Animal development is a fascinating process: starting from a single fertilized egg, an embryo grows and embryonic cells progressively differentiate into the diverse cell types and organs that make up an adult body. All this happens autonomously according to an intrinsic blueprint of development written in the four-letter alphabet of the genomic DNA sequence.
The genome not only encodes all developmentally important genes, but also carries the information necessary to specify the spatio-temporal patterns of gene expression. The gene-regulatory information is contained within the sequence of defined genomic regions, so-called cis-regulatory modules (CRMs) or enhancers. These elements retain their cell-type specific activity even when placed into an artificial context, for example when combined with a minimal promoter to drive expression of a reporter gene in transgenic animals (Arnone and Davidson, 1997).
CRMs contain binding sites for specific sets of transcription factors (TFs) and are generally thought to integrate the bound factors' regulatory cues, such that enhancer activity depends on the appropriate expression of the respective TFs. The simplicity of this model is attractive and it has indeed been shown that removing TFs or disrupting their binding sites by specific mutations impairs enhancer function (Arnone and Davidson, 1997). Despite its apparent simplicity, this model implies an underlying regulatory code that determines the exact requirements for enhancer function. A strong argument for the existence of this code would be the demonstration that enhancer activity can be predicted solely from the enhancers' TF-binding patterns. Ideally, enhancers with known activities could be used to learn rules that would be able to correctly predict the activity of novel enhancers.
In a recent study, Eileen Furlong and colleagues follow exactly this line of reasoning to show that the combinatorial binding of TFs is highly predictive of spatio-temporal enhancer activity in vivo (Zinzen et al, 2009).
Using ChIP-chip assays (chromatin immunoprecipitation combined with microarray analysis), they determine the genome-wide distribution of DNA-binding sites for five key TFs of mesoderm and muscle development in the Drosophila embryo: Twist, Tinman, Mef2, Bagpipe, and Biniou (Figure 1). At five time points during embryogenesis, they find a total of 19 522 binding sites that cluster into 8008 CRMs. Among this extensive set of CRMs, 310 overlap with known enhancers, for which independent data about their activity are available. Of these, 87 fall into one of five exclusive mesodermal expression categories: early mesoderm, visceral (gut) muscle, somatic muscle (analogous to the vertebrate skeletal muscle), and the combined categories mesoderm and somatic muscle, and visceral and somatic muscle.
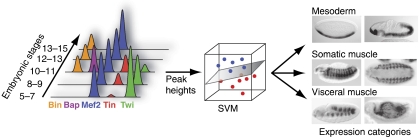
Figure 1.
Predicting the expression category for a CRM based on its temporal transcription factor binding profile. Peak heights for five factors at five developmental time points (15 conditions total) are sufficient to predict the expression category of a mesodermal enhancer with a >70% success rate. Shown is a schematic representation of the temporal binding profile of a single CRM and a support vector machine (SVM) used for the predictions, as well as the categories mesoderm, somatic muscle, and visceral muscle with example expression patterns. Photomicrographs reproduced from Zinzen et al (2009).
Using an established machine learning method (a support vector machine (SVM)), the authors predict the category of a CRM solely based on the patterns of TF occupancy as estimated by ChIP-peak heights. First, they test the approach on the 310 known CRMs: they exclude each CRM in turn for testing, train the SVM on the remaining ones, and evaluate whether the category for the test CRM is correctly predicted. This leave-one-out cross-validation protocol works surprisingly well, demonstrating that the SVM is able to learn rules from the ChIP data that are sufficiently general to correctly predict the activity of previously unseen CRMs. Indeed, when the authors apply the trained SVMs to all 8008 CRMs and test in vivo several predictions from each expression category, 71% of the predictions turn out to be exactly correct: the enhancers drive expression of transgenic reporters specifically in the predicted regions and not in other mesodermal tissues. The success rate even reaches 86% for enhancers that are exclusively active in the early mesoderm.
The predictions in each category are characterized on average by rather simple signatures: predicted mesodermal enhancers exhibit strong binding of Twist, while enhancers predicted to be active in visceral muscles are predominantly bound by Biniou. Interestingly, the dominant factors correspond to the respective known key regulators of these tissues, showing that the unbiased data-driven approach correctly recapitulates the results from genetic experiments (Furlong, 2004). Successful predictions (especially of the early mesoderm category) often largely match the factors' expression domains, reminiscent of single input modules suggesting that additionally bound factors might be neutral or might merely tune the activity. This might indicate that mesodermal/muscle CRMs differ from those in the early Drosophila embryo, for which predictions of activity relied on TF concentrations and DNA-binding affinities, possibly because these CRMs need to read TF gradients (e.g. Janssens et al, 2006; Segal et al, 2008).
Despite the simple average signatures, neither the known mesodermal CRMs nor the validated predictions show uniform binding profiles. Although all visceral muscle enhancers are bound by Biniou, some are also strongly bound by other factors. For example, two CRMs are bound by Twist but are nevertheless predicted correctly to show activity in visceral muscles but not in the early mesoderm. On the one hand, this suggests that the regulatory code is complex, flexible and not merely additive. On the other hand, the success of the predictions implies the existence of common features in enhancers with similar activities: if each enhancer reached activity through entirely different means, predictions that rely on common features would not be possible.
The accuracy of the predictions is especially surprising considering the simplicity of the approach, which relies exclusively on TF occupancy data and does not take into account multiple binding sites for one TF, nor the arrangements of binding sites, i.e. their order and spacing, or any feature of the CRM sequence. In addition, it appears that scoring TF binding only qualitatively (i.e. binary bound versus non-bound) performs almost as well.
There are several possible interpretations to this observation, with different implications for our understanding of CRMs and their functional architecture. Features such as binding site arrangement might simply not be relevant in the context of this class of enhancers as would be expected, for example, from the billboard enhancer model (Arnosti and Kulkarni, 2005). Alternatively, they could be crucial for TF binding, but not for activity once the factors are bound. Lastly, they might contribute to future improvements of such approaches. We anticipate improvements, for example, when larger training sets will become available or when more TFs will be profiled. For example, 23% of the known mesodermal/muscle enhancers were excluded from this study as they appeared not to be bound by any of the surveyed TFs, and the authors speculate that predictions for somatic muscles might improve once characteristic TFs are included.
Given that TF binding predicts enhancer function, how far are we from reading enhancer activity directly from the DNA sequence? This study confirms the prevalent observation that many binding sites (but not all) coincide with conserved TF-binding motifs. However, it also highlights some of the limitations of sequence-based predictions: enhancer activity seems to strongly depend on the dynamics of DNA binding, which is neither reflected in the DNA sequence nor simply explained by changes in TF expression. Nevertheless, sequence-based analyses have been successful in identifying enhancers driving expression in Drosophila muscle founder cells (Philippakis et al, 2006), and if we manage to bridge the gap between sequence and TF binding, we might soon be able to predict enhancer activity from the sequence in mesodermal or other defined cell types.
The study by the Furlong group makes us confident that a regulatory code exists and determines spatio-temporal enhancer activity. Given the availability of in vivo binding data for an increasing number of TFs (Celniker et al, 2009; MacArthur et al, 2009), similar approaches might in the future help map the majority of functional enhancers and explain the molecular basis of cell-type specific gene expression, differentiation, and development.
Footnotes
The author declares that he has no conflict of interest.
References
- Arnone MI, Davidson EH (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864 [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM (2005) Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH (2009) Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EE (2004) Integrating transcriptional and signalling networks during muscle development. Curr Opin Genet Dev 14: 343–350 [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J (2006) Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet 38: 1159–1165 [DOI] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis AA, Busser BW, Gisselbrecht SS, He FS, Estrada B, Michelson AM, Bulyk ML (2006) Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Comput Biol 2: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U (2008) Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE (2009) Combinatorial binding predicts spatio-temporal cisregulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]