RELBs, a constituent of the NF-κB complex (Bours et al, 1994) and I-κBα (Brown et al, 1993), TNFAIP3 (Krikos et al, 1992), IER3 (Osawa et al, 2003), and TRAF4 (Glauner et al, 2002) are NF-κB targets. c-Jun/AP-1 targets included c-Jun itself (Yazgan and Pfarr, 2002), CD69 (Castellanos et al, 1997), CD83 (Kim et al, 2004), TNFAIP3 (Hayakawa et al, 2004) and ATF3 (Lu et al, 2007). We concluded that this clique is probably composed of targets of two major transcriptional activities, controlled either separately or together by the transcription factors c-Jun/AP-1 and NF-κB.
Merged clique 2 (15 genes/transcripts)
Gene Set Enrichment Analysis (GSEA) of MC2 using GeneTrail (see Bioinformatic methods) revealed a strong enrichment of p53 targets (and only p53 targets; P=1.3357e−06). In fact eleven are well known p53 targets: BTG2 (Rouault et al, 1996), KIAA0247 (Staib et al, 2005), Fas (Li et al, 2004), CCNG1 (Bates et al, 1996), SESN1 (Velasco-Miguel et al, 1999), TRIM22 (Obad et al, 2007), CDKN1A (Sugihara et al, 2004), RPS27L (Li et al, 2007), DRAM (Kerley-Hamilton et al, 2007), BIK (Marko et al, 2003) and TNFSF10B. TRED did not identify an enrichment in p53-binding sites in the promoter regions, but many p53 genes are regulated outside conventional promoters (Wei et al, 2006). Ingenuity pathway analysis of the gene ontology of components of this group also demonstrated a significant (P<0.05) link with both the p53 pathway and apoptosis. Combined with previous study (Barenco et al, 2006), this strongly suggested that clique 2 represents p53 activity. An additional gene in this list, ASCC3, was previously predicted by HVDM and verified as p53 target after knockdown experiments (Barenco et al, 2006).
Merged clique 3 (8 genes/probe sets)
This clique comprises eight genes. Although neither GeneTrail nor TRED revealed enrichment for particular sequence motifs or known transcription factor targets, Ingenuity Pathway Analysis revealed that all these genes have a significant role in the mitotic phase of the cell cycle (P<0.05). CDK1/CDC2 induces entry into the mitosis (Lee et al, 1988). The products of three other genes are the microtubule-associated proteins, NUSAP (Raemaekers et al, 2003; Ribbeck et al, 2007), TASTIN/TROAP (Yang et al, 2008), both required for the mitotic-spindle organization and assembly, and KIF2C/MCAK (Kim et al, 1997; Ganguly et al, 2008), a kinesin-related motor protein, known to have an important role in spindle assembly and correcting errors in mitotic chromosome alignment. Finally, TOPK/PBK was also found in this clique, which is a MAPKK-like mitotic kinase involved in the spindle midzone formation and cytokinesis (Matsumoto et al, 2004; Abe et al, 2007). These members suggest that the activity profile captured by this merged clique is associated with the G2/M cell cycle phase and cytokinesis. One of the responses associated with DNA damage is cell cycle arrest at the G2/M transition (Vousden, 2000). This could explain the initial drop in transcript concentration. The subsequent rise probably results from a synchronized re-entry into the cell cycle of a surviving subpopulation of irradiated cells.
Transcription factor target predictions using global activity profiles
Two of the activity profiles we discovered were associated with three distinct transcription factors. Having identified the likely activities, we then developed a procedure to screen all the differentially expressed genes to predict which were direct targets of those activities. At first, we simply computed the correlation coefficient between individual Gj profiles and global activity profiles. Genes that yielded a high correlation were deemed to be targets of the transcription factor corresponding to these activities. However, we rejected this approach on the grounds that the technical error associated with expression measurements of individual genes was insufficiently taken into account (see Supplementary information). To overcome this problem, we devised a model-based approach that allows for principled testing and ranking of potential targets. It is noteworthy that although this decision was taken purely on principle, it is more effective, in terms of performance (see section ‘By incorporating known degradation rates, GWTM improves on existing methods').
The equation for the model we used to fit each individual gene expression Xj is shown below. The two parameters to be estimated are B′j and Sj:
where
and f is the global activity profile corresponding to the transcription factor under review. A detailed technical description of how we arrived at these formulas, along with the physical interpretation of the various components of the model can be found in the Supplementary information.
After screening (see Mathematical methods), this model was applied to the 246 most significantly upregulated genes. In the screening procedure, each gene was fitted with the model above, replacing f with the global activity profile under review. To be a predicted target a gene j had to fulfill two conditions. First, either the model score, that is, the squared sum of the residuals between model and data, had to be smaller than 100, or that sum had to represent less than 30% of the one obtained with the null model (that is, without a varying activation term). Second, the sensitivity Sj had to be significantly different from zero. As a measure of the robustness of this important parameter, we used the Z-score (that is, the reciprocal of the relative estimation error). Genes that passed the model score condition with a sensitivity Z-score greater than 6 were predicted to be targets. Each list of predicted genes was ranked according to descending sensitivity Z-score.
This resulted in a list of 57 genes predicted to be targets of NF-κB or c-Jun/AP-1, 93 genes were predicted to be targets of p53, and 17 genes were associated with the cell cycle principal activity (see Supplementary tables IV–VI). In total, predicted genes covered 70% of the 200 most upregulated genes that were screened in this procedure.
This screening step can also help to validate global activity discovery of the previous step, especially if, in the context of a less well-documented biological system, bioinformatic identification of those activities is more difficult. It may happen, for example, that a given activity is divided in the clustering step (see Supplementary information). In such cases however, it is likely that the subsequently predicted target lists will overlap in a significant manner. Such example is demonstrated in the Supplementary information (Supplementary Figure S3). In our case, the overlap between the three lists were minimal: six genes were part of both the p53 and NF-κB or c-Jun/AP-1 predicted sets and another three were part of both the p53 and cell cycle predicted sets. Generally speaking, having two steps to generate predictions allows more flexibility and quality in the outcome. This is detailed in the Supplementary information section ‘Comparison of GWTM with Graph Based Clustering of expression values'.
Experimental verification of GWTM predictions
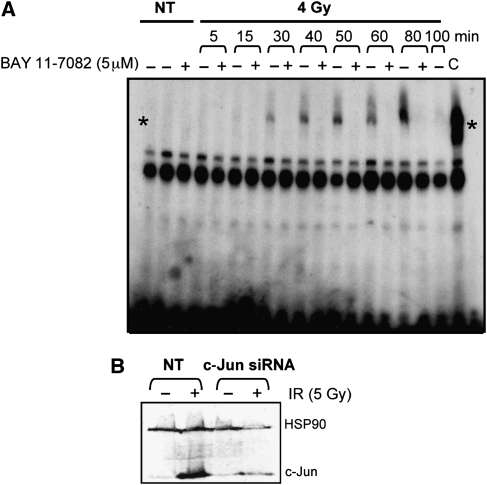
To verify our predictions, we used independent knockdown experiments for each of three transcription factors under review to identify their targets. We carried out verifications at 2 or 4 h after IR. Although the majority of genes induced in this time frame are likely to be direct targets, there remains a possibility that the list contains a proportion of indirect targets. MOLT4 cells were transfected with either siRNAc-Jun, siRNAp53 or treated with BAY 11-7082, a specific inhibitor of NF-κB (Pierce et al, 1997; Mori et al, 2002). After irradiation, cells were assessed for the effectiveness of the knock down. For NF-κB, using chromatin IP, we verified that the compound inhibited nuclear localization and DNA binding of NF-κB. (Figure 3A). We used western blots to evaluate the levels of remaining c-Jun protein after transfection with siRNAc-Jun following irradiation (Figure 3B). The effectiveness of siRNAp53 in reducing p53 activity has been published previously (Barenco et al, 2006).
Figure 3.
Experimental inhibition of transcription factors identified by GWTM. (A) Electrophoretic mobility shift assay showing precipitation of DNA bound NF-κB (*) after irradiation (4 Gy; NT, not treated) in the presence (+) or absence (−) of the specific NF-κB inhibitor BAY 11-7082 (C, control). (B) Western blot showing levels of c-Jun before and after irradiation (5 Gy) in the presence or absence of transfected siRNAc-Jun. (NT, not treated; HSP90, loading control).
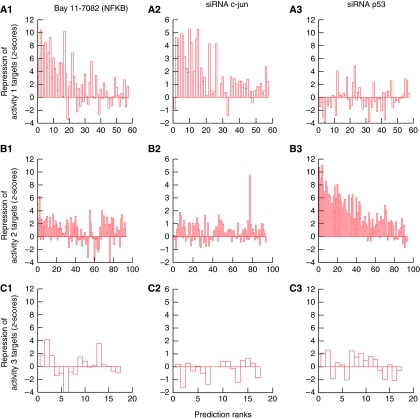
Each of these experiments involved four microarrays and the result was summarized into a verification Z-score (see Supplementary information on how these verification scores were obtained). The results of the screening and subsequent verification are described below for each global activity profile (Figure 4).
Figure 4.
Experimental verification of GWTM predictions. MOLT4 cells were irradiated in the presence of specific inhibitors of the three transcription factors identified as regulating the two major activities predicted by GWTM. Values on the y axis are Z scores representing the degree to which transcription of a target is reduced as a result of addition of the given inhibitor. Rank of GWTM prediction for each activity is represented on the x axis. (A: 1–3) Global activity 1 is inhibited principally by the NF-κB inhibitor, BAY 11-7082, but also to a slightly lesser degree by siRNAc-Jun. (B: 1–3) Global activity 2 is inhibited only by siRNAp53. (C: 1–3) Global activity 3 is unaffected by specific inhibitors of NF-κB, c-Jun or p53. The decreasing Z-scores with rank indicate the accuracy of model predictions and their ranking.
Global activity profile 1: NF-κB and c-Jun/AP-1
NF-κB activity was inhibited using the specific inhibitor, BAY 11-7082, after irradiation with 4-Gy γ-irradiation. We then calculated a Z-score that represents the degree to which BAY 11-7082 reduces irradiation-induced transcription of all target genes. The resulting list of 69 genes (verification Z-score threshold >2) was then compared with a ranked list of 69 predictions made by GWTM for activity profile 1. The upregulation of 23 of the top 30 GWTM predictions was found to be strongly inhibited as a result of NF-κB inhibition (Figure 4, A1). About 56% of the total number of verified genes were predicted in activity 1. If the verification threshold was set to 4 (25 genes), this percentage was 64%. It was noted that BAY11-7082 also reduced, but to a lesser degree, the upregulation of genes across the experiment, suggesting a minor non-specific affect on transcription.
Bioinformatic analysis indicated that c-Jun/AP-1 was also a potential activity responsible for global activity profile 1. A Z-score was calculated representing the degree to which siRNAc-Jun reduces irradiation-induced transcription of all target genes. The resulting list of 26 genes (verification Z-score threshold >2) was then compared with a ranked list of 69 predictions made by GWTM for activity profile 1. (Figure 4, A2). About 62% of total verified genes were predicted in activity 1, this value increases to 92% if the verification threshold is set to 3 (13 genes). This was a particularly specific knockdown as no other activities showed significant levels of reduction in response.
The subsequent analysis of transcripts, IR-induced upregulation of which was inhibited by c-Jun knockdown revealed that although a few of the global activity 1 transcripts were direct c-Jun/AP-1 targets, many of the established NF-κB targets were also partially reduced by c-Jun/AP-1 inhibition (Figure 4, A2). This suggests that many of the genes in this category are in fact co-regulated by NF-κB and c-Jun/AP-1.
Global activity profile 2: p53
The p53 activity was knocked down using siRNA for p53 after irradiation with 5-Gy γ-irradiation. The data for this knockdown have been previously published (Barenco et al, 2006).
The resulting list of 95 transcripts knocked down by siRNAp53 was then compared with a ranked list of the 93 predictions made by GWTM for activity profile. Upregulation of 42 of the top 50 GWTM predictions was found to be strongly inhibited as a result p53 inhibition (Figure 4, B3); in total, 64% of the verified genes were also predicted. The overlaps between verified and predicted genes are highly significant given the relatively low number of such genes in the whole of a microarray experiment; such high percentages (see also Figure 8) are unlikely to be obtained by chance.
Cross-verifications
Although predictions of the genes associated with the third principal activity were carried out, we did not run a verification experiment for this set of genes, as a single transcription factor could not be unequivocally identified in this case. However, it is noteworthy that the verification scores for the three transcriptions factors under review are, as should be expected, generally poor for this particular set (Figure 4, C1–C3 in contrast with, respectively, A1 and A2 and B3). The same observation holds for the p53 targets verification scores of which are poor for NF-κB (Figure 4; B1, contrast with A1) and c-Jun/AP-1 (Figure 4; B2, contrast with A2). Conversely, the predicted targets of the first principal activity show little p53 inhibition (Figure 4; A3, contrast with B3).
Ranking of predictions is effective
Individual predictions in each category were ranked according to descending robustness of the sensitivity parameter. This ranking system is effective as the top genes in the lists tend also to have the highest verification scores (Figure 4; A1, A2, B3).
Transcript degradation rates have a significant effect on resulting expression profiles
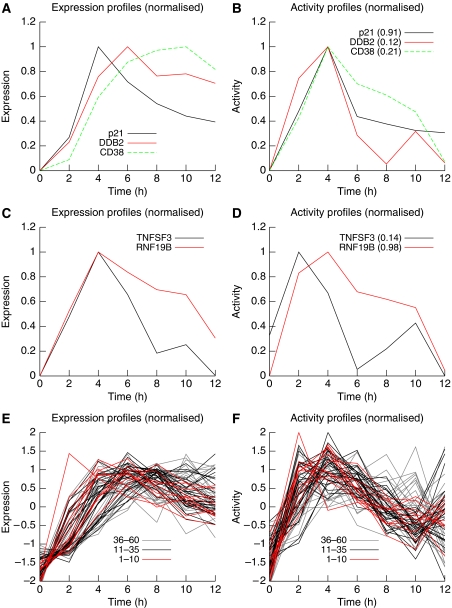
Using GWTM predictions, measured degradation data and experimental inhibition of the chief activities, we were able to verify that degradation rates significantly contribute to transcript time profiles as initially predicted (Figure 1). We observed that transcripts with very different time profiles could be targets of the same transcriptional activity. For example, p21, DDB2 and CD38 are transcripts which peak at 4, 6 and 10 h respectively after irradiation, yet because their degradation rates differ, they are in fact predicted and verified targets of the same transcription factor, p53 (Figure 5A and B).
Figure 5.
Expression profiles and individual activity profiles for selected genes. (A) The normalized expression profiles of three verified p53 targets. Transcripts with a slower turnover rate (DDB2 and CD38) accumulate slowly and peak later than other genes (p21) for which degradation rate is higher. (B) Subtracting the degradation component reveals the production component. The individual activity profiles are similar because the three genes under review are activated by the same transcription factor, p53. (C) Despite having different activators (NF-κB and p53, respectively), genes TNFSF3 and RNF19B exhibit similar expression profiles. (D) TNFSF3 expression results from the combination of a fast onset activation (controlled by NF-κB) with a relatively ‘slow' degradation component. In contrast, RNFB19B is activated by a ‘slower' transcription factor (p53) but tends to track its movement more closely because of a relatively high transcript turnover rate. (E, F) are the same as (A, B), respectively, but include more predicted p53 targets, the top 60 (A, B represent the top three predictions). Contrasting the profiles shown in (E, F) helps creating a more coherent image. Ranking of the genes in the prediction list has been colour-coded and shows that lower ranked genes exhibit a more noisy profile, calling for a principled way to attribute genes individually to global activities.
Similarly, transcripts with very similar profiles can be the targets of different activities. Lymphotoxin-β (TNFSF3) and RNF19B share a similar expression profile (Figure 5C), peaking at 4 h and declining steadily to 12 h. GWTM using measured degradation rates predicted correctly that despite these similar profiles, Lymphotoxin-β is a target of NF-κB, whereas RNF19B is a p53 target gene (Figure 5D).
The same observations can be made on a large scale. The first 60 predicted targets of p53 tend to peak during a wide range of time points (from 4 to 12 h after irradiation; Figure 5E), whereas their activity profiles are more coherent (Figure 5F). It is noteworthy, however, that a principled approach to individual gene attribution to activities is indeed necessary (see ‘Transcription factor target predictions using global activity profiles' section above), as lower ranked genes (36–60) tend to have a noisier profile (Figure 5F).
By incorporating known degradation rates, GWTM improves on existing methods
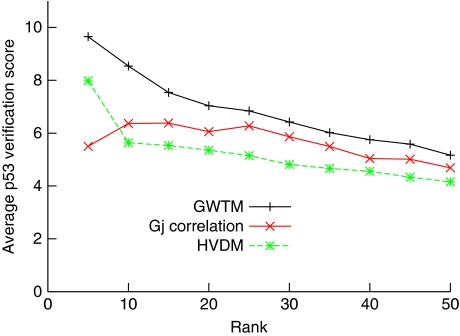
Although no previous knowledge of p53 targets was used in the modelling, GWTM was successful in identifying verifiable targets. When this list is compared with the list generated by HVDM, we found that an overlap of more than 60% in the most upregulated genes (33 of 50). The difference between the two originates, in part, from a stricter filtering applied in GWTM (see Materials and methods), which better identifies unreliable measurement of transcript values. Unsurprisingly, excluded genes had low, sometimes negative, verification scores. More importantly, knowledge and incorporation of the degradation rate permitted a re-ranking of predictions. Most of the ‘newcomers' in the top 50 GWTM list are genes with a lower dynamic range in the signal, which led to wider confidence intervals in the HVDM estimation. Inclusion of the known degradation rate caused a slight deterioration in the model fit, but greatly improved the robustness of the sensitivity rate, which we used in both methods to rank the predictions. Overall, this change benefits the quality of predictions (Figure 6, green curve). For the top 50 genes, the verification scores are better (22% higher on average) and higher verification scores tend to be ranked more accurately. If only the top 25 genes are compared, the verification scores are on average 30% higher with GWTM. This shows that taking into account extra information, in the form of individual transcript degradation rates, can provide perceivable benefits for the prediction quality. Finally, it is noteworthy that basing predictions and ranking of putative targets only on the correlation coefficient also delivers inferior results (Figure 6, red curve).
Figure 6.
Comparison of various methods for predicting p53 targets. Each curve represents the average p53 validation score up to the rank indicated on the horizontal axis. +/black, GWTM; */green, HVDM; x/red, using correlation of individual Gj profiles with the p53 global activity profile ranked by descending correlation coefficient.
Main activity predictions cover a large majority of the most upregulated genes in the DNA-damage response system
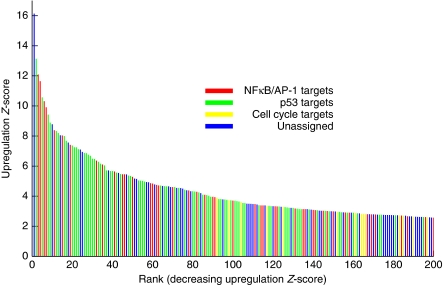
The 246 radiation-responsive genes were ranked by descending upregulation Z-score. The transcriptional activity responsible for more than 65% of these transcripts (Figure 7) could be discovered using GWTM. This percentage improves for most upregulated genes, with GWTM explaining 76% of the 100 most upregulated genes and 82% for the first 50 (Figure 8, black curve).
Figure 7.
GWTM predicts a large majority of the upregulated transcripts. A Z-score representing degree of DNA damage-induced upregulation was computed for each differentially expressed gene. GWTM-predicted targets of each of the three principal activities are highlighted (red, NF-κB- or c-Jun/AP-1-predicted targets; green, p53-predicted targets; yellow, cell cycle-predicted targets). The blue coloured bars correspond to genes that were not predicted to be direct targets of any of the three activities. Some of those may be co-regulated targets of more than one activity. For example, the most highly regulated gene, IER3, is a target of both NF-κB and p53.
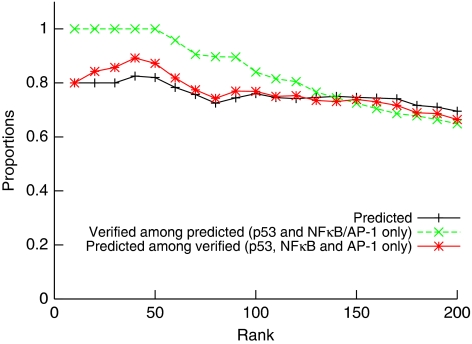
Figure 8.
Cumulative accuracy of GWTM. +/Black curve: cumulative fraction of the upregulated genes that are predicted by GWTM to be targets of one of the three main activities. Green: proportion of GWTM predicted genes that were verified by experimental knockdown of transcription factor (p53 and NF-κB /c-Jun only). Red: proportion of genes verified by experimental knockdown of transcription factor (only p53 and NF-κB or c-Jun/AP-1) that were also predicted by GWTM.
In a similar manner, figures tend to be better when one considers the proportion of predicted genes that are verified (or the converse) through the complementary experiments previously described. For example, 100% of the predicted genes that are among the 50 most upregulated genes were also verified, but that figure decreases as lower ranked genes are included (Figure 8, green curve). Conversely, 87% of the verified genes among the first 50 most upregulated genes were correctly predicted by GWTM (Figure 8, red curve).
Discussion
We have created an analytical workflow for the analysis of complex transcriptional data sets. GWTM uses a mathematical model to split the transcriptional response into its component parts, then applies a clustering method to group transcripts with similar behaviour. The factors controlling the predicted activities were identified using bioinformatics and verified experimentally. The method not only associates a target with its controlling transcription factor but also defines the confidence with which this association is made, allowing subsequent analysis of a gene list to be prioritized in a rational manner.
We have previously shown that by linking transcript levels measured in a time series, a dynamic picture of network activity can be created. We developed a mathematical modelling approach called HVDM that incorporates RNA production and degradation terms, and allows transcription factor activity to be deduced from microarray data. We term this activity the hidden variable because information needed to derive it is hidden in the transcript values reported in the array data. Derived activity profiles can then be used to predict and rank other putative targets of the same transcription factor, thereby creating a quantitative model of a transcriptional network. We applied HVDM to predict a ranked list of p53 targets that were experimentally validated by depleting cells of p53 using RNAi, with a high success rate.
HVDM, however, is limited to systems in which prior information about transcript control is available, and can only model one behaviour at a time. The key to the success of GWTM was the observation that global measurement of mRNA degradation rates could extract extensive hidden information regarding the entire transcriptional network from microarray data in the absence of prior biological knowledge of the system. Recently, strong evidence has been presented emphasizing on the importance of including degradation rates when modelling gene expression profiles (Yang et al, 2003; Perez-Ortin, 2007; Perez-Ortin et al, 2007; Molina-Navarro et al, 2008). While studying the transcriptional response to stress in yeast, Shalem et al (2008) found co-ordinated mRNA production and degradation such that greater message stability could lead to a sustained level of transcript despite a short production period. Very recently, Matsushita et al (2009) have demonstrated a critical role for an RNAse in regulating the immune response in mice through mediating mRNA degradation of a set of inflammatory genes.
We used microarrays to measure the mRNA degradation rates of all genes in the human T cell line, MOLT4, after treatment with ionizing radiation. By incorporating degradation data into a rearranged HVDM model equation, we were able to isolate production terms for all the transcripts. The time course data yielded transcript production profiles, which we are able to cluster on the basis of correlation. Then by applying a graph representation, we identified three clusters of genes with similar production profiles. By definition, transcript production is controlled by transcription factors, either alone or in combination. The three clusters were, therefore, likely to correspond to three global activity profiles influencing transcript production during the response to ionizing radiation.
We then used bioinformatics approaches to predict the identity of the factors controlling the major activities. The two most dominant activities contained genes known to be regulated by NF-κB, c-Jun/AP-1 and p53. The first activity is an early response that peaks 2 h after treatment with ionizing radiation. The model was unable to distinguish between NF-κB and c-Jun/AP-1 as candidate targets for this activity. When we verified model predictions for activity 1 by inhibiting NF-κB or c-Jun transcription factors using the NF-κB inhibitor, BAY 11-7082, and siRNAcJun, respectively, in the context of ionizing radiation, our results clearly showed a very high degree of crosstalk between the two factors. With c-Jun itself being a target of NF-κB, a likely explanation is that many of these genes are targets of targets such that NF-κB activates c-Jun, which activates its own targets. The kinetics of this pathway must be rapid as GWTM recognizes this regulation as one activity. It may be possible to separate these activities by taking closer time points during the first 2 h after irradiation.
The data from IPA and GSEA indicated that the second global activity identified by GWTM was controlled by p53, in agreement with our previous results obtained using HVDM. Examination of genes comprising the third global activity seems to indicate a portion of cells re-entering the cell cycle in the latter stages of the experiment. E2F is a possible candidate for controlling this activity. However, it is not clear whether this population represents cells surviving irradiation (unlikely at the dose of irradiation applied), or cells aberrantly re-entering cycle before undergoing apoptosis.
The generation of confidence limits for predictions means that appropriate cutoffs can be applied when deciding the importance or biological significance of transcriptional changes in a microarray experiment. Overall, our results predict the controlling dynamics behind the majority of genes whose expression increases as a result of irradiation-induced DNA damage. A total of 76 of the 100 most confidently changing genes were assigned by the model to a controlling factor and verified experimentally. This number falls, as would be expected, with the addition of genes with lower confidence of irradiation-induced change, or with lower confidence of model prediction. Use of confidence limits can, therefore, ensure the maximal cost–benefit analysis of microarray data because although a lower than expected proportion of the behaviour was explained by the model, additional experiments could be performed to extend the proportion of explicable behaviour.
Although it is ideal if the transcription factors controlling the discovered activities can be identified, this may not always be possible due to limitations in the bioinformatics databases. However, by synthesizing the causes of a complex response involving hundreds of genes to a considerably smaller number of activities, the search for activators is substantially simplified. As GWTM attaches probability levels to each transcript in an activity, it is possible to select on a rational basis the strongest members of the group. If one can identify what regulates this subgroup of targets it is likely to be the transcription factor for all of them. Using this approach it may even be possible to analyse co-regulated genes with unknown transcription factors to identify common regulatory motifs—akin to orphan receptors—where the binding domain is known before the transcription factor that binds it.
Around 25% of the 200 most upregulated transcripts were not explained by the model. Measurement error is a likely explanation for some of these discrepancies. Many of the genes at the lower end of the scale are expressed at lower levels in which the limiting factor is the microarray detection technology itself, and so there are uncertainties regarding gene detection, whether transcripts really are upregulated and whether they fit the model. One potential solution to this problem could come with a switch to a less error-prone technology. High throughput sequencing platforms, such as the Illumina GAII, ABI SOLiD and Roche GS-FLX all generate essentially digital signals, eliminating measurement errors resulting from cross-hybridization. We predict that modelling this type of data will lead to even more accurate models, and we are currently investigating this possibility.
We noticed a number of highly upregulated transcripts behaviour of which was also not predicted accurately by the model, but which were knocked down by the verification experiments. In the case of the most significantly changing gene, IER3, there is biological evidence that co-regulation between the NF-κB and p53 pathways may account for its synergistic behaviour. This is probably also the case for several other targets. Other hints to the importance of co-regulation appear in some of the results presented in the Supplementary information. For example, FAS, through distinct probe sets, is a constituent of merged cliques 1 and 2 (see Supplementary Tables I and II), and has indeed been shown to be the target of NF-κB (Kuhnel et al, 2000), as well as p53. Similarly, of the six genes that are predicted targets of global activity 1 and 2, two (CD70 and PTP4A1) present good verification scores for NF-κB and p53 dependence (Supplementary Tables IV and V). We are exploring whether introducing another term to the model could potentially account for such behaviour.
Other experimental methods like ChIP-Chip and ChIP-Seq, which measure binding of transcription factors to regulatory domains, are gaining in power and affordability, and are revealing the complexity of gene regulation (Valouev et al, 2008; Visel et al, 2009). However, ChIP experiments are typically static and require prior knowledge of the system. Combining ChIP-seq data with data-driven modelling approaches, like GWTM, will allow us to identify targets for Chip-seq, and to quantitatively relate transcriptional dynamics with the kinetics of transcription factor binding. Together these approaches could lead to a better understanding of how gene networks are regulated in complex responses.
In conclusion, the aim of GWTM is to maximize the efficient and systematic use of genomics technologies like microarrays. The approach can dissect complex transcriptional responses into their major controlling activities and identify the targets of each one. Furthermore, the method does not require complex and expensive artificial model systems, and only necessitates a short time course of microarray data measuring transcript levels and degradation rates. Our approach, therefore, offers a data-driven method for accurately identifying gene network components, establishing their connectivity and quantifying the controlling activities at the transcriptome level.
Materials and methods
Biological methods
Cell lines and reagents
Human MOLT4 cells (T cell acute lymphoblastic leukaemia) were obtained from NIBSC, UK (CFARP011) and cultured in RPMI supplemented with 10% FCS, L-glutamine and antibiotics. Functional p53 phenotype was established as previously described (Barenco et al, 2006). For c-Jun downregulation verification experiments, the following antibodies were used: anti-HSP90 (Cell Signaling Technology, ref. 4874) and anti-c-Jun (Santa Cruz Biotechnology, H-79). Proteins were detected by enhanced chemiluminescence (ECL+, GE Healthcare) and quantified by densitometry.
Microarray time course
Three time courses of radiation-induced gene expression were generated in an earlier experiment using MOLT4 cells. Data for these experiments are available at ArrayExpress (E-MEXP-549).
Transcript degradation rate estimation
MOLT4 cells in log phase (1 × 106 cells per ml) were irradiated with 4-Gy irradiation at room temperature, at a dose rate of 2.45 Gy/min using a 137Cs γ-irradiator. After 4 h, actinomycin D (5 μM) was added to block transcription. Aliquots of culture were removed at hourly intervals and RNA was extracted (TRIzol; Invitrogen). The quantity and quality of RNA and cDNA were determined by Nanodrop spectrophotometer and Bioanalyser 2100 (Agilent). Transcript levels were determined at different time points using Affymetrix Human U133A arrays.
Microarray data analysis
Microarray data was summarized using the MAS5 or PLIER algorithms (Affymetrix). Signal distribution was assessed using Genespring 7.3 (Agilent), and log-transformed for normalization to the median. Further analysis was conducted on raw, that is, non-log-transformed data. The clustering algorithm was implemented using a mixture of R code (filtering and individual activity profile computation) and C++ (graph theory algorithms and clustering) combined with a scripting language (Python). The degradation rate computation and screening procedure were implemented in R. Data are available in MAGE-ML format through ArrayExpress (E-MEXP-2176 for the c-Jun downregulation experiment, E-MEXP-2177 for the NF-κB inhibition experiment and E-MEXP-2179 for the degradation rate measurement experiment).
Inhibition of NF-κB activity
MOLT4 cells in log phase (1 × 106 cells per ml) were pre-treated for 2 h with the I-κBα phosphorylation inhibitor BAY 11-7082 (Calbiochem) at 5 μM concentration, and the control samples with corresponding volume of DMSO. Cells were then irradiated with 4 Gy irradiation at room temperature, at a dose rate of 2.45 Gy/min using a 137Cs γ-irradiator. Cells were collected 2 h after irradiation and RNA was extracted (TRIzol; Invitrogen). The quantity and quality of RNA and cDNA were determined by Nanodrop spectrophotometer and Bioanalyser 2100 (Agilent). Affymetrix human U133A arrays were hybridized as standard (www.affymetrix.com). Array quality was determined using R and Affymetrix Expression Console file values.
Preparation of nuclear extracts and gel shift to confirm NF-κB downregulation
Inhibition of NF-κB activation by BAY 11-7082 was confirmed by electrophoretic-mobility shift assay (EMSA) on nuclear extracts prepared from aliquots collected 5, 15, 30, 40, 50, 60, 80 and 100 min after irradiation. Nuclei were prepared from the irradiated cells by washing the collected cells twice in 500 μl hypotonic buffer, followed by lysis in hypotonic buffer (10 mM Tris–HCl (pH 7.8), 5 mM KCl, 2 mM MgCl2, 1 mM DTT, complete protease inhibitors (Roche Diagnostics)), supplemented with 0.25% IGEPAL CA-630 (Sigma), for 5 min on ice. Nuclei were pelleted (600 g/15 min/4°C), washed twice with 500 μl of hypotonic buffer, pelleted again (600 g/15 min/4°C), and suspended in two volumes of hypotonic buffer containing 0.3 M NaCl. The suspension was kept on ice for 30 min and agitated occasionally. Finally, nuclei were pelleted and the supernatant was used directly for EMSA (600 g, 15 min at 4°C). Nuclear extracts of Jurkat cells, stimulated with TPA (phorbol, 12-myristate, 13-acetate) and calcium ionophore (Active Motif), were used as positive control.
Nuclear extracts (5 μg) were incubated with a 32P-labelled 22-mer double-stranded oligonucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′) containing the NF-κB consensus sequence (underlined) and 1 μg of poly(dI·dC) (Amersham Biosciences) in binding buffer (10 mM Tris–HCl, 100 mM NaCl, 2 mM EDTA, 4% (w/v) Ficoll, 1 mM DTT), for 30 min at 30°C. DNA probes were prepared by end-labelling both strands of the oligonucleotide using [γ-32P]ATP (Amersham Biosciences) and T4 polynucleotide kinase (New England Biolabs). A 100-fold excess of non-radioactive probe was used as a control to specifically compete for binding. A double-stranded mutated oligonucleotide (5′-AGTTGAGATCACTGGGACAGGC-3′) was also used to examine the specificity of binding of NF-κB to the DNA (data not shown). Reaction mixtures were loaded onto a 4% non-denaturing acrylamide gel. Gels were run in 0.5 × Tris–acetate buffer for 1 h at 110 V, dried and exposed to PhosphorImager screen.
Inhibition of c-Jun/AP-1 activity
Cells were transfected with 100 nM siRNAc-Jun (Santa Cruz Biotechnology, sc-44204) using electroporation. At 48 h after transfection, cells were irradiated with 5-Gy irradiation and incubated at 37°C for 2 h. They were then collected and RNA and protein were prepared and processed for microarray analysis and western blot verification, respectively. Affymetrix Gene Array 1.0 ST arrays were hybridized as standard (www.affymetrix.com).
Bioinformatic methods
To identify the transcription factors responsible for each controlling activity, we took advantage of several bioinformatics resources. First, we analysed the promoter regions of model predicted genes using a matrix search for known transcription factor binding motifs (TRED; http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home). Second, we carried out GSEA using GeneTrail (based on the TRANSFAC database) (http://genetrail.bioinf.uni-sb.de/) on genes in both merged cliques, and in lists of model-predicted genes associated with cliques, for enrichment in target genes for known transcription factors. Next we analysed the same gene lists for their gene ontology category and for significant associations with known biochemical pathways using the commercially available package Ingenuity Pathway Analysis and the freely available DAVID.
Mathematical methods
Degradation rate measurement from microarray data
Typically gene turnover rates are measured over a short time span (Yang et al, 2003). Reasoning that this would be problematic in estimating slow decaying transcripts with sufficient precision, we decided to extend the period up to 6 h after transcription blocking. This in turn caused problems with fast-decaying transcripts. The latter were afflicted by a ‘tailing-off' effect. We solved this problem by using a model incorporating a curvature term, allowing for a much better fit of the data (see Supplementary information).
In standard microarray experiments, overall gene expression is assumed to be constant across experimental conditions and individual microarrays are normalized accordingly to some central statistic, such as the trimmed average or the median. In the context in which the overall quantity of RNA can only diminish because its production has been turned off, this type of normalization cannot be carried out. Instead, we used an iterative procedure to normalize individual chips to the slower decaying genes, assuming a small fraction of the latter had a degradation rate close to zero (see Supplementary information for more details). A similar procedure was used in the study by Yang et al (2003).
First derivative estimation from data
A detailed description of the way we obtained the matrix A mentioned in the results section is given in the Supplementary information of the study by Barenco et al (2006). We give here a simple example to illustrate how we arrive at this matrix. Imagine we are given two time points x(t1) and x(t3) and want to estimate the slope x′ at the intermediate time point between these two (t2). The simplest way is to simply taking the slope of the line passing through these points:
which can be rewritten as:
Thus, this estimation is a linear combination of the function values weighted by coefficients that depend only on the time intervals between the points. This can be generalized to using more time points to estimate the slope at an intermediate point. In this case, a polynomial of degree n−1 is used for the fit. Interestingly, the nature of the estimation is unchanged; it is still a linear combination of the function values and the coefficients involved depend only on the time intervals between points. Thus, these coefficients can be stored and re-used for different function values. In our case, we arrived at the following matrix A, which is applicable to our case in which, using seven measurements regularly spaced between 0 and 12 h, we want to estimate the first derivative at the same time points.
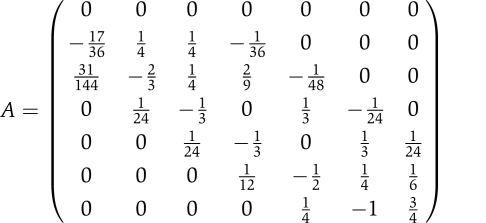 |
It should be noted that all the rows in this matrix sum up to 0, and that the first row of A is filled with zeros as we suppose that the net rate of change of every transcript is still nil at the moment (t=0) in which we apply the stimulus.
Pre-clustering filtering
Two broad criteria were used to select genes; first, they had to be upregulated and present in the expression time course, and the degradation rate measurement had to be sufficiently precise. To fulfill the first set of criteria, the upregulation Z-score with respect to the 0 h time point had to be greater than 1.5 in at least one of the six observations and the detection P-value smaller than 0.1 in one time point at least. For the second criterium, we considered only those genes with a sufficient goodness-of-fit. The sum of the squared residuals between model and data (in the degradation experiment) was compared to a chi-squared distribution with a number of degrees of freedom corresponding to the difference between the data and parameter count (recall we have two possible models). If the P-value (corresponding to the right tail of the distribution) was less than 0.01, the gene was not retained. Furthermore, we only kept those genes which had a sufficient signal intensity in the degradation experiment, this was achieved by retaining only those with an intercept, as determined by the model, greater than five. After this filtering, 828 individual activity profiles were retained.
Influence of clustering parameters
The objective of the clustering procedure is two-fold. First, we want to obtain as many distinct principal activities as possible; and second, we want the underlying merged cliques to include as many constituent genes as possible to increase the robustness of the corresponding principal activity profile, because it is obtained by averaging the normalized individual activity profiles. We varied systematically two clustering parameters to achieve these aims.
An edge is drawn between two individual genes if the correlation between their individual activity profiles is above a certain threshold, α. This is the first parameter. Choosing a high value of α tends to reduce the overall connectivity of the graph and thus satisfies the first objective (more distinct merged cliques). However, a reduced connectivity also reduces the size of the merged cliques, in contradiction with the second aim. Conversely, a low value of α increases the connectivity of the graph and thus tends to increase the size of the cliques. However, too low a value for this parameter can have the effect of agglomerating groups of genes that should not be linked. This was the case in particular for the genes associated with the first two activities.
We reasoned that this could be due to two possible effects. First, genes that are co-regulated by two distinct activities can create ‘bridges' between cliques. Second, genes that are hampered by a high level of noise could also create this bridging. Although nothing can be done against the first effect, the second possibility was handled by introducing a second parameter, β. Genes for which smaller inter-replicate correlation coefficient was below β were excluded from the analysis. Thus, a high value of β can counteract a low value for α. It should be noted that increasing β will, all other things being equal, diminish the size of merged cliques.
To determine appropriate values for α and β, we varied them systematically, observing the resulting number of merged cliques, their size and their composition. We noticed that four merged cliques emerged, but that too low a value for α (0.75), whichever value was chosen for β, caused two important merged cliques (corresponding to activities 1 and 2 in the main paper) to agglomerate into one. These four merged cliques were found, in varying size and composition, over a range of couple of values for these two parameters (for example, (0.77, 0.75), (0.8, 0.45), and (0.83, 0.3)). We determined the ‘optimal' couple of parameters (0.8, 0.45) by choosing the situation in which the size of these four main cliques was larger.
Next we looked for enriched GO terms for each of the four merged cliques (see main article). Although for three of these main cliques it was easy to identify the underlying activity, we could not find enriched terms for the fourth. This set of genes was also very heterogeneous. We thus proceeded to check whether there was a more technical explanation behind the appearance of this fourth clique and found out that the expression profiles of genes in that merged clique were all highly correlated with the rescaling factors in the experiment. This merged clique being a clear technical artefact caused by probe saturation, we eliminated it. Further, we also eliminated from the subsequent screening all those genes for which expression values were too highly correlated with the scaling factors.
Filtering the genes before screening
Before screening the genes against each of the three activity profiles, we performed the following filtering steps. We retained only those genes that are significantly upregulated; we applied the Benjamini–Hochberg criteria (with controlled false discovery rate of 5%) on the maximal upregulation P-value computed on all six time time points after system activation following the control time point (Benjamini and Hochberg, 1995). Similarly, only those genes that were detected at least once (Affymetrix, P-value<0.1) during the time course were retained. Next, to avoid a biased degradation rate measurement, we kept only those genes for which the intercept value calculated on the degradation time course was between 5 and 1500. We reasoned that outside this range the degradation rate was more likely to be underestimated because of compression effects. Finally, we noticed that some genes were highly correlated with the scaling factors used in the experiment. To eliminate them, we retained only those for which correlation with these scaling factors was smaller than 0.6. In total, 246 genes were retained for the screening.
Supplementary Material
Supplementary material, Supplementary figures S1–4, Supplementary tables S1–3
Supplementary tables referred to in the main manuscript
Acknowledgments
The authors wish to thank Nipurna Jina and UCL Genomics for help running the microarrays. This study was funded by the BBSRC (grant number BB/E008488/1).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe Y, Takeuchi T, Kagawa-Miki L, Ueda N, Shigemoto K, Yasukawa M, Kito K (2007) A mitotic kinase TOPK enhances Cdk1/cyclin B1-dependent phosphorylation of PRC1 and promotes cytokinesis. J Mol Biol 370: 231–245 [DOI] [PubMed] [Google Scholar]
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP (2007) GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res 35: W186–W192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenco M, Papouli E, Shah S, Brewer D, Miller CJ, Hubank M (2009) rHVDM: an R package to predict the activity and targets of a transcription factor. Bioinformatics 25: 419–420 [DOI] [PubMed] [Google Scholar]
- Barenco M, Tomescu D, Brewer D, Callard R, Stark J, Hubank M (2006) Ranked prediction of p53 targets using hidden variable dynamic modeling. Genome Biol 7: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Rowan S, Vousden KH (1996) Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13: 1103–1109 [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Bours V, Azarenko V, Dejardin E, Siebenlist U (1994) Human RelB (I-Rel) functions as a kappa B site-dependent transactivating member of the family of Rel-related proteins. Oncogene 9: 1699–1702 [PubMed] [Google Scholar]
- Bron C, Kerbosch J (1973) Finding all cliques of an undirected graph. Commun ACM 16: 575–577 [Google Scholar]
- Brown K, Park S, Kanno T, Franzoso G, Siebenlist U (1993) Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc Natl Acad Sci USA 90: 2532–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho DM, Collins JJ (2009) Systems biology strikes gold. Cell 137: 24–26 [DOI] [PubMed] [Google Scholar]
- Carter GW, Galas DJ, Galitski T (2009) Maximal extraction of biological information from genetic interaction data. PLoS Comput Biol 5: e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos MC, Munoz C, Montoya MC, Lara-Pezzi E, Lopez-Cabrera M, de Landazuri MO (1997) Expression of the leukocyte early activation antigen CD69 is regulated by the transcription factor AP-1. J Immunol 159: 5463–5473 [PubMed] [Google Scholar]
- Dojer N, Gambin A, Mizera A, Wilczynski B, Tiuryn J (2006) Applying dynamic Bayesian networks to perturbed gene expression data. BMC Bioinformatics 7: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Bhattacharya R, Cabral F (2008) Cell cycle dependent degradation of MCAK: evidence against a role in anaphase chromosome movement. Cell Cycle 7: 3187–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner H, Siegmund D, Motejadded H, Scheurich P, Henkler F, Janssen O, Wajant H (2002) Intracellular localization and transcriptional regulation of tumor necrosis factor (TNF) receptor-associated factor 4 (TRAF4). Eur J Biochem 269: 4819–4829 [DOI] [PubMed] [Google Scholar]
- Hayakawa J, Mittal S, Wang Y, Korkmaz KS, Adamson E, English C, Ohmichi M, McClelland M, Mercola D (2004) Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol Cell 16: 521–535 [DOI] [PubMed] [Google Scholar]
- Husmeier D (2003) Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics 19: 2271–2282 [DOI] [PubMed] [Google Scholar]
- Jiang C, Xuan Z, Zhao F, Zhang MQ (2007) TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res 35: D137–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerley-Hamilton JS, Pike AM, Hutchinson JA, Freemantle SJ, Spinella MJ (2007) The direct p53 target gene, FLJ11259/DRAM, is a member of a novel family of transmembrane proteins. Biochim Biophys Acta 1769: 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IG, Jun DY, Sohn U, Kim YH (1997) Cloning and expression of human mitotic centromere-associated kinesin gene. Biochim Biophys Acta 1359: 181–186 [DOI] [PubMed] [Google Scholar]
- Kim JW, Park HY, Lee MJ, Jang MJ, Lee SY, Park YM, Son DH, Chang YC, Bae YS, Kwak JY (2004) Phosphatidic acid and tumor necrosis factor-alpha induce the expression of CD83 through mitogen activated protein kinase pathway in a CD34+ hematopoietic progenitor cell line, KG1. Int Immunopharmacol 4: 1603–1613 [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM (1992) Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 267: 17971–17976 [PubMed] [Google Scholar]
- Kuhnel F, Zender L, Paul Y, Tietze MK, Trautwein C, Manns M, Kubicka S (2000) NFkappaB mediates apoptosis through transcriptional activation of Fas (CD95) in adenoviral hepatitis. J Biol Chem 275: 6421–6427 [DOI] [PubMed] [Google Scholar]
- Lee MG, Norbury CJ, Spurr NK, Nurse P (1988) Regulated expression and phosphorylation of a possible mammalian cell-cycle control protein. Nature 333: 676–679 [DOI] [PubMed] [Google Scholar]
- Li J, Tan J, Zhuang L, Banerjee B, Yang X, Chau JF, Lee PL, Hande MP, Li B, Yu Q (2007) Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res 67: 11317–11326 [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP (2003) Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci USA 100: 15522–15527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Chen J, Hai T (2007) The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem J 401: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko NF, Dieffenbach PB, Yan G, Ceryak S, Howell RW, McCaffrey TA, Hu VW (2003) Does metabolic radiolabeling stimulate the stress response? Gene expression profiling reveals differential cellular responses to internal beta versus external gamma radiation. FASEB J 17: 1470–1486 [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Abe Y, Fujibuchi T, Takeuchi T, Kito K, Ueda N, Shigemoto K, Gyo K (2004) Characterization of a MAPKK-like protein kinase TOPK. Biochem Biophys Res Commun 325: 997–1004 [DOI] [PubMed] [Google Scholar]
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S (2009) Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Mo ML, Palsson BO (2009) Understanding human metabolic physiology: a genome-to-systems approach. Trends Biotechnol 27: 37–44 [DOI] [PubMed] [Google Scholar]
- Molina-Navarro MM, Castells-Roca L, Belli G, Garcia-Martinez J, Marin-Navarro J, Moreno J, Perez-Ortin JE, Herrero E (2008) Comprehensive transcriptional analysis of the oxidative response in yeast. J Biol Chem 283: 17908–17918 [DOI] [PubMed] [Google Scholar]
- Mori N, Yamada Y, Ikeda S, Yamasaki Y, Tsukasaki K, Tanaka Y, Tomonaga M, Yamamoto N, Fujii M (2002) Bay 11-7082 inhibits transcription factor NF-kappaB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood 100: 1828–1834 [DOI] [PubMed] [Google Scholar]
- Obad S, Olofsson T, Mechti N, Gullberg U, Drott K (2007) Regulation of the interferon-inducible p53 target gene TRIM22 (Staf50) in human T lymphocyte activation. J Interferon Cytokine Res 27: 857–864 [DOI] [PubMed] [Google Scholar]
- Osawa Y, Nagaki M, Banno Y, Brenner DA, Nozawa Y, Moriwaki H, Nakashima S (2003) Expression of the NF-kappa B target gene X-ray-inducible immediate early response factor-1 short enhances TNF-alpha-induced hepatocyte apoptosis by inhibiting Akt activation. J Immunol 170: 4053–4060 [DOI] [PubMed] [Google Scholar]
- Perez-Ortin JE (2007) Genomics of mRNA turnover. Brief Funct Genomic Proteomic 6: 282–291 [DOI] [PubMed] [Google Scholar]
- Perez-Ortin JE, Alepuz PM, Moreno J (2007) Genomics and gene transcription kinetics in yeast. Trends Genet 23: 250–257 [DOI] [PubMed] [Google Scholar]
- Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME (1997) Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem 272: 21096–21103 [DOI] [PubMed] [Google Scholar]
- Raemaekers T, Ribbeck K, Beaudouin J, Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg J, Carmeliet G (2003) NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol 162: 1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW (2007) A role for NuSAP in linking microtubules to mitotic chromosomes. Curr Biol 17: 230–236 [DOI] [PubMed] [Google Scholar]
- Rouault JP, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud JP, Ozturk M, Samarut C, Puisieux A (1996) Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet 14: 482–486 [DOI] [PubMed] [Google Scholar]
- Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y (2008) Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Shamir R (2000) CLICK: a clustering algorithm with applications to gene expression analysis. Proc Int Conf Intell Syst Mol Biol 8: 307–316 [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article 3 [DOI] [PubMed] [Google Scholar]
- Staib F, Robles AI, Varticovski L, Wang XW, Zeeberg BR, Sirotin M, Zhurkin VB, Hofseth LJ, Hussain SP, Weinstein JN, Galle PR, Harris CC (2005) The p53 tumor suppressor network is a key responder to microenvironmental components of chronic inflammatory stress. Cancer Res 65: 10255–10264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara T, Magae J, Wadhwa R, Kaul SC, Kawakami Y, Matsumoto T, Tanaka K (2004) Dose and dose-rate effects of low-dose ionizing radiation on activation of Trp53 in immortalized murine cells. Radiat Res 162: 296–307 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A (2008) Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Miguel S, Buckbinder L, Jean P, Gelbert L, Talbott R, Laidlaw J, Seizinger B, Kley N (1999) PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene 18: 127–137 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH (2000) p53: death star. Cell 103: 691–694 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T (2006) A systems approach to mapping DNA damage response pathways. Science 312: 1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr (2003) Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Liu X, Yin Y, Fukuda MN, Zhou J (2008) Tastin is required for bipolar spindle assembly and centrosome integrity during mitosis. FASEB J 22: 1960–1972 [DOI] [PubMed] [Google Scholar]
- Yazgan O, Pfarr CM (2002) Regulation of two JunD isoforms by Jun N-terminal kinases. J Biol Chem 277: 29710–29718 [DOI] [PubMed] [Google Scholar]
- Zou M, Conzen SD (2005) A new dynamic Bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics 21: 71–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material, Supplementary figures S1–4, Supplementary tables S1–3
Supplementary tables referred to in the main manuscript