Abstract
The phototrophic bacterium Chloroflexus aurantiacus uses a yet unsolved 3-hydroxypropionate cycle for autotrophic CO2 fixation. It starts from acetyl-CoA, with acetyl-CoA and propionyl-CoA carboxylases acting as carboxylating enzymes. In a first cycle, (S)-malyl-CoA is formed from acetyl-CoA and 2 molecules of bicarbonate. (S)-Malyl-CoA cleavage releases the CO2 fixation product glyoxylate and regenerates the starting molecule acetyl-CoA. Here we complete the missing steps devoted to glyoxylate assimilation. In a second cycle, glyoxylate is combined with propionyl-CoA, an intermediate of the first cycle, to form β-methylmalyl-CoA. This condensation is followed by dehydration to mesaconyl-C1-CoA. An unprecedented CoA transferase catalyzes the intramolecular transfer of the CoA moiety to the C4 carboxyl group of mesaconate. Mesaconyl-C4-CoA then is hydrated by an enoyl-CoA hydratase to (S)-citramalyl-CoA. (S)-Citramalyl-CoA is cleaved into acetyl-CoA and pyruvate by a tri-functional lyase, which previously cleaved (S)-malyl-CoA and formed β-methylmalyl-CoA. Thus, the enigmatic disproportionation of glyoxylate and propionyl-CoA into acetyl-CoA and pyruvate is solved in an elegant and economic way requiring only 3 additional enzymes. The whole bicyclic pathway results in pyruvate formation from 3 molecules of bicarbonate and involves 19 steps but only 13 enzymes. Elements of the 3-hydroxypropionate cycle may be used for the assimilation of small organic molecules. The 3-hydroxypropionate cycle is compared with the Calvin–Benson–Bassham cycle and other autotrophic pathways.
Keywords: autotrophy, acetyl-CoA carboxylase, Calvin cycle
Organisms that can grow using CO2 as the sole carbon source are called “autotrophs.” Their energy source may be light, as is the case in plants, algae, cyanobacteria, and various other phototrophic bacteria. An autotrophic symbiotic cyanobacterium conferred the CO2 fixation machinery on a eukaryotic cell giving rise to the chloroplasts of plant cells. Alternatively, energy can derive from chemical reactions using inorganic substrates, as in chemolithoautotrophic Bacteria and Archaea. Autotrophs are responsible for generating the biomass on which humans, animals, and many microbes thrive, and they play a major role in the nitrogen and sulfur cycles of this planet. The Darwin year reminds us that the development of a primordial metabolism required the synthesis of organic building blocks from inorganic carbon.
In general terms, the endergonic assimilation of CO2 into cellular building blocks requires reducing equivalents and an input of energy. An organic molecule serves as CO2 acceptor, which becomes carboxylated by a carboxylase enzyme. The CO2 acceptor molecule needs to be regenerated in an autocatalytic cycle in which reduction steps are needed to reduce CO2 (oxidation state +4) to the level of cell carbon (average oxidation state 0). Highly endergonic steps of the pathway are driven by ATP hydrolysis; reduction steps are driven by low-potential reduced coenzymes, normally NADPH, and, rarely, by reduced ferredoxin. The product that can be obtained from such a metabolic cycle should be a central cellular metabolite, from which all building blocks for polymers can be derived.
In nature 6 mechanisms to assimilate CO2 into cell material have been established (1). In the reductive pentose phosphate (Calvin–Benson–Bassham) cycle discovered about 50 years ago, CO2 reacts with a 5-carbon sugar yielding 2 carboxylic acids from which the sugar is regenerated (2). This cycle operates in plants, algae, cyanobacteria, and in aerobic or facultative anaerobic proteobacteria. The presence of the key enzyme, ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO), is often considered to be an indication of autotrophy. In 1966, Evans et al. (3) proposed another autotrophic cycle for the green sulfur bacterium Chlorobium limicola, the reductive citric acid (Arnon-Buchanan) cycle (4). This cycle, which consumes less energy, involves enzymes that are sensitive to oxygen, and therefore this cycle is found only in anaerobes or in aerobes growing at very low oxygen tensions. These organisms include some members of the alpha-, delta- and epsilon-proteobacteria, green sulfur bacteria, and microaerophilic bacteria belonging to the early branching bacterial phylum Aquificae. At the beginning of the 1980s, a third autotrophic pathway was found in certain Gram-positive bacteria and methane-forming archaea, the reductive acetyl-CoA (Wood-Ljungdahl) pathway (5–8). In these strictly anaerobic organisms that now also include some proteobacteria, planctomycetes, spirochetes, and Euryarchaeota, 1 CO2 molecule is reduced to CO and 1 CO2 molecule is reduced to a methyl group (bound to a carrier); subsequently, acetyl-CoA is synthesized from CO and this methyl group. This pathway is the most favorable in terms of energy, but it requires many trace metals (Fe, Co, Ni, Mo, or W) as constituents of enzymes or coenzymes. Furthermore, the key enzyme, CO dehydrogenase/acetyl-CoA synthase, is among the most oxygen-sensitive enzymes known.
Recently 2 more autotrophic cycles were discovered in Archaea, the dicarboxylate/4-hydroxybutyrate cycle and the 3-hydroxypropionate/4-hydroxybutyrate cycle (9, 10). The latter uses acetyl-CoA and propionyl-CoA carboxylation, as also is known from the fourth autotrophic cycle in Chloroflexus aurantiacus, the 3-hydroxypropionate cycle (also referred to as the “3-hydroxypropionate/malyl-CoA cycle”). This filamentous anoxygenic phototroph is thermophilic and facultative autotrophic. It lives in hot, slightly alkaline, nutrient-poor, shallow springs where it forms visible orange microbial mats together with cyanobacteria and other bacteria (11, 12). It grows heterotrophically in the dark by aerobic respiration, but it has the capability of fixing inorganic carbon in the light. C. aurantiacus is of particular interest in the study of the evolution of photosynthesis and of its autotrophic carbon-fixation cycle, which has not been completely elucidated (13–23).
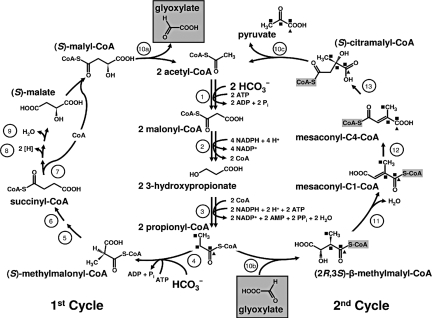
In brief, in a first cycle, 1 acetyl-CoA molecule and 2 bicarbonate molecules are converted to (S)-malyl-CoA (Fig. 1). Bicarbonate fixation proceeds via acetyl-CoA and propionyl-CoA carboxylation. (S)-Malyl-CoA then is cleaved to glyoxylate and acetyl-CoA, thus closing the first cycle. A second cycle has been postulated by which glyoxylate and propionyl-CoA are converted to acetyl-CoA and pyruvate, thus regenerating acetyl-CoA and producing pyruvate as universal precursor for biosynthesis (Fig. 1). This second cycle starts with the condensation of glyoxylate and propionyl-CoA to a C5-dicarboxylic acid CoA thioester. As can be seen in Fig. 1, in the course of the conversion of this intermediate to citramalyl-CoA, the CoA moiety somehow must be transferred from the “right” to the “left” carboxyl group of the C5-dicarboxylic acid. The last proven step of the glyoxylate assimilation cycle is the formation of mesaconyl-C1-CoA (2-methylfumaryl-CoA) (reaction 11 in Fig. 1).
Fig. 1.
The complete 3-hydroxypropionate cycle, as studied in C. aurantiacus. [1] Acetyl-CoA carboxylase, [2] malonyl-CoA reductase, [3] propionyl-CoA synthase, [4] propionyl-CoA carboxylase, [5] methylmalonyl-CoA epimerase, [6] methylmalonyl-CoA mutase, [7] succinyl-CoA:(S)-malate-CoA transferase, [8] succinate dehydrogenase, [9] fumarate hydratase, [10 a, b, c] (S)-malyl-CoA/β-methylmalyl-CoA/(S)-citramalyl-CoA (MMC) lyase, [11] mesaconyl-C1-CoA hydratase (β-methylmalyl-CoA dehydratase), [12] mesaconyl-CoA C1-C4 CoA transferase, [13] mesaconyl-C4-CoA hydratase. Carbon-labeling patterns during the interconversion of propionyl-CoA plus glyoxylate to pyruvate plus acetyl-CoA via C5 compounds are shown. 14C carbon atoms derived from [1-14C]propionyl-CoA are marked by ▴, and 13C carbon atoms derived from [1,2,3-13C]propionyl-CoA are marked by ■. Note that the cleavage of citramalyl-CoA requires that the CoA moiety be shifted finally from the “right” carboxyl group of β-methylmalyl-CoA to the “left” carboxyl group of citramalyl-CoA. This shifting is accomplished by an intramolecular CoA transfer (reaction 12). Otherwise, citramalyl-CoA cleavage into pyruvate and acetyl-CoA would not be feasible.
Here, we demonstrate missing enzymes and intermediates, completing this autotrophic CO2 fixation cycle. This cycle involves an intramolecular CoA transferase that catalyzes the unprecedented transfer of the CoA moiety from the C1-carboxyl group to the C4-carboxyl group of the dicarboxylic acid. A common enoyl-CoA hydratase forms (3S)-citramalyl-CoA, the intermediate to be cleaved to pyruvate and acetyl-CoA. The advantages of this cycle compared with the ubiquitous Calvin–Benson–Bassham cycle are discussed. Under oligotrophic aquatic conditions, elements of the 3-hydroxypropionate cycle may be used by other bacteria for the assimilation of small organic molecules.
Results
Conversion of Mesaconyl-C1-CoA to an Unknown CoA Thioester by Cell Extracts.
We used 3 recombinant enzymes in an assay to synthesize labeled mesaconyl-C1-CoA from labeled propionyl-CoA and glyoxylate to be used as substrate for the next missing enzyme of the cycle (Fig. 2, see Fig. 1 for labeling). Extracts of autotrophically and heterotrophically grown cells transformed mesaconyl-C1-CoA (2-methylfumaryl-CoA) at a rate of 2 μmol min−1 (U) mg−1 protein and 0.6 U mg−1 protein (55 °C), respectively. The product eluted under different HPLC conditions after mesaconyl-C1-CoA. Approximately 94% of labeled propionyl-CoA was converted to mesaconyl-C1-CoA and the observed product. The equilibrium concentrations of mesaconyl-C1-CoA and its product were nearly identical, indicating a freely reversible reaction with an equilibrium constant near 1.
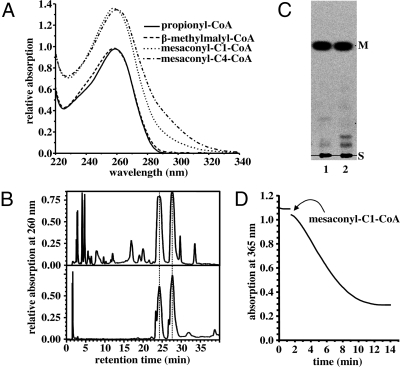
Fig. 2.
Characterization of substrates and products of the conversion of mesaconyl-C1-CoA to mesaconyl-C4-CoA. (A) UV spectra of propionyl-CoA, β-methylmalyl-CoA, mesaconyl-C1-CoA, and mesaconyl-C4-CoA. (B) HPLC separation of 2 chemically synthesized isomeric forms of mesaconyl-CoA carrying the CoA moiety randomly at either C1 or C4 (Bottom) compared with the separation of enzymatically formed mesaconyl-C1-CoA and its product (Upper). A 40-mL gradient from 2% to 10% acetonitrile in 40 mM K2HPO4/HCOOH buffer (pH 4.2), with a flow rate of 1 mL min−1, was used with a reversed-phase column. Detection was at 260 nm. (C) TLC separation of labeled products after alkaline hydrolysis of the CoA thioester products formed from [1-14C]propionyl-CoA and glyoxylate. Detection was by phosphoimaging. Lane 1, mesaconyl-C1-CoA; lane 2, observed product. S, start; M, mesaconate. (D) Spectrophotometric assay used for the characterization of the mesaconyl-CoA C1-C4 CoA transferase. Mesaconyl-C4-CoA hydratase, MMC lyase, and lactate dehydrogenase were used as coupling enzymes. The reaction could be started by either mesaconyl-C1-CoA or mesaconyl-CoA C1-C4 CoA transferase. In the presence of mesaconyl-CoA C1-C4 CoA transferase mesaconyl-C1-CoA (here 0.2 mM) is converted to mesaconyl-C4-CoA, which is hydrated to (S)-citramalyl-CoA by mesaconyl-C4-CoA hydratase. (S)-Citramalyl-CoA subsequently is cleaved into acetyl-CoA and pyruvate by MMC lyase. Pyruvate then is reduced by lactate dehydrogenase to lactate under NADH consumption, followed spectrophotometrically at 365 nm.
Surprisingly, the same product was formed when cell extract was omitted but a different preparation of the recombinant trifunctional (S)-malyl-CoA/β-methylmalyl-CoA/(S)-citramalyl-CoA (MMC) lyase from C. aurantiacus was used. This enzyme preparation was derived from the expression of a DNA fragment of C. aurantiacus in Escherichia coli that contained both the gene coding for the MMC lyase and the upstream neighbor gene coding for a putative CoA transferase. The transferase protein had been ignored because it was expressed only in trace quantities.
Identification of the Unknown CoA Thioester as Mesaconyl-C4-CoA.
The product formed from mesaconyl-C1-CoA was a CoA thioester as well, based on its spectral properties (Fig. 2A), migration in HPLC (Fig. 2B), and alkali sensitivity (Fig. 2C). Electrospray ionization mass spectrometry (ESI-MS) determined a mass of 878.0 Da (negative ion mode) and 879.8 Da (positive ion mode), corresponding to the mass of mesaconyl-C1-CoA or an isomer. However, after alkaline hydrolysis of the 2 labeled CoA esters, only mesaconate (methylfumarate) was obtained (Fig. 2C); no other isomer, such as itaconate (methylenesuccinate) or citraconate (methylmaleate), was formed. Interestingly, the same product mixture was obtained when mesaconyl-CoA was synthesized chemically (Fig. 2B). One would expect the chemical synthesis to yield similar amounts of mesaconyl-C1-CoA and mesaconyl-C4-CoA. Therefore, the product may represent mesaconyl-C4-CoA (3-methylfumaryl-CoA); indeed, this possibility was verified by NMR spectroscopy (see SI Text).
Identification of the CoA Transferase Forming Mesaconyl-C4-CoA and Characterization of the Enzyme.
The enzyme transforming mesaconyl-C1-CoA to mesaconyl-C4-CoA obviously was a type of CoA transferase. A search for putative CoA transferase genes in the genome led to an ORF (Caur_0175), which clustered with MMC lyase (Caur_0174) (as mentioned above) and other genes required for the conversion of propionyl-CoA plus glyoxylate to pyruvate plus acetyl-CoA (Fig. 3). The gene was cloned, expressed in E. coli as an N-terminal His10-tagged protein (47 kDa), and the recombinant protein was purified. It catalyzed the expected reversible transformation of mesaconyl-C1-CoA to mesaconyl-C4-CoA (reaction 12 in Fig. 1) at high rates (Vmax of 520 U mg−1 at 55 °C, turnover number 840 s−1) with an apparent Km for mesaconyl-C1-CoA of 0.24 mM. No CoA transferase activity was observed with itaconate, mesaconate, (R)-/(S)-malate, and (R)-/(S)-citramalate as CoA acceptors when acetyl-CoA or succinyl-CoA as CoA were used as CoA donors. The enzyme exhibited a broad pH optimum at pH20 °C of 7.5–7.8 (extrapolated pH55 °C of 7.1–7.4), with half maximal activity at pH20 °C 6.6. It did not require free mesaconate as a CoA acceptor, because the enzyme did not form [14C]mesaconyl-CoA from [14C]mesaconate plus unlabeled mesaconyl-C1-CoA. Correspondingly, it did not form [14C]mesaconate from [14C]mesaconyl-C1-CoA plus unlabeled mesaconate.
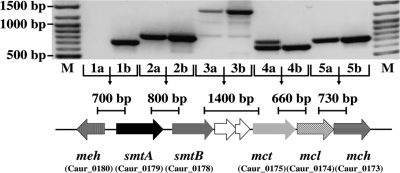
Fig. 3.
Organization of genes involved in the glyoxylate assimilation cycle. For catalyzed reactions (in parentheses) see Fig. 1. To amplify the intergenic regions of the cluster shown below, standard PCRs were performed with cDNA (a) and genomic DNA (b) as control. The positions of the amplified fragments are indicated by bars, and their expected sizes are given in bp. Lane M contained a 100-bp DNA ladder. meh, mesaconyl-C4-CoA hydratase (mesaconyl-C4-CoA (enoyl-CoA) hydratase) (reaction 13); smtAB, succinyl-CoA:(S)-malate CoA transferase subunits A and B (reaction 7); mct, mesaconyl-CoA C1-C4 CoA transferase (reaction 12); mcl, trifunctional (S)-malyl-CoA/β-methylmalyl-CoA/(S)-citramalyl-CoA (MMC) lyase (reactions 10 a, b, c); mch, mesaconyl-C1-CoA hydratase (β-methylmalyl-CoA dehydratase) (reaction 11). Between the genes of smtB and mct there are 2 ORFs of unknown function (no similar proteins are found in the database). For primers, see Table S1.
The UV-visible spectrum of the purified recombinant enzyme showed an absorption band only at 280 nm. Gel filtration indicated a molecular mass of about 73 ± 4 kDa, suggesting a homodimeric structure. The high rate of amino acid sequence similarities/identities (ca. 42%/25%) (Table S2 and Fig. S1) with enzymes belonging to class III CoA transferases (24) suggested that an aspartate residue in the active site forms an acid anhydride with the formerly CoA-activated acid. Although this anhydride intermediate should react with borohydride and hydroxylamine, resulting in enzyme inactivation, these enzymes exhibit very different sensitivities to these compounds (25–30). The CoA transferase was tested for inactivation in the presence of its substrate; the enzyme was only partially inactivated by hydroxylamine (residual activity about 60%), and sodium borohydride had no effect at all. This lack of effect may be caused by a closure of the active site upon substrate binding. Such a conformational change has been observed for the crotonobetainyl-CoA:carnitine CoA transferase (CaiB) from E. coli (31). This change in the conformation of the enzyme may prevent the inactivator molecules from entering the active site and is consistent with the observation that no mesaconate is released during catalysis.
Further Transformation of Mesaconyl-C4-CoA Involving an Enoyl-CoA Hydratase.
A coupled photometric assay was developed to measure the further transformation of mesaconyl-C4-CoA to acetyl-CoA and pyruvate in cell extracts at 45 °C. The specific activities (extrapolated to 55 °C) in extracts of autotrophically and heterotrophically grown cells were 1.2 and 0.5 U mg−1, respectively. This reaction sequence needed no cofactor other than Mg2+. Citramalyl-CoA was observed as an intermediate (as in Fig. 4), indicating that mesaconyl-C4-CoA was hydrated to citramalyl-CoA, followed by citramalyl-CoA cleavage to acetyl-CoA and pyruvate. The extrapolation of specific activities to 55 °C was based on the assumption that the reaction rate doubles with an increase in temperature of 10 °C.
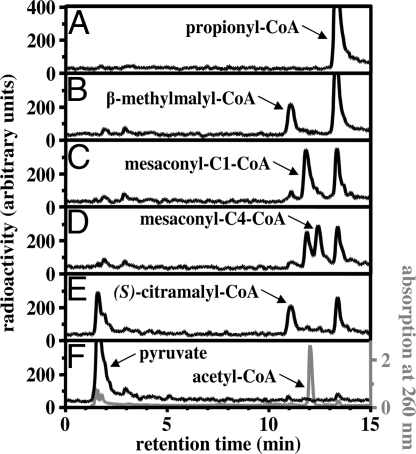
Fig. 4.
HPLC separation of 14C-labeled products formed from [1-14C]propionyl-CoA and glyoxylate by different purified recombinant enzymes. (A) Before addition of enzyme. (B) After the addition of MMC lyase. (C) As B, plus mesaconyl-C1-CoA hydratase. (D) As C, plus mesaconyl-CoA C1-C4 CoA transferase. (E) As D, plus mesaconyl-C4-CoA hydratase. (F) As E: formation of non-labeled acetyl-CoA and 14C-pyruvate after additional incubation time. A reversed-phase column was developed for 7 min under isocratic conditions with 100 mM NaH2PO4 (pH 4.0) in 7.5% methanol (vol/vol), followed by a linear 10-min gradient from 0% to 60% of 100 mM sodium acetate (pH 4.6) in 90% methanol (vol/vol) at a flow rate of 1 mL min−1. Acetyl-CoA was detected by diode array detection because of its absorption at 260 nm. Radioactivity was detected by solid-state scintillation counting.
The next missing enzyme, mesaconyl-C4-CoA hydratase, was purified from autotrophically grown cells (Table S3) SDS-PAGE revealed a polypeptide of 30 kDa, and peptide mass fingerprint analysis identified the corresponding ORF (Caur_0180). It was located directly adjacent to a cluster of genes coding for several enzymes of the 3-hydroxypropionate cycle (Fig. 3). The gene was cloned and expressed in E. coli as an N-terminal His10-tagged protein (33 kDa); the recombinant enzyme was purified, and its activity was measured at 45 °C. It catalyzed the expected hydration of mesaconyl-C4-CoA to (S)-citramalyl-CoA (reaction 13 in Fig. 1). The enzyme exhibited a specific activity of 950 U mg−1 (extrapolated to 55 °C, turnover number 1,045 s−1) with an apparent Km value for mesaconyl-C4-CoA of 75 μM and was inhibited by substrate concentrations higher than 0.3 mM. Gel filtration indicated a molecular mass of about 52 ± 5 kDa, suggesting a homodimeric structure. The amino acid sequence had similarities to a group of uncharacterized conserved proteins (COG3777) with a C-terminal hotdog-fold domain and possibly to an itaconyl-CoA hydratase (AAX86477) from Pseudomonas sp. L1 described by Kornberg and associates (32, 33). This group of enzymes apparently forms a distinct subclass of the enoyl-CoA hydratase superfamily, based on sequence comparison.
Cleavage of (S)-Citramalyl-CoA into Acetyl-CoA and Pyruvate by Recombinant MMC Lyase and Reconstitution of the Second Cycle from Purified Enzymes.
(S)-Citramalyl-CoA cleavage required another enzyme. To assay for this enzyme activity, chemically synthesized mesaconyl-CoA (both C1- and C4-CoA) was incubated at 45 °C with mesaconyl-CoA C1-C4 CoA transferase, and mesaconyl-C4-CoA hydratase to form (S)-citramalyl-CoA. Addition of cell extracts led to a rapid formation of acetyl-CoA and pyruvate. Extrapolated to 55 °C, the rate with extracts of autotrophically and heterotrophically grown cells was 2 U mg−1 and 0.6 U mg−1, respectively. Recombinant MMC lyase has been known to catalyze the cleavage of (S)-citramalyl-CoA into acetyl-CoA and pyruvate (22). We therefore studied the recombinant His10-tagged lyase and found specific activities for (S)-malyl-CoA cleavage into acetyl-CoA and glyoxylate of 1.1 U mg−1 (reaction 10a in Fig. 1), for propionyl-CoA condensation with glyoxylate to β-methylmalyl-CoA of 10 U mg−1 (reaction 10b), and for (S)-citramalyl-CoA cleavage into acetyl-CoA and pyruvate of 24 U mg−1 (reaction 10c). (R)-Malyl-CoA and (R)-citramalyl-CoA were not used. The enzyme catalyzed the reverse reaction, the condensation of pyruvate and acetyl-CoA to (S)-citramalyl-CoA. However, the equilibrium of this reaction was strongly in favor of acetyl-CoA and pyruvate (50:1), although pyruvate was added in 10-fold excess to acetyl-CoA.
The entire glyoxylate assimilation route (glyoxylate + propionyl-CoA → acetyl-CoA + pyruvate) involving 5 enzymatically catalyzed reactions could be reconstituted in vitro by using the 4 recombinant enzymes MMC lyase, mesaconyl-C1-CoA hydratase, mesaconyl-CoA C1-C4 CoA transferase, and mesaconyl-C4-CoA hydratase (Fig. 4).
Cluster of Genes Involved in the Second Cycle.
All genes coding for the enzymes required for the glyoxylate assimilation route plus the 2 genes coding for the subunits of succinyl-CoA:(S)-malate CoA transferase (Caur_0178, Caur_0179) were clustered in the genome (Fig. 3). A similar gene cluster is present in the related Chloroflexi strains Chloroflexus aggregans, Roseiflexus castenholzii, and Roseiflexus sp. RS-1 (34). Two additional small ORFs (Caur_0176, Caur_0177), which probably do not have a function in this pathway, were present in C. aurantiacus only. Activity measurements indicated that the encoded enzymes were up-regulated to a similar extent under autotrophic growth conditions. Of these 6 genes, 5 are oriented in the same direction. Neighboring genes are co-transcribed, as evidenced by RT-PCR of mRNA (Fig. 3), and may constitute an operon. The sixth gene coding for mesaconyl-C4-CoA hydratase (meh) is orientated in the opposite direction. Thus the intergenic region between these clusters may harbor the binding sites for regulatory proteins.
Discussion
This work has completed the 3-hydroxypropionate autotrophic carbon fixation cycle, the fourth autotrophic pathway (Table S4). All characteristic enzymes have been identified and purified. The cycle results in the net fixation of 3 molecules of bicarbonate into 1 molecule of pyruvate, which can be used to produce all cellular building blocks. This pathway is an intertwined bicycle that schematically resembles a pretzel (Fig. 1).
In a first cycle starting from acetyl-CoA, 2 bicarbonate molecules are fixed to form 1 molecule of glyoxylate. In a second cycle, glyoxylate plus propionyl-CoA are disproportionated to acetyl-CoA plus pyruvate without any redox step or additional coenzyme participation. Propionyl-CoA is derived from acetyl-CoA and bicarbonate, as in the first cycle. The reaction sequence glyoxylate + propionyl-CoA → acetyl-CoA + pyruvate is associated with a standard free energy change ΔG°′ of −14 kJ mol−1 (35), resulting in an equilibrium constant Kequ [acetyl-CoA] [pyruvate]/[glyoxylate] [propionyl-CoA] = 300 and rendering the second cycle almost unidirectional toward pyruvate formation. The reaction sequence requires only 3 additional enzymes and solves a mechanistically intriguing problem, the shift of the CoA moiety, in a most economical and elegant way. An unparalleled attribute of the complete pathway is the crossing of the 2 cycles, because acetyl-CoA conversion to propionyl-CoA is common to both. The stoichiometry, cofactor specificity, energy demand, and other features of the cycle now can be compared with other CO2 fixation mechanisms.
Comparison with Other Autotrophic Carbon Fixation Pathways.
The stoichiometry of the 3-hydroxypropionate cycle follows the equation: 3 HCO3− + 5 ATP + 6 NADPH + 1 quinone → 1 pyruvate + 6 NADP + 1 quinoneH2 + 3 ADP + 3 phosphate + 2 AMP + 2 pyrophosphate; this input of energy corresponds to 7 ATP equivalents for pyruvate formation, as required in the Calvin cycle. In terms of the energy demand of an autotrophic pathway, the costs for synthesizing all CO2-fixation–related enzymes are important also. Poor catalytic efficiency of RubisCO, for instance, not only leads to the loss of energy associated with photorespiration but also pushes the organisms to use special carbon-concentrating mechanisms. Moreover, carboxylases with low catalytic efficiency, such as RubisCO, need to be synthesized in large quantities. In other words, the synthesis of the catalyst itself may devour a huge amount of energy as well as nitrogen and sulfur sources. Therefore, the expenditures for the synthesis of all additional enzymes of an autotrophic carbon fixation pathway actually may determine its energy cost. The other tradeoff is the necessity for low-potential electron donors for autotrophic pathways that produce acetyl-CoA or pyruvate directly. Balancing that need against the number of unique proteins required for integrating the 3-hydroxypropionate cycle (relatively high) or the Calvin cycle (only 2 proteins, but many of them, are needed) into central metabolism may determine whether an organism that uses the pathway comes to dominate a given ecosystem. For discussion of other autotrophic pathways, notably those requiring anaerobic conditions, see ref. 36. A comparison of the respective features of all currently known CO2 fixation pathways is provided in Table S4.
Advantages of the 3-Hydroxypropionate Bicycle and Its Elements.
Given that organisms with the Calvin cycle have come to dominate most ecosystems, they presumably have some advantages. In microorganisms the Calvin cycle is found solely in aerobic and facultative aerobic Bacteria, notably in Cyanobacteria. Those bacteria are less energy limited, and therefore the high ATP costs of the Calvin cycle may be of minor importance. In bacteria the need for sugar phosphates in biosynthesis of cell wall and lignin precursors is much less than in plants. The main metabolic fluxes are diverted from acetyl-CoA, pyruvate, oxaloacetate, and 2-oxoglutarate, and their synthesis from 3-phosphoglycerate is partly connected with a loss of CO2. Therefore, in bacteria, autotrophic pathways directly yielding acetyl-CoA or pyruvate are more economical, and this statement holds true for all alternative carbon-fixation pathways, including the 3-hydroxypropionate bicycle. Also, the 2 autotrophic cycles differ with respect to the inorganic carbon species used: CO2 in the Calvin cycle versus HCO3− in the 3-hydroxypropionate bicycle. Because the bicarbonate concentration in slightly alkaline water, where Chloroflexus lives, is much higher than the concentration of dissolved CO2 (the apparent acid dissociation association constant of HCO3−/CO2 is 6.3), Chloroflexus may profit from using bicarbonate instead of CO2.
Most Chloroflexi probably grow as mixotrophs. Why might their pathway be better than the Calvin cycle in this regard? A complete or even a rudimentary 3-hydroxypropionate cycle allows co-assimilating trace amounts of organic compounds even under oxic conditions (no enzyme of the cycle is oxygen sensitive), an ability that may be advantageous in oligotrophic aquatic habitats. Examples of such substrates are the fermentation products acetate, propionate, and succinate (including their corresponding alcohols), which may be formed by cyanobacteria and possibly by fermenting bacteria associated with the Chloroflexus mats. Numerous other compounds that are metabolized via acetyl-CoA or propionyl-CoA are included in this substrate spectrum as well. Notably, 3-hydroxypropionate is a very common metabolite that can be assimilated by this mechanism. It is an intermediate in the metabolism of dimethylsulfoniopropionate (37–40), a ubiquitous osmoprotectant and antioxidant of algae (41, 42); other small sources of 3-hydroxypropionate are pyrimidines and β-alanine (43, 44).
However, as discussed later, the limited distribution of the characteristic enzymes/genes of the 3-hydroxypropionate cycle suggests that these advantages may become effective only in a limited set of natural niches where Chloroflexi compete successfully against other bacteria and cyanobacteria. Another reason for the limited occurrence of this cycle may be a late and singular invention in the Chloroflexi. Interestingly, the autotrophic aerobic Crenarchaeota use a similar mechanism of converting acetyl-CoA to succinyl-CoA via 3-hydroxypropionate (10, 45). However, this crenarchaeal autotrophic pathway probably has evolved independently, and the involved enzymes show little or no similarity to the Chloroflexus enzymes (10).
Occurrence of the 3-Hydroxypropionate Bicycle and Its Elements.
The completely sequenced genomes indicate a similar pathway may operate in Chloroflexus aggregans, Roseiflexus castenholzii, and Roseiflexus sp. RS-1 (Chloroflexaceae), which contain all postulated genes of the cycle. Erythrobacter sp. NAP1 and some phototrophic gamma-Proteobacteria (NOR5–3, NOR51-B) harbor the Chloroflexus-type genes required for the conversion of acetyl-CoA to succinyl-CoA but lack the other genes of the 3-hydroxypropionate bicycle. Likewise, the heterotrophic Congregibacter litoralis and Nitrococcus mobilis and the photolithoautotrophic Chloroherpeton thalassium contain the propionyl-CoA synthase gene only. These bacteria may use a rudimentary cycle for the mixotrophic assimilation of acetate, 3-hydroxypropionate, and/or propionate under oligotrophic (e.g., marine) conditions. Interestingly, the gamma-proteobacterium strain HTCC2080 possesses the genes for a chimeric 3-hydroxypropionate/4-hydroxybutyrate cycle that even may allow autotrophic growth: Genes required for the conversion of acetyl-CoA plus 2 bicarbonate molecules to succinyl-CoA are of the Chloroflexus type. In contrast, the regeneration of acetyl-CoA from succinyl-CoA seems to correspond to the pathways found in autotrophic Crenarchaeota (9, 10, 46).
Streamlining of the Pathway and Its Genes.
The whole cycle requires 19 chemical steps, but only 13 enzymes are involved, suggesting that several enzymes are multifunctional (e.g. bifunctional malonyl-CoA reductase, trifunctional propionyl-CoA synthase, and trifunctional (S)-malyl-CoA/β-methylmalyl-CoA/(S)-citramalyl-CoA lyase). Succinyl-CoA:(S)-malate CoA transferase also catalyzes 2 formal steps, the release of succinate and the activation of (S)-malate. The streamlined pathway and the compact genetic organization of most of its genes indicate an advanced and successful adaptation of the organism to its natural niche that is not occupied by other autotrophic bacteria or algae. In 4 members of Chloroflexaceae (see earlier discussion) a similar cluster of genes required for the second glyoxylate assimilation cycle and for the last reactions of the first glyoxylate formation cycle is present (Fig. 3; for catalyzed reactions, see Fig. 1). Because the encoded enzymes are up-regulated under autotrophic conditions, we expect a common regulator.
An Internal CoA Transferase and an Enoyl-CoA Hydratase.
We have identified and characterized an enzyme of the CoA transferase family (class III) (E. C. 2.8.3.x.) (24) that functions in an intramolecular CoA transfer between 2 carboxyl groups (Fig. 1). Its suggested systematic name is “mesaconyl-CoA C1-C4 CoA transferase.” The amino acid sequence identity and similarity values for different representatives of this family are supplied in Table S2. A highly conserved aspartate residue (Asp 169 in the CaiB nomenclature), which is located in the active site and binds the organic acid in an anhydride bond (47, 48), is conserved in this CoA transferase. Other residues that are important for folding are conserved as well. Regarding the inhibition experiments, the catalytic mechanism of this enzyme is enigmatic in view of the presently considered mechanisms (28). Furthermore, no free carboxylic acid is involved, nor does free mesaconate exchange with mesaconyl-CoA. Additional studies are required to address this tantalizing mechanistic problem.
Mesaconyl-C4-CoA is hydrated by a rather conventional enoyl-CoA hydratase to (S)-citramalyl-CoA (Fig. 1) that differs strongly from mesaconyl-C1-CoA hydratase (23). Although both enzymes are members of the superfamily of hotdog domain-containing proteins (49), their amino acid sequences are not significantly similar. Enoyl-CoA hydratases (E. C. 4.2.1.17) catalyze reversible reactions of the type (3S)-3-hydroxyacyl-CoA → trans-2(or 3)-enoyl-CoA + H2O. Therefore the formation of the (S)-stereoisomer of citramalyl-CoA, as observed experimentally, was expected.
Open Questions.
The demonstration of all enzyme activities of the CO2 fixation bicycle, their whole-cell regulation, and the organization of the genes provide final evidence for the operation of the autotrophic CO2 fixation bicycle. The roles of 2 other enzymes, succinyl-CoA:(R)-citramalate CoA transferase and (R)-citramalyl-CoA lyase, which act on the (R)-stereosisomers of (citra)malate (21, 24) and which also are up-regulated under autotrophic conditions, are still unclear. Their specific activities are an order of magnitude lower than those of the (S)-citramalate–specific CoA transferase and lyase. They may be required to recycle (R)-citramalate or (R)-malate formed incidentally by side reactions. The regulation of acetyl-CoA carboxylase, which is the carboxylase of the 3-hydroxypropionate cycle and the initial enzyme in lipid biosynthesis, poses another problem that may prevent this cycle from being introduced into an organism that uses the Calvin cycle. How fatty acid biosynthesis and acetyl-CoA carboxylase are regulated separately, depending on the needs of the cell, is not known. Furthermore, it still is unknown if propionyl-CoA carboxylation also is catalyzed by a bifunctional enzyme, as is the case in the crenarchaeal 3-hydroxypropionate/4-hydroxybutyrate cycle (10).
Materials and Methods
C. aurantiacus strain OK-70-fl (DSMZ 636) was grown anaerobically and phototrophically either under autotrophic conditions with H2 and CO2 (80:20, vol/vol) or under heterotrophic conditions with casamino acids and yeast extract as described elsewhere (20). The genome sequence of the strain J-10-fl was used for primer design. Virtually no differences in the genes of the 2 strains were observed. The exact procedures of cloning and heterologous expression of genes from C. aurantiacus in E. coli, enzyme purification, enzyme measurements, syntheses, identification of compounds, and all other methods are described in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
G.F. acknowledges the great impact of Professor Achim Trebst on his interest in autotrophy, which traces back to his stay in the Lehrstuhl fuer Biochemie der Pflanzen at Bochum University. This work was supported by Deutsche Forschungsgemeinschaft. The authors thank Nasser Gad'on and Christa Ebenau-Jehle (Freiburg) for invaluable expert technical assistance. Thanks also are owed to Ansgar Schlichting, Silke Friedmann, and Birgit E. Alber (Freiburg), who performed preliminary experiments with the CoA transferase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.A.B. is a guest editor invited by the Editorial Board.
See Commentary on page 21015.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908356106/DCSupplemental.
References
- 1.Thauer RK. A fifth pathway of carbon fixation. Science. 2007;318:1732–1733. doi: 10.1126/science.1152209. [DOI] [PubMed] [Google Scholar]
- 2.Calvin M, Bassham JA. The Photosynthesis of Carbon Compounds. New York: W. A. Benjamin, Inc.; 1962. [Google Scholar]
- 3.Evans MC, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci USA. 1966;55:928–934. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan BB, Arnon DI. A reverse KREBS cycle in photosynthesis: Consensus at last. Photosynth Res. 1990;24:47–53. [PubMed] [Google Scholar]
- 5.Utter MF, Wood HG. Mechanisms of fixation of carbon dioxide by heterotrophs and autotrophs. Adv Enzymol Relat Areas Mol Biol. 1951;12:41–151. doi: 10.1002/9780470122570.ch2. [DOI] [PubMed] [Google Scholar]
- 6.Ljungdahl L, Irion E, Wood HG. Total synthesis of acetate from CO2. I. Co-methylcobyric acid and CO-(methyl)-5-methoxybenzimidazolylcobamide as intermediates with Clostridium thermoaceticum. Biochemistry. 1965;4:2771–2780. doi: 10.1021/bi00888a030. [DOI] [PubMed] [Google Scholar]
- 7.Ljungdahl L, Wood HG. Incorporation of C14 from carbon dioxide into sugar phosphates, carboxylic acids, and amino acids by Clostridium thermoaceticum. J Bacteriol. 1965;89:1055–1064. doi: 10.1128/jb.89.4.1055-1064.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerby R, Zeikus JG. Growth of Clostridium thermoaceticum on H2/CO2 or CO as energy source. Curr Microbiol. 1983;8:27–30. [Google Scholar]
- 9.Huber H, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 11.Pierson BK, Castenholz RW. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen and sp nov. Arch Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 12.Castenholz RW, Pierson BK. In: Anoxygenic Photosynthetic Bacteria. Blankenship RE, Madigan MT, Bauer CE, editors. Dordrecht, Germany: Kluwer Academic Publishers; 1995. pp. 87–103. [Google Scholar]
- 13.Holo H, Grace D. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch Microbiol. 1989;151:252–256. [Google Scholar]
- 14.Strauss G, Eisenreich W, Bacher A, Fuchs G. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing Archaebacterium Thermoproteus neutrophilus and in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur J Biochem. 1992;205:853–866. doi: 10.1111/j.1432-1033.1992.tb16850.x. [DOI] [PubMed] [Google Scholar]
- 15.Eisenreich W, Strauss G, Werz U, Fuchs G, Bacher A. Retrobiosynthetic analysis of carbon fixation in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur J Biochem. 1993;215:619–632. doi: 10.1111/j.1432-1033.1993.tb18073.x. [DOI] [PubMed] [Google Scholar]
- 16.Strauss G, Fuchs G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur J Biochem. 1993;215:633–643. doi: 10.1111/j.1432-1033.1993.tb18074.x. [DOI] [PubMed] [Google Scholar]
- 17.Herter S, et al. Autotrophic CO2 fixation by Chloroflexus aurantiacus: Study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J Bacteriol. 2001;183:4305–4316. doi: 10.1128/JB.183.14.4305-4316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herter S, Busch A, Fuchs G. L-malyl-coenzyme A lyase/beta-methylmalyl-coenzyme A lyase from Chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO2 fixation. J Bacteriol. 2002;184:5999–6006. doi: 10.1128/JB.184.21.5999-6006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herter S, Fuchs G, Bacher A, Eisenreich W. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J Biol Chem. 2002;277:20277–20283. doi: 10.1074/jbc.M201030200. [DOI] [PubMed] [Google Scholar]
- 20.Hügler M, Menendez C, Schägger H, Fuchs G. Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J Bacteriol. 2002;184:2404–2410. doi: 10.1128/JB.184.9.2404-2410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alber BE, Fuchs G. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J Biol Chem. 2002;277:12137–12143. doi: 10.1074/jbc.M110802200. [DOI] [PubMed] [Google Scholar]
- 22.Friedmann S, Alber BE, Fuchs G. Properties of R-citramalyl-coenzyme A lyase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J Bacteriol. 2007;189:2906–2914. doi: 10.1128/JB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarzycki J, et al. Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria. J Bacteriol. 2008;190:1366–1374. doi: 10.1128/JB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heider J. A new family of CoA-transferases. FEBS Lett. 2001;509:345–349. doi: 10.1016/s0014-5793(01)03178-7. [DOI] [PubMed] [Google Scholar]
- 25.Friedmann S, Alber BE, Fuchs G. Properties of succinyl-coenzyme A:D-citramalate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J Bacteriol. 2006;188:6460–6468. doi: 10.1128/JB.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedmann S, Steindorf A, Alber BE, Fuchs G. Properties of succinyl-coenzyme A:L-malate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus. J Bacteriol. 2006;188:2646–2655. doi: 10.1128/JB.188.7.2646-2655.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leutwein C, Heider J. Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: An enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J Bacteriol. 2001;183:4288–4295. doi: 10.1128/JB.183.14.4288-4295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthold CL, Toyota CG, Richards NG, Lindqvist Y. Reinvestigation of the catalytic mechanism of formyl-CoA transferase, a class III CoA-transferase. J Biol Chem. 2008;283:6519–6529. doi: 10.1074/jbc.M709353200. [DOI] [PubMed] [Google Scholar]
- 29.Dickert S, Pierik AJ, Linder D, Buckel W. The involvement of coenzyme A esters in the dehydration of (R)-phenyllactate to (E)-cinnamate by Clostridium sporogenes. Eur J Biochem. 2000;267:3874–3884. doi: 10.1046/j.1432-1327.2000.01427.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Darley D, Selmer T, Buckel W. Characterization of (R)-2-hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of L-leucine to isocaproate by Clostridium difficile. Appl Environ Microbiol. 2006;72:6062–6069. doi: 10.1128/AEM.00772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan ES, Li Y, Iannuzzi P, Cygler M, Matte A. Crystal structure of Escherichia coli crotonobetainyl-CoA: Carnitine CoA-transferase (CaiB) and its complexes with CoA and carnitinyl-CoA. Biochemistry. 2005;44:5728–5738. doi: 10.1021/bi047656f. [DOI] [PubMed] [Google Scholar]
- 32.Cooper RA, Itiaba K, Kornberg HL. The utilization of aconate and itaconate by Micrococcus sp. Biochem J. 1965;94:25–31. doi: 10.1042/bj0940025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper RA, Kornberg HL. The utilization of itaconate by Pseudomonas sp. Biochem J. 1964;91:82–91. doi: 10.1042/bj0910082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klatt CG, Bryant DA, Ward DM. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environmental Microbiology. 2007;9:2067–2078. doi: 10.1111/j.1462-2920.2007.01323.x. [DOI] [PubMed] [Google Scholar]
- 35.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs G. In: Biology of Autotrophic Bacteria. Schlegel HG, Bowien B, editors. Madison, WI: Science Tech Publishers; 1989. pp. 365–382. [Google Scholar]
- 37.Ansede JH, Pellechia PJ, Yoch DC. Metabolism of acrylate to beta-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl Environ Microbiol. 1999;65:5075–5081. doi: 10.1128/aem.65.11.5075-5081.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansede JH, Pellechia PJ, Yoch DC. Nuclear magnetic resonance analysis of [1-13C]dimethylsulfoniopropionate (DMSP) and [1-13C]acrylate metabolism by a DMSP lyase-producing marine isolate of the alpha-subclass of Proteobacteria. Appl Environ Microbiol. 2001;67:3134–3139. doi: 10.1128/AEM.67.7.3134-3139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd JD, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–669. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- 40.Yoch DC. Dimethylsulfoniopropionate: Its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol. 2002;68:5804–5815. doi: 10.1128/AEM.68.12.5804-5815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston AW, et al. Molecular diversity of bacterial production of the climate-changing gas, dimethyl sulphide, a molecule that impinges on local and global symbioses. J Exp Bot. 2008;59:1059–1067. doi: 10.1093/jxb/erm264. [DOI] [PubMed] [Google Scholar]
- 42.Sunda W, Kieber DJ, Kiene RP, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 43.Andersen G, et al. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J Mol Biol. 2008;380:656–666. doi: 10.1016/j.jmb.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 44.Loh KD, et al. A previously undescribed pathway for pyrimidine catabolism. Proc Natl Acad Sci USA. 2006;103:5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishii M, et al. Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch Microbiol. 1997;166:368–371. doi: 10.1007/BF01682981. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-Vera WH, Berg IA, Fuchs G. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J Bacteriol. 2009;191:4286–4297. doi: 10.1128/JB.00145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jonsson S, Ricagno S, Lindqvist Y, Richards NG. Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes. J Biol Chem. 2004;279:36003–36012. doi: 10.1074/jbc.M404873200. [DOI] [PubMed] [Google Scholar]
- 48.Stenmark P, Gurmu D, Nordlund P. Crystal structure of CaiB, a type-III CoA transferase in carnitine metabolism. Biochemistry. 2004;43:13996–14003. doi: 10.1021/bi048481c. [DOI] [PubMed] [Google Scholar]
- 49.Dillon SC, Bateman A The hotdog fold: Wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.