Abstract
Circadian rhythms in mammals are generated by a negative transcriptional feedback loop in which PERIOD (PER) is rate-limiting for feedback inhibition. Casein kinases Iδ and Iε (CKIδ/ε) can regulate temporal abundance/activity of PER by phosphorylation-mediated degradation and cellular localization. Despite their potentially crucial effects on PER, it has not been demonstrated in a mammalian system that these kinases play essential roles in circadian rhythm generation as does their homolog in Drosophila. To disrupt both CKIδ/ε while avoiding the embryonic lethality of CKIδ disruption in mice, we used CKIδ-deficient Per2Luc mouse embryonic fibroblasts (MEFs) and overexpressed a dominant-negative mutant CKIε (DN-CKIε) in the mutant MEFs. CKIδ-deficient MEFs exhibited a robust circadian rhythm, albeit with a longer period, suggesting that the cells possess a way to compensate for CKIδ loss. When CKIε activity was disrupted by the DN-CKIε in the mutant MEFs, circadian bioluminescence rhythms were eliminated and rhythms in endogenous PER abundance and phosphorylation were severely compromised, demonstrating that CKIδ/ε are indeed essential kinases for the clockwork. This is further supported by abolition of circadian rhythms when physical interaction between PER and CKIδ/ε was disrupted by overexpressing the CKIδ/ε binding domain of PER2 (CKBD-P2). Interestingly, CKBD-P2 overexpression led to dramatically low levels of endogenous PER, while PER-binding, kinase-inactive DN-CKIε did not, suggesting that CKIδ/ε may have a non-catalytic role in stabilizing PER. Our results show that an essential role of CKIδ/ε is conserved between Drosophila and mammals, but CKIδ/ε and DBT may have divergent non-catalytic functions in the clockwork as well.
Keywords: casein kinase I delta, casein kinase I epsilon, dominant-negative mutant, PERIOD
Circadian rhythms are prevalent among organisms, and they are regulated by endogenous molecular oscillators called circadian clocks (1, 2). In mammals, important daily activities such as sleep/wake cycles and metabolic homeostasis are governed by the endogenous circadian clock (3–5). Available data suggest that the major driving force of the molecular clock is the transcriptional negative feedback loop containing CLOCK (or its paralog, NPAS2), BMAL1, PERIOD (PER), and CRYPTOCHROME (CRY). The CLOCK (or NPAS2):BMAL1 heterodimer activates transcription of the negative elements, Per and Cry, as well as circadian output genes, through E-box enhancer elements (1, 3, 6, 7). As PER levels increase in the cytoplasm, PER associates with CRY, and the complex enters the nucleus to shut down transcription driven by CLOCK:BMAL1. Thus, temporal accumulation and degradation rates of PER predominate in determining the timing of the negative feedback loop.
PER proteins are progressively phosphorylated and disappear over a circadian day (8, 9). Numerous studies using biochemical and genetic approaches showed that CKIδ/ε can phosphorylate PER in vitro and in cultured cells (10–15). Phosphorylation of PER can affect its cellular location and stability (10, 12–14, 16). In Drosophila, genetic studies have demonstrated that DOUBLE-TIME (DBT), an ortholog of CKIδ/ε, is required for normal phosphorylation and turnover of dPER, and for behavioral circadian rhythms (17, 18). However, in mammals, the known mutations in CKIε or CKIδ, including null mutations (11, 19–21), do not substantially disrupt the molecular oscillator and circadian rhythms to the extent seen in Drosophila mutants carrying the dbtP or dbtAR allele (17, 18, 22), suggesting that the two mammalian enzymes are at least partially redundant, or there are other kinases that can compensate for the loss of CKIδ/ε. In mutant mammals carrying mutations in CKIε or CKIδ, PER still oscillates in abundance and phosphorylation. Interestingly, a CKIδ null mutation produced more severe phenotypes than did a CKIε null mutation, suggesting that they may not be equally redundant (20).
Dominant-negative approaches have been successfully used to disrupt endogenous casein kinase 2 (CK2) and DBT activities in vivo in the Drosophila clockwork (23, 24). In mammalian cells, as in Drosophila, it appears that general reduction of CKIδ/ε activities by kinase inhibitor drugs or the K38R mutant CKIε results in a slower oscillator (25–27). However, unlike in Drosophila, it has not been shown whether PER phosphorylation and circadian rhythms are completely disrupted when CKIδ/ε activities are severely compromised. Because a mouse with null mutations for both CKIδ and ε is not viable, we turned to the dominant-negative approach for testing whether CKIδ/ε are essential for clock function. However, the dominant-negative approach can be compromised by potential redundancy between CKIδ and ε in the mammalian system, since a higher dose of a dominant negative mutant may be required to stoichiometrically dominate two endogenous kinases, as compared to the single kinase in Drosophila. According to our estimation, CKIδ is twice as abundant as CKIε in vivo, at least in MEFs (see below). We therefore used the dominant-negative approach to disrupt CKIε in a CKIδ null genetic background. Our present study showed that the general scheme of PER regulation by CKIδ/ε (or DBT) may be conserved between Drosophila and mammals, but regulation of the mammalian clock is much more complicated due to partial (not complete) redundancy between PER1 and 2, and between CKIδ and ε. Furthermore, CKIδ/ε may have non-catalytic, yet essential roles in the clockwork as has been shown recently in Drosophila (28), and these roles may have evolved separately between the insect and mammalian lineages.
Results and Discussion
DN-CKIδ/ε Lengthen Circadian Period in Per2Luc MEFs.
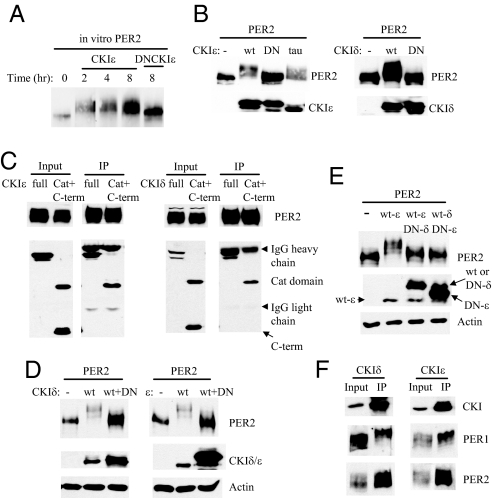
The K38R mutant form of CKIε retains the ability to bind to PER but lacks any kinase activity (14, 23, 29); it thus acts as an ideal dominant-negative mutant. We confirmed that the dominant negative CKIε (DN-CKIε) did not noticeably phosphorylate PER2 in vitro and in cultured cells, in contrast to wild-type (wt) CKIε and CKIε with the tau (gain-of-function) mutation (Fig. 1 A and B). The K38R mutant form of the homologous CKIδ also failed to phosphorylate PER2 in cultured cells (Fig. 1B). Because both DBT and CKIε bind PER through their conserved catalytic domain (10, 30) and both CKIδ and ε are 97% identical in the catalytic domain (10), we expected that CKIδ would also bind PER through its catalytic domain. Our binding assays confirmed that PER2 binds the catalytic domain (and not the C terminus) of both CKI δ and ε (Fig. 1C). Consistent with these binding assays, each DN mutant kinase could effectively inhibit both wild-type kinases in phosphorylating PER2, when the DN mutant kinases were expressed in molar excess relative to their wt counterparts (Fig. 1 D and E). Importantly, we showed in our MEFs that endogenous CKIδ/ε associate with endogenous PER1 and 2 (Fig. 1F) as has been shown in intact mice (8). Predominantly hyperphosphorylated PER1 interacted with the kinases while PER2 in various phosphorylation states associated with the kinases (Fig. 1F), as previously shown in liver (8).
Fig. 1.
DN-CKIδ or ε disrupt activities of both kinases since the kinases bind PER through their conserved catalytic domain. Blots representative of three experiments are shown. (A) In vitro kinase assay resolved by Western blot: high levels of PER2 phosphorylation cause slower mobility in SDS/PAGE. PER2 was highly phosphorylated by CKIε, but not by DN-CKIε. The reactions were stopped at the indicated times (in hours) by adding 2× sample buffer. Lane 1 represents PER2 alone. (B) Cell culture kinase assay resolved by immunoblot using anti-PER2 and anti-CKIδ/ε antibodies. In NIH 3T3 cells, the DN-CKIδ/ε do not phosphorylate PER, while wt CKIδ/ε and tau CKIε do. Both wt and DN-CKIε have a C-terminal MYC tag, while tau CKIε has no tag. Both wt- and DN-CKIδ have a FLAG tag. (C) PER2 was coexpressed with a full-length CKIδ/ε, or N-terminal (Catalytic domain; amino acid 1–277) + C-terminal (aa278–416 for CKIε and aa278–415 for CKIδ) peptides in HEK293 cells. Cell extracts were subjected to immunoprecipitation (IP) for PER2. Note that only catalytic domains were copurified with PER2. Both full-length and truncated mutant CKIδ/ε have an N-terminal FLAG tag, which was detected on the immunoblots. Note that in the rightmost panel, the full length CKIδ-FLAG comigrates with the IgG heavy chain. (D) The DN-CKIδ(ε) inhibits wt-CKIδ(ε)-dependent phosphorylation of PER2. NIH 3T3 cells were transfected with PER-V5 and pcDNA3.1, wt-CKI, or a 1:10 ratio of wt and DN-CKI. The cell lysates were subjected to immunoblotting using anti-PER2 and anti-CKIδ or ε antibodies. Both wt and DN-CKIδ have an N-terminal FLAG tag. wt- and DN-CKIε have a MYC and a FLAG tag, respectively. (E) DN-CKIδ(ε) can inhibit wt-CKIε(δ)-mediated PER2 phosphorylation. wt-CKIδ and DN-CKIε or wt-CKIε and DN-CKIδ were transfected into NIH 3T3 cells in 1:10 ratio as above. The kinases are the same as in (D). To detect both CKIδ and ε on the same blot, two antibodies were used at the same time. (F) MEF extracts were subjected to IP for CKIδ or ε, and assayed by immunoblot for PER1 and 2.
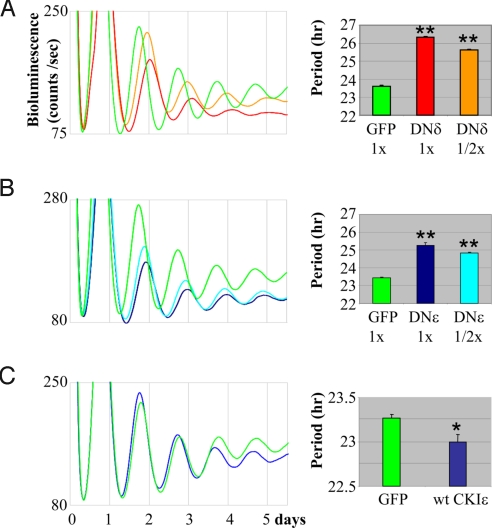
We generated adenoviral constructs to express DN-CKIδ or ε because adenovirus enables highly efficient transgene delivery into MEFs. We introduced two different titers of adenovirus expressing DN-CKIδ or ε into Per2Luc MEFs, to quantitatively measure how CKI disruption affects circadian rhythms. The adenovirus efficiently infected our MEFs (>90%) (Fig. S1A). Expression of either mutant kinase induced dramatically longer periods and reduced amplitudes of circadian bioluminescence rhythms compared to control MEFs expressing a non-relevant protein, GFP (Fig. 2 A and B). DN-CKIδ MEFs showed significantly longer periods than those of DN-CKIε MEFs (≈1 h). It is not known whether this difference is due to a difference in expression levels or non-redundant disruption of the molecular clock between two mutant kinases. Interestingly, during the course of this study, Etchegaray et al. reported that CKIδ may play a more important role than CKIε in the circadian clock, since a CKIδ null mutation can cause more severe phenotypes than a CKIε null mutation in ex vivo liver and MEFs (20). In any case, our studies demonstrated that DN-CKIδ and ε can effectively perturb circadian rhythms, consistent with previous studies showing that period is lengthened when CKIδ/ε activities are disrupted pharmacologically and genetically (20, 25–27). On the other hand, overexpression of either wt CKIε or wt CKIδ slightly shortened period by only approximately 15 min, suggesting that endogenous CKIδ/ε are near saturation levels compared to clock protein substrates such as PER (Fig. 2C and Fig. S1B). Since the adenoviral infection efficiency is not approximately 100% and uniform among MEFs, the above results could be more dramatic if single cells expressing high levels of the exogenous proteins were analyzed.
Fig. 2.
The DN-CKIδ/ε lengthened the period of bioluminescence (PER2:Luc) rhythms in MEFs. (A–C) MEFs were infected with adenovirus expressing DN-CKIδ (A), DN-CKIε (B,) or wt CKIε (C) for 2 h, serum-shocked for 2 h and placed into the real-time luminometer. The 3XFLAG tag added to the N-termini of wt and DN kinases was used to ensure similar expression among different adenoviruses. ½× represents half titer of 1×. The numbers are shown as mean ± SEM of triplicate samples. The results are representative of several experiments. *, P < 0.05; **, P < 0.01.
Consistent with still robust circadian rhythms in DN-CKIε MEFs, both PER1 and 2 were similarly rhythmic in the MEFs compared to GFP MEFs (Fig. S2A) (PER2 in DN-CKIδ MEFs is shown in Fig. S2B). However, it is apparent that phase of PER2 phosphorylation/abundance is a little delayed compared to control cells. Although bioluminescence rhythm amplitudes in DN-CKIδ/ε MEFs damped more dramatically in later cycles after serum shock, protein rhythms in these later cycles could not be compared because they were hardly detectable even in control samples. It is noteworthy that amplitudes in PER rhythms also significantly dampen in CKIδ-deficient mice (20). When endogenous CKIδ and ε levels were quantified by comparing signal intensities between endogenous kinases in MEFs and in vitro translated kinases, as has been done previously (8), CKIδ was found to be approximately 2-fold more abundant than CKIε (Fig. S2C), suggesting that the more severe phenotype in CKIδ-deficient tissue (20) and in our DN-CKIδ MEFs, relative to their CKIε counterparts, may be partly due to the stoichiometry. The levels of overexpressed DN-CKIδ/ε were at least 5- to 10-fold higher than their endogenous counterparts, more abundant than the combined levels of both endogenous kinases (Fig. S2D).
The DN-CKIε interacted with endogenous PER (Fig. S2E). It seems that the affinity of DN-CKIε for PER2 may not be as strong as that of endogenous CKIε, since the DN-CKIε/endogenous-CKIε ratio after copurification with PER2 was much lower than their ratio in straight extracts (Fig. S2E and Fig. S2F; the ratio was >10 in straight extracts vs. 2–3 in PER2-copurified samples). This was at least partly due to subcellular localization of DN-CKIε. Both endogenous PER and CKIδ/ε were predominantly nuclear (Fig. S3A), whereas DN-CKIε was widely distributed between cytoplasm and nucleus (Fig. S3B). The different subcellular localization of DN-CKIε could be due to overexpression relative to endogenous PER, since subcellular localization of CKIδ/ε is affected by PER in vivo (8, 31), or due to the lack of kinase activity. It could not be determined how much DN-CKIδ relative to endogenous CKIδ was bound to PER since DN-CKIδ could not be separated from the IgG heavy chain (see Fig. 1). Consistent with the overall rhythmic PER oscillations, PER reached its maximally phosphorylated status and progressively disappeared in the presence of DN-CKIε (Fig. S4A) after cycloheximide treatment at T24 (24 h after serum shock). Although there seemed to be a subtle difference between DN-CKIε and GFP MEFs in terms of PER phosphorylation and degradation, it was difficult to quantify the subtle difference by the immunoblotting method. To quantify how DN-CKIδ/ε affect PER2:Luc degradation, we measured the degradation rate of PER2:Luc in GFP, DN-CKIδ or ε MEFs at the peak of PER2:Luc expression after cycloheximide treatment, as has been done by Meng et al. (19). When half-lives of PER2:Luc were compared between control and DN-CKIδ/ε MEFs, they were significantly lengthened in DN-CKIδ/ε-expressing MEFs compared to control MEFs (Fig. S4 B and C). The small but significant difference in PER2:Luc half-life relative to the period lengthening is consistent with the findings in liver explants derived from CKIδ-deficient mice (20).
CKIδ-Deficient MEFs Exhibit Longer but Robust Circadian Rhythms.
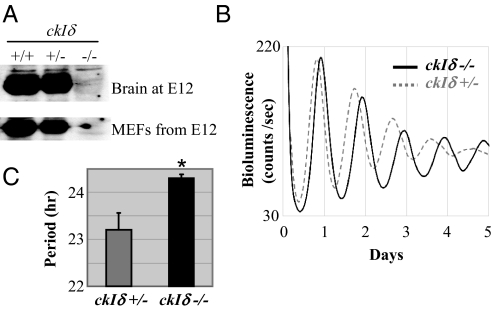
If CKIδ and ε are essential kinases for the clock, then it is likely that MEFs expressing DN-CKIε or δ are still rhythmic because of incomplete stoichiometric dominance of the exogenous kinases and redundancy between the two endogenous kinases. We tested higher titers of adenovirus hoping that expression of DN-CKIδ/ε would be proportionally increased. However, higher titers did not result in higher levels of expression, but instead caused cytotoxicity. To improve the relative stoichiometry, we decided to use our dominant negative approach in cells from CKIδ-deficient mice (21). CKIδ is more abundant than CKIε (Fig. S2C), so the elimination of CKIδ is more favorable to our aims than the elimination of CKIε. We generated CKIδ-deficient MEFs in the Per2Luc background (Fig. 3A), and thus were able to quantitatively assess circadian clock function by measuring bioluminescence rhythms. As expected based on our hypothesis of CKI redundancy, the mutant MEFs exhibited robust rhythmicity, but approximately 1 h longer period than wt MEFs or heterozygote MEFs derived from littermate embryos (Fig. 3 B and C). During the course of this study, Etchegaray et al. tested a different CKIδ mutant mouse and reported similar findings: liver explants and primary MEFs derived from the mutant mouse exhibit approximately 2 and 1.5 h longer periods, respectively, compared to wt controls (20). These genetic studies strongly suggest that CKIδ and ε are indeed redundant in PER phosphorylation and circadian rhythm generation (19, 20).
Fig. 3.
Circadian period is lengthened in ckΙδ−/− MEFs. (A) Brain extracts at embryonic day 12 and MEF extracts from the embryos were immunoblotted for CKIδ. (B) Bioluminescence (PER2:Luc) rhythms in MEFs isolated from homozygote and heterozygote ckΙδ mutant littermate embryos. (C) Periods are shown as mean ± SEM of triplicate samples. These data are representative of at least three experiments. *, P < 0.05
Overexpression of DN-CKIε Disrupts Circadian Rhythms and Compromises Oscillations in PER Phosphorylation and Abundance in the CKIδ-Deficient MEFs.
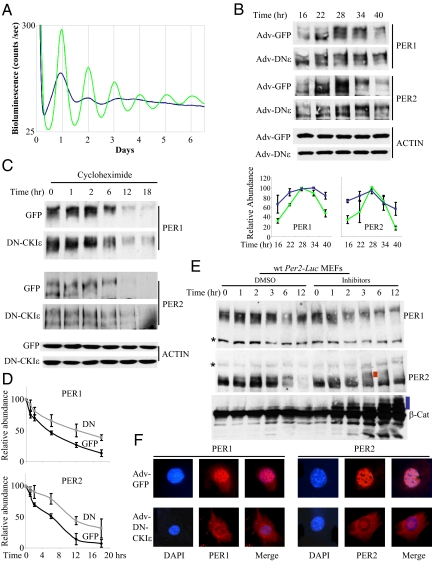
When CKIδ-deficient MEFs were infected with the same titers of GFP and DN-CKIε adenovirus as above (Fig. 2), Adv-DN-CKIε MEFs exhibited almost complete disruption of bioluminescence rhythms, while GFP MEFs showed a normal circadian rhythm (Fig. 4A and Fig. S4D). Consistent with the redundancy between CKIδ and ε in vitro, overexpression of DN-CKIδ also severely disrupted the bioluminescence rhythms (Fig. S5A). However, these results cannot rule out the possibility that the two kinases have distinct roles/substrates in the clockwork.
Fig. 4.
Overexpression of DN-CKIε completely disrupts circadian rhythms and compromises the molecular clock in ckΙδ−/− MEFs. (A) Bioluminescence rhythms were measured in GFP (green) and DN-CKIε (blue) MEFs as above. Rhythms could not be detected after a very weak second peak in DN-CKIε cells when analyzed using the Clocklab software. The results are representative of three experiments. (B) PER1 and 2 rhythms were measured in GFP and DN-CKIε MEFs as above. Bottom graphs are quantification of PER abundance indicated as mean ± SEM of three experiments. (C) CHX was added to MEFs and the cells were harvested as in Fig. S4A. (D) The results from (C) were quantified as above by densitometric scanning. The numbers are shown as mean ± SEM of three experiments. (E) wt MEFs were treated with DMSO or protease inhibitors, MG132+PSI, at T24 and harvested at indicated times. * indicates non-specific bands, the red bar indicates extra slow-migrating PER2, and the blue bar indicates multiubiquitinated β-Catenin. Note that the extra slow-migrating PER2 isoforms are not observed in control MEFs. Data are representative of at least three experiments. (F) PER1 and 2 were stained in GFP and DN-CKIε ckIδ−/− MEFs fixed at T24 after serum shock. Representative cells are shown from three independent experiments. PERs were predominantly (>90%) nuclear in Adv-GFP, CKIδ-deficient cells whereas they were observed in both compartments in most (>90%) Adv-DN-CKIε, CKIδ-deficient cells; more cells are shown in Fig. S6.
Although there is an initial peak of the bioluminescence (PER2:Luc) after serum shock in Adv-DN-CKIε and Adv-DN-CKIδ MEFs, its shape was not normal, and the rhythm rapidly degrades. Consistent with these data, PER1 and 2 rhythms in abundance and phosphorylation were also severely disrupted (Fig. 4B). We believe that weakly rhythmic PER2 phosphorylation and presence of hyperphosphorylated PER may be due to still incomplete stoichiometric dominance of DN-CKIε over the endogenous counterpart in CKIδ-deficient MEFs. Another possibility for this incomplete inhibition of PER phosphorylation would be that other kinases such as CK2 or GSK3β may compensate for the loss of CKIδ/ε activities, as these kinases have also been implicated in the phosphorylation of PER (32–34). Nevertheless, as in the arrhythmic Drosophila mutant dbtAR showing both hypo- and hyperphosphorylated dPER (22), circadian rhythms were severely compromised. To measure if PER phosphorylation is indeed delayed in this condition, Adv-DN-CKIε MEFs were treated with cycloheximide and phosphorylation/degradation rates were measured (Fig. 4 C and D). The maximal phosphorylated forms were reached for PER1 between 6 and 12 h after the treatment in control CKIδ-deficient cells, as in wt MEFs. However, in Adv-DN-CKIε cells, there were still some hypophosphorylated isoforms even 18 h after the treatment (see Fig. S5B for side-by-side comparison). More interestingly, PER2 in these cells remained in hypo- and hyperphosphorylated groups and their levels decreased progressively, unlike PER2 in control cells, which reaches the maximally phosphorylated state between 2 and 6 h after the treatment (see Fig. S5B for side-by-side comparison). There was also a significant delay in PER degradation in Adv-DN-CKIε, CKIδ-deficient MEFs when measured by the immunoblotting method (Fig. 4D). Since it has been suggested that PER phosphorylated by CKIδ/ε can be targeted for proteasome-mediated degradation (25, 35), we hypothesized that primarily hyperphosphorylated PER1 is targeted for degradation while both hypo- and hyperphosphorylated forms of PER2 are targeted for degradation. This is consistent with our coimmunoprecipitation data showing that predominantly hyperphosphorylated forms of PER1 interact with CKIδ/ε, but both hyper- and hypophosphorylated PER2 associates with the kinases (Fig. 1F). When proteasome inhibitors were added to wt MEFs, predominantly slow-migrating (presumably hyperphosphorylated) PER1 accumulated, consistent with our hypothesis (Fig. 4E). It is not known why de novo synthesized hypophosphorylated PER1 failed to accumulate in the proteasome-treated cells. For PER2, extra slow-migrating PER2 (indicated by the red bar in Fig. 4E), which cannot be seen in control cells, was observed between 3 and 6 h after the cells were treated with proteasome inhibitors, suggesting that hyperphosphorylated PER2 species may be substrates for the proteasome. Interestingly, from 3 h since the cells were treated with proteasome inhibitors, hypophosphorylated PER2 accumulated and these species did not become hyperphosphorylated even 12 h after the treatment, suggesting that hypophosphorylated PER2 may be also substrate of the proteasome and inhibition of the proteasome pathway somehow disrupts normal phosphorylation of PER2. Unlike β-Catenin, which accumulated dramatically with discrete multiubiquitinylated species (indicated by the blue bar in Fig. 4E) (36–38), PER1 and 2 neither accumulated dramatically nor exhibited the distinctive pattern of multiubiquitinylation. Thus, if PER is degraded by the ubiquitin-proteasome system, then in our conditions, multiubiquitinylated species could not be resolved perhaps due to heterogeneous phosphorylation and high molecular weight of PER. Alternatively or in addition, PER degradation may occur through non-proteasomal pathways. Our results are different from studies using transiently expressed PER and showing dramatic increase of PER levels or readily visible multiubiquitinated PER after proteasome treatment (14, 25). It has been well-known that stability of endogenous clock proteins including PER is regulated by physical interaction with other clock proteins (3, 8, 39, 40). Thus, PER that is stoichiometrically overexpressed relative to endogenous clock proteins may not follow normal physiological degradation pathways.
We also measured how disruption of CKIδ/ε-mediated PER phosphorylation affects subcellular distribution of PER in these MEFs (Fig. 4F and Fig. S6). Both PER1 and 2 were predominantly nuclear when their levels were high in control CKIδ −/− MEFs. However, both proteins were localized between cytoplasm and nucleus when CKIε activity was disrupted by DN-CKIε in CKIδ-deficient MEFs, suggesting that PER phosphorylation by CKIδ/ε is required for normal cellular localization.
Disruption of PER:CKIδ/ε Interaction Destabilizes PER and Compromises Circadian Rhythms.
Our data so far strongly suggest that regulation of PER by CKIδ/ε is an essential feature of the circadian clock by showing that rhythms of PER phosphorylation/abundance and subcellular localization are disrupted and bioluminescence rhythms are almost completely compromised by DN-CKIε combined with CKIδ deficiency. However, these phenotypes could have resulted indirectly from disruption of some unknown clock protein(s), for example, through interaction of DN-CKIε with the protein(s). To address this issue, we sought to disrupt the specific interaction between PER and CKIδ/ε and evaluate the effect on clock function.
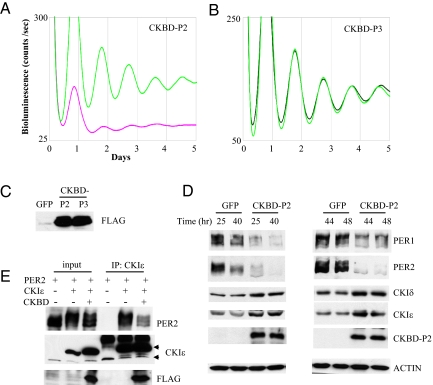
It is plausible that PER phosphorylation by CKIδ/ε can be effectively and specifically disrupted in vivo by overexpressing the previously characterized CKIδ/ε-binding domain (CKBD) of PER (10, 14, 41). PER1 and 2—but not PER3—bind the kinases. However, when CKBD is swapped between PER2 and 3, then the chimeric PER2 no longer binds CKIε while chimeric PER3 can bind CKIε (41). We tested if the clock can be disrupted when PER interaction with CKIδ/ε is specifically disrupted by overexpression of CKBD from PER2 in MEFs. As shown in Fig. 5A, PER2:Luc rhythms were severely disrupted when CKBD-P2 was overexpressed, but circadian rhythms were not affected when the corresponding domain from PER3 was overexpressed (Fig. 5B). Expression levels in MEFs were comparable between CKBD-P2 and P3 (Fig. 5C). Furthermore, basal levels of the bioluminescence rhythms were much lower in CKBD-P2-MEFs compared to GFP- or CKBD-P3-MEFs, suggesting that PER2:Luc levels are constitutively low in CKBD-P2 MEFs. Indeed, immunoblot data confirmed that PER1 and PER2:Luc levels are unusually low in CKBD-P2 MEFs (Fig. 5D and Fig. S7A). On the other hand, levels of CKIδ/ε were increased in these cells. Similarly, overexpression of PER2 increases CKIδ/ε levels, while Per1 and Per2 deficiency results in lower levels of CKIδ/ε (Fig. S7 B and C). Thus, it seems that the PER1/2 CKBD increases the stability of CKIδ/ε by direct binding. We speculate that the low levels of PER are due to 1) cytoplasmic retention and premature degradation of PER, resulting from inhibition of CKIδ/ε-induced phosphorylation and 2) reduction in the physical protection or stabilization offered by CKIδ/ε as the physical interaction between PER and CKIδ/ε is disrupted. It has been suggested that proteasomal degradation of PER primarily occurs in the cytoplasm (19, 26, 33), and physical interaction between PER and CRY stabilizes PER (8, 40). We confirmed by IP assays that CKBD-P2 can effectively disrupt the interaction between PER2 and CKIε in MEFs (Fig. 5E). The binding assays were performed using transiently overexpressed PER2, CKIε and CKBD-P2 in MEFs to compensate for the low levels of endogenous PER2 in the presence of CKBD-P2. Per1, Per2, and dbp mRNA levels were comparable between CKBD-P2 and GFP MEFs, confirming that the low levels of PER are mainly due to posttranscriptional regulation of PER by CKBD (Fig. S7D). We argue that the rhythm phenotype caused by CKBD-P2 is due to specific disruption of the physical interaction between PER and CKIδ/ε, not due to a non-specific effect of CKBD-P2, because similarly overexpressed CKBD-P3 did not have any effect on circadian rhythms.
Fig. 5.
Disruption of PER interaction with CKIδ/ε by CKBD-P2 abolishes circadian rhythms and destabilizes PER in MEFs. (A and B) Casein kinase binding domain from PER2 (CKBD-P2) (A) and the corresponding domain from PER3 (CKBD-P3) (B) were overexpressed in wt MEFs and bioluminescence rhythms were measured as above. The FLAG tag added to the N-termini of both CKBDs was used to ensure similar expression levels. Note that the basal line for CKBD-P2 is extremely low. This was consistently observed in several experiments. (C) MEFs from (A) and (B) were immunoblotted for FLAG. (D) GFP and CKBD-P2 MEFs at indicated times were harvested and the extracts were immunoblotted. Note that endogenous CKIδ/ε levels were increased in CKBD-P2 MEFs. A dark exposure for PER2 blot in the left panels is shown in Fig. S7A to demonstrate that low levels in CKBD-P2 MEFs are not due to smearing of PER2 band. Blots are representative of three experiments. (E) PER2, CKIε and/or CKBD-P2 were overexpressed in wt MEFs, the cells were harvested 24 h after the infection and the extracts were subjected to IP for CKIε. The resulting immunocomplexes were immunoblotted for PER2, CKIε, and FLAG. Top and bottom arrow indicate exogenous and endogenous CKIε, respectively. Blots are representative of four experiments.
Since circadian rhythms were completely disrupted by two different approaches targeting the kinase activities and specific interaction between the kinases and the substrate (PER), we argue that CKIδ/ε are essential for rhythm generation as is the case in Drosophila. However, the CKBD approach revealed an unexpected role of CKIδ/ε, in stabilization of PER. This role apparently does not require normal kinase activity since PER levels are not significantly changed in DN-CKIδ/ε-expressing MEFs, where kinase activity is greatly reduced but physical interaction is still intact. In Drosophila, dPER is more stable in the dbtP mutant flies where dbt expression is greatly reduced (18), suggesting that regulation of PER stability through physical interaction with CKIδ/ε is not conserved in Drosophila. However, recent studies showed that DBT also has a non-catalytic role in the clockwork. DBT acts as a scaffolding protein to recruit an inhibitor(s) into the dPER inhibitor complex to phosphorylate and inactivate the dCLOCK transcription factor (28). Thus, mammalian CKIδ/ε and DBT may have separately evolved essential roles beyond their catalytic activity in the generation of circadian rhythms.
Experimental Procedures
Animals, Cells, and Antibodies.
All animals were maintained and used according to the FSU Animal Care and Use Committee's guidelines. The Per1/2 mutant and matching mice were described in ref. 42. The ckIδ mutant mouse was kindly provided by Dr. Louis Ptáček and Dr. Ying-Hui Fu (UCSF). Wild-type MEFs used in Figs. 2, 4, 5, and Figs. S2 and S4 were isolated from homozygous Per2Luc mice (43) and immortalized by retroviral transduction of a dominant-negative mutant p53 (GSE56) (44). ckIδ hetero and homozygote mutant MEFs in Figs. 3 and 4 were isolated from littermate embryos at E12 and immortalized as above. Antibodies to clock proteins (PER1–1-R, PER2–1-R, CKIε-GP, and CKIδ-GP) were described in refs. 8 and 41.
In Vitro Translation (IVT) and Immunocytochemistry.
IVT was performed using TnT rabbit recticulocyte extract (Promega) in the presence of L-35S methionine to enable quantification of the labeled product (8). In vitro translated proteins were quantified according to the manufacturer's protocol. Immunocytochemistry in MEFs was done as described in ref. 45. MEFs were fixed for immunocytochemistry 24 h after viral infection and serum shock. Anti-FLAG antibody was used to detect DN-CKIε.
In Vitro Kinase Assay.
The in vitro kinase assay in Fig. 1A was performed as described in ref. 41. Briefly, PER2, wtCKIε and DN-CKIε were synthesized in vitro as above and mixed with 1:3 molar ratio of PER2 to wt or DN-CKIε as indicated in Fig. 1A.
See the SI Text for additional experimental procedures.
Supplementary Material
Acknowledgments.
We thank Jiangqin Lai for excellent technical assistance during the project. We thank Dennis Chang for assistance with manuscript revisions. This work was supported by National Institutes of Health Grant NS-053616 (to C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906651106/DCSupplemental.
References
- 1.Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005;6:S9–13. doi: 10.1038/sj.embor.7400424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 4.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 5.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuninkova L, Brown SA. Peripheral circadian oscillators: Interesting mechanisms and powerful tools. Ann N Y Acad Sci. 2008;1129:358–370. doi: 10.1196/annals.1417.005. [DOI] [PubMed] [Google Scholar]
- 7.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 9.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 10.Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keesler GA, et al. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport. 2000;11:951–955. doi: 10.1097/00001756-200004070-00011. [DOI] [PubMed] [Google Scholar]
- 13.Camacho F, et al. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- 14.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 16.Blau J. PERspective on PER phosphorylation. Genes Dev. 2008;22:1737–1740. doi: 10.1101/gad.1696408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloss B, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 18.Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 19.Meng QJ, et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etchegaray JP, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 22.Rothenfluh A, Abodeely M, Young MW. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr Biol. 2000;10:1399–1402. doi: 10.1016/s0960-9822(00)00786-7. [DOI] [PubMed] [Google Scholar]
- 23.Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith EM, Lin JM, Meissner RA, Allada R. Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS Genet. 2008;4:e12. doi: 10.1371/journal.pgen.0040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanselow K, et al. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton KM, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Thera. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 28.Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 30.Preuss F, et al. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- 32.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 33.Maier B, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchiya Y, et al. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 35.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 36.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitagawa M, et al. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 39.Kondratov RV, et al. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagita K, et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 43.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ossovskaya VS, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.