Abstract
A new class of small RNAs (endo-siRNAs) produced from endogenous double-stranded RNA (dsRNA) precursors was recently shown to mediate transposable element (TE) silencing in the Drosophila soma. These endo-siRNAs might play a role in heterochromatin formation, as has been shown in S. pombe for siRNAs derived from repetitive sequences in chromosome pericentromeres. To address this possibility, we used the viral suppressors of RNA silencing B2 and P19. These proteins normally counteract the RNAi host defense by blocking the biogenesis or activity of virus-derived siRNAs. We hypothesized that both proteins would similarly block endo-siRNA processing or function, thereby revealing the contribution of endo-siRNA to heterochromatin formation. Accordingly, P19 as well as a nuclear form of P19 expressed in Drosophila somatic cells were found to sequester TE-derived siRNAs whereas B2 predominantly bound their longer precursors. Strikingly, B2 or the nuclear form of P19, but not P19, suppressed silencing of heterochromatin gene markers in adult flies, and altered histone H3-K9 methylation as well as chromosomal distribution of histone methyl transferase Su(var)3–9 and Heterochromatin Protein 1 in larvae. Similar effects were observed in dcr2, r2d2, and ago2 mutants. Our findings provide evidence that a nuclear pool of TE-derived endo-siRNAs is involved in heterochromatin formation in somatic tissues in Drosophila.
Keywords: RNAi, nucleus, viruses
Recent deep sequencing efforts have provided critical information on Drosophila small RNA repertoires in various tissues and during distinct developmental stages (1–7). Four classes of small RNAs mediate posttranscriptional gene silencing in Drosophila: i) ≈22-nt miRNAs are processed from stem-loop precursors by Dicer-1 and repress mRNA expression; ii) ≈25-nt piRNAs are produced from transposable element (TE) transcripts in gonads where they silence TEs through a feedback regulatory mechanism involving the PIWI subfamily of Argonautes (2, 3, 8–11); iii) 21-nt siRNAs are processed from long dsRNAs by Dicer-2 and trigger RNAi, for instance in response to viral infection (12–14); and iv) recently discovered 21-nt endo-siRNAs are processed from endogenous dsRNA precursors by Dicer-2 and silence TEs, and possibly endogenous mRNA in somatic tissues (1, 5–7, 15).
In S. pombe, siRNAs produced from repetitive sequences in chromosome pericentromeres direct heterochromatin formation and transcriptional gene silencing. As in S. pombe (16), Drosophila heterochromatin is prominent in pericentromeric regions, mostly comprised of short satellite repeats and TEs, and is associated with histone H3 methylation on lysine 9 (H3K9) by the histone methylase Su(var)3–9 (Clr4 in S pombe). This allows recruitment of the Heterochromatin Protein HP1 (SWI6 in S. pombe) to maintain and spread heterochromatin to nearby genes (17). Despite these analogies, the evidence supporting a role of small RNAs in heterochromatin formation and transcriptional gene silencing in Drosophila remain indirect (18, 19). Mutants for the Argonautes Piwi and Aubergine or for the RNA helicase Spindle-E exhibit decreased H3-K9 methylation, altered recruitment of HP1 and decreased silencing of heterochromatin markers and of several classes of TEs (20–22); Piwi was shown to interact directly with HP1 (23). In addition, it is noteworthy that these data point out piRNAs that are mostly produced in gonads, suggesting that this class of small RNAs play an initiator role in heterochromatin establishment in the germ line.
Here, we show that another class of siRNA derived from TE transcripts, endo-siRNAs, plays a role in heterochromatin formation in somatic tissues during larval development and in adults. Our data strongly suggest that proper nuclear localization of these siRNAs is essential to regulate chromatin dynamics in Drosophila.
Results and Discussion
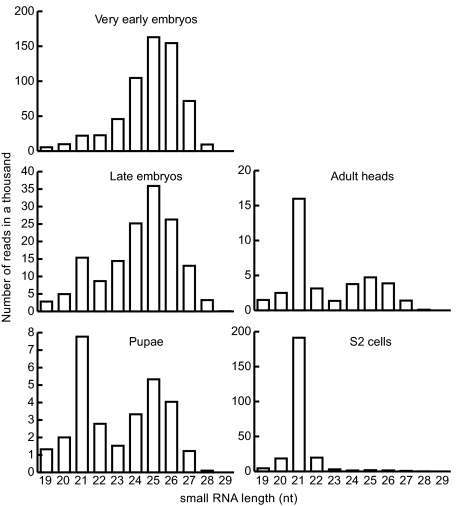
We examined the length distribution of TE-matching small RNAs in publicly available small RNA libraries from the fly soma (4), see Materials and Methods). We found that a dramatic shift in the size of repeat-derived small RNAs occurs during development: the greater population of ≈25 nt species detected in very early embryos, largely composed of maternally deposited TE-derived piRNAs (24), is replaced by a population of ≈21 nt species in pupae, adult heads and S2 cells (Fig. 1). This size shift is consistent with previous observations indicating that TE-derived siRNAs are produced in somatic tissues (1). Whether these endogenous TE-derived siRNAs are involved in heterochromatin formation in the soma, however, remains unanswered (25). To address this question, viral proteins known to counteract antiviral RNAi were expressed in flies and their effects on endogenous TE-derived siRNAs were assessed in the soma. The Tombusvirus P19 and Flock House virus B2 proteins suppress antiviral RNAi in plants and insects, respectively (26). P19 forms a head-to-tail homodimer that specifically sequesters siRNA duplexes (27–29), whereas B2 forms a four-helix bundle that binds to one face of an A-form RNA duplex, independent of its length. As a consequence, and unlike P19, B2 prevents the processing of long dsRNAs into siRNAs by the Drosophila Dicer-2 (30–33). We found that silencing of endogenous white or EcR genes by inverted-repeat constructs (34, 35) is suppressed in transgenic adults expressing B2 or P19 in the eye (Fig. S1 A–B). In contrast, P19 fused to a nuclear localization peptide (NLS-P19) (Fig. S2A) barely suppresses white and EcR RNAi, in agreement with siRNA-mediated target cleavage taking place in the cytoplasm. Development of B2, P19 and NLS-P19 transgenic animals was not altered, nor was the repression of a sensor construct reporting bantam activity (36), indicating no obvious interference with the miRNA pathway by either viral protein (Fig. S1C).
Fig. 1.
The length of TE-matching small RNAs varies during Drosophila development. Length distribution of the TE-matching small RNAs sequences from very early embryos, early embryos, pupae, adult heads and S2 cell libraries. Numbers of reads are normalized to the sequencing deep of each library. The peak of ≈25nt small RNAs in the head library likely corresponds to piRNAs from contaminating ovaries in the sample (4).
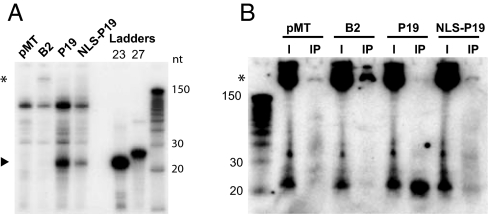
Having established that constitutive expression of B2 and P19 specifically impairs hairpin-induced RNAi in living flies, we tested whether they altered the endogenous siRNA pathway. Epitope-tagged B2, P19, or NLS-P19 were transiently expressed in S2 cells (Fig. S2A); 3′-end RNA labeling revealed the presence of ≈21nt RNAs in immunoprecipitates of P19 and, to a lesser extent, of NLS-P19 (Fig. 2A, arrowhead). Among them, siRNAs antisense to the LTR-retrotransposons HMS-Beagle (Fig. 2B) and to transcripts of the roo TE (Fig. S2D) were detected. By contrast, B2 immunoprecipitates contained barely detectable amounts of siRNAs (Fig. 2 A and B). Rather, long (>200nt) RNA species were detected (Fig. 2A, asterisk), including HMS-Beagle and roo antisense transcripts (Fig. 2B and Fig. S2D, asterisk), suggesting that B2 binds long double-stranded TE RNAs and likely compromises small RNA biogenesis.
Fig. 2.
P19 and B2 respectively sequester endogenous TE-matching siRNAs or longer precursors in S2 cells. (A) Immunoprecipitated P19 and NLS-P19 sequester ≈21 nt RNAs that migrate as ≈22–23 nt species after 3′pCp labeling (arrowhead) whereas larger RNA species are sequestered exclusively by B2 (*). Control immunoprecipitation (pMT) was performed using S2 cells transfected with the empty expression vector pMT-DEST48 (B) A sense HMS-Beagle probe revealed enriched siRNAs in Northern blots of P19 and NLS-P19 immunoprecipitates (IP) and longer RNA species (*) in B2 immunoprecipitate. I, corresponds to total RNA input material.
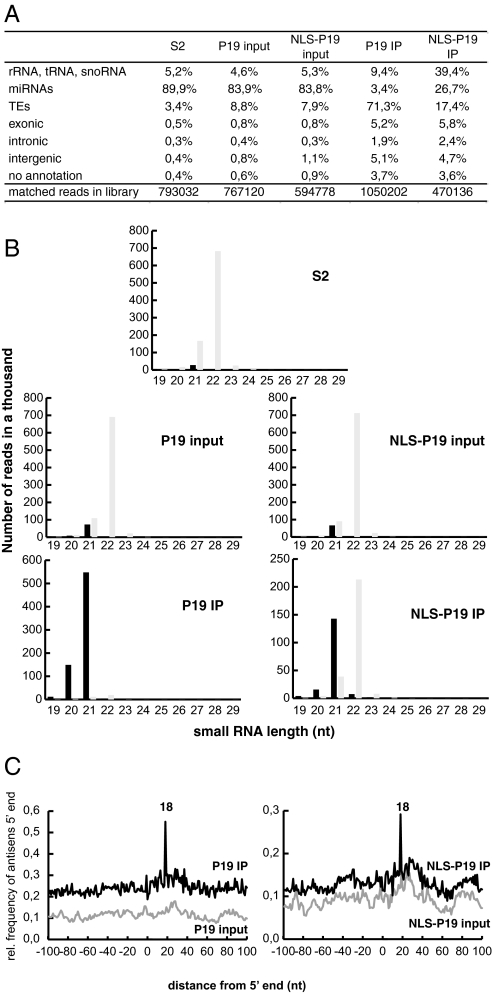
To characterize the small RNAs bound in vivo by P19 and NLS-P19, we performed high-throughput sequencing by Solexa methodology of small RNA libraries from S2 cells expressing P19 or NLS-P19 (P19 and NLS-P19 inputs), as well as from small RNAs libraries coimmunoprecipitated with P19 or NLS-P19, respectively (P19 IP and NLS-P19 IP). We also generated a small RNA library from nontransfected cells (S2) as a control. Deep sequencing of each of the five libraries yielded from 470,136 to 1,050,202 19–29nt RNA reads that matched the D. melanogaster genome. These reads fell into the various RNA classes depicted in Fig. 3A. The proportion of 21nt TE-matching siRNAs ranged from 3.4% to 8% in the libraries from control, P19- and NLS-P19-expressing S2 cells. However, these species were strikingly enriched to 71.3% in the library immunoprecipitated from P19-bound RNAs (Fig. 3 A and B). Although to a lesser extent (17.4%), TE-matching siRNAs were also significantly enriched in immunoprecipitated NLS-P19-bound RNAs. We noted that the proportion of rRNA-matching products increased in parallel in this specific library (Fig. 3A) whereas there was an enrichment in exon-matching small RNAs in P19 and NLS-P19 bound RNAs (Fig. 3A, raw “exonic”). Genome mapping of these small RNAs indicated that they mostly correspond to previously described endo-siRNAs produced from cis-natural antisense transcripts (1, 5, 6, 37). In contrast to endo-siRNAs, the proportion of miRNAs found in P19 and NLS-P19 immunoprecipitates decreased dramatically (3.4% and 26.7%, respectively), compared with that found in libraries from control, P19- and NLS-P19-expressing cells (≈85%) (Fig. 3 A and B).
Fig. 3.
Characterization of small RNAs bound by P19 and NLS-P19. (A) Annotation of small RNAs isolated from S2 cells, S2 cells expressing P19 (P19 input) or NLS-P19 (NLS-P19), and P19 (P19 IP) or NLSP19 (NLS-P19 IP) immunoprecipitates. (B) Length profiles of miRNAs (gray bars) and TE-matching small RNAs (black bars) in small RNA libraries. Numbers of reads are normalized to the sequencing deep of each library. (C) Frequency maps, for the P19 input, P19 IP, NLS-P19 input and NLS IP libraries, of the separation of TE-matching siRNAs mapping to opposite genomic strands. The spike at position 18 indicates the position of maximal probability of finding the 5′ end of a complementary siRNA, which corresponds to a 19nt offset (graphs start at 0).
The P19 protein specifically binds 21bp double-stranded siRNAs with 2nt overhangs (27, 28). We thus expected an enrichment of siRNA duplexes showing perfect strand complementarity over 19nt in the libraries from P19 IP and NLS-P19 IP bound RNAs. To test this notion we plotted the distance between each TE-matched siRNA 5′ end in the genome and the 5′ end of its neighbors on the opposite strand over a window of −100/+100nt. We found that the relative probability of finding an siRNA partner whose 5′ end can be mapped 19nt away on the complementary strand dramatically increased in the P19 IP and NLS-P19 IP libraries compared with the S2, P19 input and NLS-P19 input libraries (Fig. 3C). Likewise, we observed a dramatic increase in the probability of 19bp duplex formation when the analysis was restricted to the siRNAs matching the 297 retrotransposon only (Fig. S3).
Altogether, these results demonstrate that P19 and NLS-P19 sequester endo-siRNA duplexes, which correspond in their majority to TE-derived siRNAs. It is noteworthy that both in Northern experiments and high-throughput sequencing, the fraction of endo-siRNAs bound by NLS-P19 appeared reduced as compared with the one bound by P19, which is nearly exclusively cytoplasmic (Fig. S2A). Nonetheless, the nature of TEs giving rise to the most abundant endo-siRNAs did not change among the 5 libraries, with 297, blood and 1731 retrotransposons invariably accounting for ≈60% of the TE-matching siRNAs (Fig. S4A). In addition, genomic matches of the TE-derived siRNAs did not vary significantly between libraries (Fig. S4B). Therefore, the respective affinity of P19 and NLS-P19 for TE-derived siRNAs does not appear to be biased for particular classes of TEs. Because the two proteins immunoprecipitated at similar levels (Fig. S2C), the results thus suggest the existence of an abundant, cytoplasmic pool of endo-siRNA in S2 cells, in addition to a moderately abundant pool of nuclear endo-siRNAs.
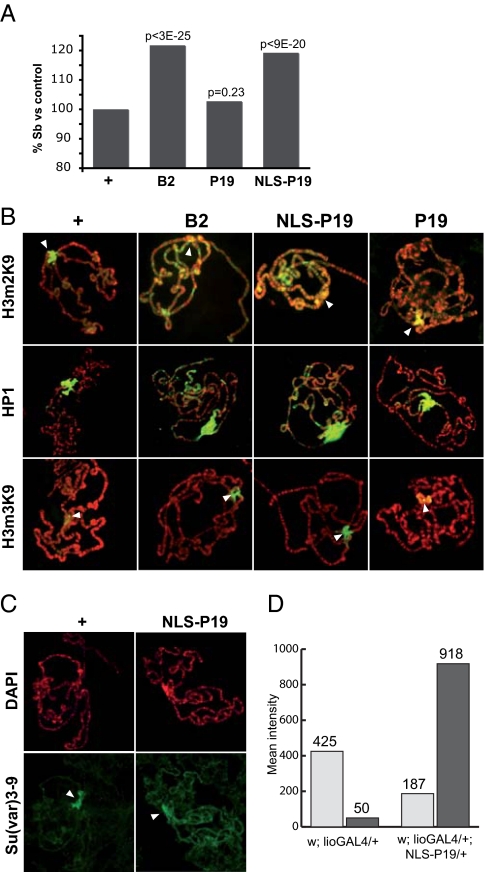
We next tested whether the effects of P19, NLS-P19 and B2 on endo-siRNAs impinged on heterochromatin formation and distribution in adult flies. To this end, we measured their effect on Position Effect Variegation (PEV), a process by which heterochromatin invasion of a marker gene causes its silencing. The T(2;3)SbV translocation relocates a dominant mutant allele of Stubble (Sb1) from its normal position on chromosome 3R to the 2R pericentromere. The ensuing heterochromatic silencing of SbV results in longer, nearly wild type bristles (38). Ubiquitous expression of B2 and NLS-P19 relieved SbV silencing in adult flies, indicating that B2 and NLS-P19 suppress PEV; in contrast, P19 had no effect (Fig. 4A).
Fig. 4.
Viral RNAi suppressors affect heterochromatin. (A) Ubiquitous expression of the viral protein B2 or NLS-P19 suppresses Sb1 variegation whereas P19 has no effect. Bristles were scored in the non-TM3 Ser progeny of homozygous Act5C-GAL4>B2, Act5C-GAL4>P19 or Act5C-GAL4>NLS-P19 females crossed to T(2;3)Sbv, In(3R)Mo, Sb1 sr1/TM3 Ser males. Statistical significance indicated above the bars was assessed by pair wise Chi-2 tests with the control score observed in the non-TM3 Ser progeny (+) of homozygous Act5C-GAL4 females crossed to T(2;3)Sbv, In(3R)Mo, Sb1 sr1/TM3 Ser males. (B) Expression in salivary glands of NLS-P19 and B2, but not P19, induces ectopic deposition of H3m2K9 across chromosome arms and decreased levels at the pericentromere (Top, arrowheads point to pericentromere), ectopic HP1 localization across chromosome arms (Middle), and H3m3K9 spreading across the pericentromere (Bottom, arrowheads point to chromocenter core). (C) NLS-P19 impairs Su(var)3–9 recruitment at the pericentromere and relocates the protein across chromosome arms. (D) Signal count of images in C shows a 2-fold decrease of Su(var)3–9 levels across the pericentromere (gray bars), for an 18-fold increase across chromosome arms (black bars). Mean intensity values are given as arbitrary units. Antibodies: green; DAPI: red.
When expressed in larval salivary glands, B2 was found in the cytoplasm and only faintly detected in the nucleus or in perinuclear regions; P19 remained cytoplasmic and was enriched at the cytoplasmic membrane whereas NLS-P19 was exclusively nuclear (Fig. S5A). In polytene chromosomes from wild type salivary glands (Fig. 4B), dimethylation of histone H3-K9 residue (H3m2K9) typically covers heterochromatin in the pericentromere, telomeres, and a few loci along chromosome arms. In contrast, ≈60% of polytene chromosomes from larvae expressing NLS-P19 had poor H3m2K9 labeling at the pericentromere, but showed, in contrast, strong labeling spread across chromosome arms. Chromosomes from B2-expressing animals displayed similarly altered H3m2K9 patterns, albeit less frequently (≈30%). Furthermore, we found that NLS-P19 and B2 strongly increased the pericentromeric distribution of H3m3K9 (Fig. 4B), another heterochromatic mark that normally accumulates at the chromocenter core, and only weakly at the pericentromere in a Su(var)3–9-dependent manner (39). NLS-P19 and B2 also affected the distribution of Heterochromatin Protein 1 (HP1), normally concentrated at the pericentric heterochromatin and the fourth chromosome. Indeed, paralleling the spreading of H3m2K9, strong ectopic HP1 labeling was detected on the arms of NLS-P19 and B2 polytene chromosomes (Fig. 4B), in agreement with a role for the H3m2K9 mark in recruiting HP1 (40).
In accordance with the lack of P19 effect on PEV, distribution of H3m2K9, H3m3K9 and HP1 was unaltered in flies expressing P19 at the same levels as NLS-P19 (Fig. 4B and Fig. S5B). This result indicates that the P19 effect on heterochromatin entails its nuclear localization, as had been previously shown in plants (41). Finally, we tested the distribution of Su(var)3–9, a major and well characterized Drosophila H3-K9 methyltransferase that locates prominently at the pericentromere (39). In animals expressing NLS-P19 in salivary glands, Su(var)3–9 labeling was reduced at the pericentromere and accumulated ectopically along chromosome arms (Fig. 4C), mirroring the unusual H3m2K9 patterns induced by NLS-P19 and B2. Because neither B2 nor NLS-P19 associates directly with chromosomes, these results strongly suggest that sequestering siRNAs or their precursors is sufficient to generate aberrant H3K9 methylation patterns and ectopic HP1 localization on chromosomes. Collectively, the data suggests that endo-siRNAs are required for heterochromatin silencing in the adult soma and for proper targeting of Su(var)3–9 and H3K9 methylation at the pericentromere.
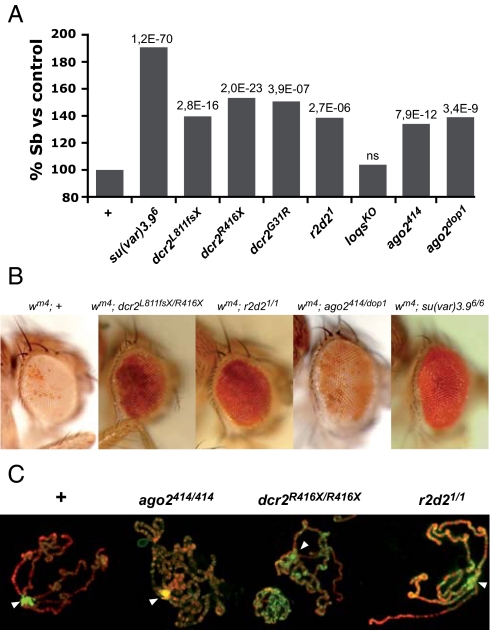
To test this model further, we analyzed the effect of mutations in RNAi pathway components on PEV. These mutations are expected to alter the biogenesis or activity of endo-siRNAs (1, 2). The Su(var)3-9 mutation, used as a reference, eliminated SbV silencing in the T(2;3)SbV test strain. This silencing was also significantly compromised in heterozygous dcr2, r2d2, and ago2 mutants but not in loqs mutant (Fig. 5A). Because RNAi mutations are exclusively of paternal origin in this experiment, the result indicates that Dcr2, Ago2, and R2d2 are zygotically required for SbV repression. Notably, the inhibiting effect of the dcr2G31R mutation, which specifically inactivates the nuclease function of Dcr2 while presumably keeping its dsRNA binding property intact (42), suggests that processing of siRNAs, is mandatory for SbV silencing. We carried out similar analyses on the white-mottled 4 rearrangement, which juxtaposes the euchromatic white gene next to pericentric heterochromatin on chromosome X (43). The silencing, in this case, is manifested by mottling of the eye color. As observed with the effects of viral silencing suppressors on SbV, wm4 silencing was relieved in dcr2, r2d2, and ago2 homozygous male mutant flies (Fig. 5B). We conclude that the RNAi pathway is involved in the spreading of heterochromatin onto variegating reporters in the adult soma.
Fig. 5.
Mutations of RNAi pathway components affect heterochromatin. (A) Sbv silencing in wild type and heterozygous mutant contexts. The number of Sb bristles in Sbv/+ males heterozygous for the indicated mutant allele was expressed as a percentage relative to the Sbv/+ control males (+). Statistical significance indicated above the bars was assessed by pair wise Chi-2 tests with the control. (B) Eyes from wm4 males carrying the given mutant alleles are compared with wm4; + control males. See Fig. S7 for the design of genetic tests. Su(var)3-9 mutant is shown for comparison. (C) ago2, dcr2, and r2d2 mutants show aberrant H3m2K9 patterns (green) in chromosomes from salivary glands similar to those found in NLS-P19- and B2-expressing animals.
Having established that RNA silencing mutations compromise PEV, we further analyzed heterochromatic mark deposition on polytene chromosomes from homozygous ago2, dcr2, and r2d2 mutants. All three mutations caused an aberrant H3m2K9 pattern similar to the one observed in NLS-P19- and B2-expressing animals (Fig. 5C), in agreement with the defects in heterochromatin formation previously reported in other tissues of dcr2 and ago2 mutants (44, 45). Moreover, ectopic HP1 labeling was detected on the arms of ago2 mutant chromosomes whereas HP1 staining strikingly decreased at the pericentromere of dcr2 mutant chromosomes (Fig. S6). Altogether, these results strongly suggest that siRNAs, Ago2, Dcr2, and R2D2 are somatically involved in the targeting of Su(var)3–9 and subsequent H3K9 methylation and HP1 recruitment at the pericentromere.
The present study implicates components of the RNAi pathway in heterochromatin silencing during late Drosophila development. The study also provides correlative evidence supporting a functional link between endo-siRNAs and the formation or maintenance of somatic heterochromatin in flies. The viral proteins NLS-P19 and B2 suppress the silencing of PEV markers and induce aberrant distribution of H3m2K9 and H3m3K9 heterochromatic marks as well as histone H3 methylase Su(var)3–9 in larval tissues. Dcr2 and Ago2 mutations have similar effects. In striking contrast, cytoplasmic P19 has no noticeable effect on chromatin. We propose that B2 inhibits Dcr2-mediated processing of double-stranded TE read-through transcripts in the cytoplasm; we further propose that NLS-P19 sequesters TE-derived siRNA duplexes. This model implies that part of the cytoplasmic pool of TE-derived endo-siRNA (which might be involved in PTGS events) is translocated back into the nucleus to exert chromatin-based functions. In C. elegans, silencing of nuclear-localized transcripts involves nuclear transport of siRNAs by an NRDE-3 Argonaute protein (46). A similar siRNA nuclear translocation system, possibly mediated by Ago2, may also exist in flies. Alternatively, an as yet unidentified siRNA duplex transporter may be involved. Deep sequencing analyses show that the fraction of siRNAs sequestered by NLS-P19 is smaller as compared with the one bound by P19 in the cytoplasm. Thus, the poor effects of P19 on nuclear gene silencing may be explained if the cytoplasmic pool of siRNA competes with the pool of siRNA to be translocated in the nucleus.
The Dcr-1 partner Loquacious (Loqs), but not the Dcr-2 partner R2D2, was unexpectedly found to be required for biogenesis of siRNA derived from fold-back genes that form dsRNA hairpins (6, 7, 15). By contrast, it is noteworthy that loqs mutations had little or no impact on the accumulation of siRNA derived from TE (6, 7). Our finding that r2d2 but not loqs mutation suppresses the silencing of PEV reporters and delocalizes H3m2K9 and H3m3K9 heterochromatic marks agrees with these results and further suggests that siRNA involved in heterochromatin formation and siRNA derived from endogenous hairpins arise from distinct r2d2- and loqs-dependent pathways, respectively. One possible mechanism by which TE- or repeat-derived endo-siRNAs could promote heterochromatin formation is by tethering complementary nascent TE transcripts and guiding Su(var)3–9 recruitment and H3K9 methylation. Identifying which enzymes tether siRNAs to chromatin in animals is a future challenge. In addition, some endo-siRNAs could also impact on heterochromatin formation by posttranscriptionaly regulating the expression of chromatin modifiers, such as Su(var)3–9. In any case, our results demonstrate the value of viral silencing suppressor proteins in linking siRNAs to heterochromatin silencing in the fly soma, as established in S. pombe and higher plants (25, 47). Because silencing suppressors are at the core of the viral counterdefensive arsenal against antiviral RNA silencing in fly (12–14, 26), whether they also induce epigenetic changes in chromatin states during natural infections by viruses deserves further investigation.
Materials and Methods
Suppressor Transgenic Constructs.
NLS-P19 DNA was obtained by fusion PCR between P19 and Transformer nuclear localization peptide sequences. B2, P19 and NLS-P19 DNAs were cloned into pPWF (for expression of FLAG tagged proteins in transgenic lines), pMT-DEST48 (for copper-inducible expression of V5 tagged proteins in S2 cells) or pAWH (for constitutive expression of HA tagged proteins in S2 cells) using the Gateway system (Invitrogen). See SI Text for detailed DNA cloning procedures.
Transgenic and Mutant Stocks.
We obtained the GMR>IR[w] transgenic line from R. Carthew, the GMR>GAL4 driver (n°1104) and UAS>GFP (n°9258) lines from the Bloomington Stock Center, the engrailed>GAL4, Tubulin>GFP and Tubulin>GFP-ban transgenic stocks from S. Cohen, the lio>GAL4 driver line (48) from J.-M. Dura, the UAS>H2b-YFP line from Y. Bellaiche (49) and the Act5C>GAL4 17a driver line (n°U192) from the Fly stocks of National Institute for Genetics from Japan.
The following mutant fly stocks were used at 25 °C: [1] w1118, [2] ln (1)wm4h (50), [3] y w eyFLP; FRT42D dcr2R416X, [4] y w eyFLP; FRT42D dcr2L811fsX, [5] y w; r2d21/CyO, [6] y w; ago2414, [7] y w; ago2dop1/TM6B Tb, [8] wm4; Su(var)3-96/TM6B Tb.
The genetic crosses were performed as described in SI Text.
Immunostaining of Polytene Chromosomes.
Primary antibodies were rabbit anti-H3m2K9 (1:20) and anti-H3m3K9 (1:150) from Upstate, mouse anti-HP1 (1:50, DSHB University of Iowa) and rabbit anti-Su(var)3.9 (1:50, (40). Late third-instar larvae raised at 22 °C were dissected in PBS. Except for HP1 labeling, salivary glands were prefixed for 20 sec in solution 2 (3.7% paraformaldehyde, 1% Triton X-100 in PBS pH 7.5), fixed for 2 min in solution 3 (3.7% paraformaldehyde, 50% acetic acid) and squashed onto a polyL-lysine coated slide. Polytene spreads were then stained according to http://www.epigenome-noe.net/researchtools/protocol.php?protid = 1 with overnight primary incubations at 4 °C. Secondary antibodies were FITC-anti-mouse, Cy3-anti-mouse, FITC-anti-rabbit, or Cy3-anti-rabbit (1:150, Jackson Immunoresearch).
For HP1 labeling, salivary glands were prefixed for 10 sec in solution 2, fixed for 90 sec in solution 3; polytene squashes were primarily incubated with anti-HP1 antibody for 2 h at room temperature and secondary antibody was 594-Alexa-anti-mouse (1:200, Invitrogen). Preparations were mounted in DAPI containing Vectashield and analyzed on Leica DM RXA epifluorescence and/or Apotome Coolsnap wide-field microscopes.
For HP1 staining, control salivary glands from a lio>GAL4/+; UAS>H2b-YFP/+ and salivary glands from lio>GAL4/+; UAS>B2/+, lio-GAL4/+; UAS>P19/+, lio-GAL4/+; UAS>NLS-P19/+ or mutant larvae were spread on the same slide and chromosome sets were genotyped owning to yellow fluorescence of YFP (see additional examples in Fig. S8). For Su(var)3–9 staining, salivary glands from a lio-GAL4/+; NLS-P19/+ female and a lio-GAL4/+; + male were spread on the same slide and chromosome sets identified by their X chromosome appearance. All images were taken with identical settings, allowing us to perform rough image analysis using ImageJ. We measured mean intensity values in three defined areas covering the pericentromere or chromosome arms. After background correction to eliminate signal coming from debris, we determined the mean signal intensity of five images per genotype from three independent assays.
Immunoprecipitations, RNA Labeling, and Western and Northern Blot Analyses.
For detailed information, see SI Text.
Small RNA Libraries.
Small RNA sequence files from staged collections of 0–1 h (GSM180330) and 12–24 h (GSM180333) embryos, pupae (GSM180336), adult heads (GSM180328) and S2 cells (GSM180337) were downloaded from GEO under accession nos. GPL5061 and GSE7448. P19 and NLS-P19 bound RNAs as well as small RNAs from S2 cells and stably transformed P19 and NLS-P19 S2 cells were cloned using the DGE-Small RNA Sample Prep Kit and the Small RNA Sample Prep v1.5 Conversion Kit from Illumina, following manufacturer instructions. Libraries were sequenced using the Illumina Genome Analyzer II and submitted to the National Center for Biotechnology Information Small Read Archive (SRA) under the accession SRP001090. Informatic analysis of sequence data are detailed in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank D. Kirschner for DNA constructs and the Plate-Forme Imagerie Dynamique for technical help; S. Ronsseray (institut Jacques Monod, Paris, France), H. Siomi (Keio University School of Medicine, Tokyo, Japan), R. Carthew (Northwestern University, Evanston, IL). P Zamore (University of Massachusetts Medical School, Worchester, MA), C. Vaury (Facult́e de Medecine, Clermont-Ferrand, France) and M. Delattre (University of Geneva, Geneva, Switzerland) for providing materials; and C. Saleh and M. Vignuzzi for critical discussions. This work was supported by fellowships from the Fondation pour la Recherche Médicale (to D.F.) and the Lebanese Centre National de la Recherche Scientifique (to B.B.) and an Agence Nationale de Recherche grant (project AKROSS) (to C.A. and O.V.).
Footnotes
This article contains supporting information online at www.pnas.org/cgi/content/full/0809208106/DCSupplemental.
References
- 1.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 6.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 9.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 10.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 13.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloc A, Martienssen R. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–517. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 18.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josse T, et al. Telomeric trans-silencing: An epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–1643. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klenov MS, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 23.Brower-Toland B, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huisinga KL, Elgin SC. Small RNA-directed heterochromatin formation in the context of development: What flies might learn from fission yeast. Biochim Biophys Acta. 2009;1789:3–16. doi: 10.1016/j.bbagrm.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargason JM, Szittya G, Burgyan J, Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 28.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nature Structural & Molecular Biology. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host & Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 35.Schubiger M, Carre C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 36.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 37.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nature Structural & Molecular Biology. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore GD, Sinclair DA, Grigliatti TA. Histone Gene Multiplicity and Position Effect Variegation in Drosophila melanogaster. Genetics. 1983;105:327–344. doi: 10.1093/genetics/105.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebert A, et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schotta G, et al. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papp I, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 43.Tartof KD, Hobbs C, Jones M. A structural basis for variegating position effects. Cell. 1984;37:869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- 44.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshpande G, Calhoun G, Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guang S, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 48.Simon AF, Boquet I, Synguelakis M, Preat T. The Drosophila putative kinase linotte (derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mech Dev. 1998;76:45–55. doi: 10.1016/s0925-4773(98)00104-x. [DOI] [PubMed] [Google Scholar]
- 49.Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol. 2001;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- 50.Reuter G, Wolff I. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet. 1981;182:516–519. doi: 10.1007/BF00293947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.