Abstract
In mammals, the circadian oscillator generates approximately 24-h rhythms in feeding behavior, even under constant environmental conditions. Livers of mice held under constant darkness exhibit circadian rhythm in abundance in up to 15% of expressed transcripts. Therefore, oscillations in hepatic transcripts could be driven by rhythmic food intake or sustained by the hepatic circadian oscillator, or a combination of both. To address this question, we used distinct feeding and fasting paradigms on wild-type (WT) and circadian clock-deficient mice. We monitored temporal patterns of feeding and hepatic transcription. Both food availability and the temporal pattern of feeding determined the repertoire, phase, and amplitude of the circadian transcriptome in WT liver. In the absence of feeding, only a small subset of transcripts continued to express circadian patterns. Conversely, temporally restricted feeding restored rhythmic transcription of hundreds of genes in oscillator-deficient mouse liver. Our findings show that both temporal pattern of food intake and the circadian clock drive rhythmic transcription, thereby highlighting temporal regulation of hepatic transcription as an emergent property of the circadian system.
Keywords: CREB, metabolism, mouse liver, circadian rhythms
The circadian system is an evolutionarily conserved mechanism that helps organisms anticipate predictable environmental changes by generating near-24-h rhythms in behavior and physiology and synchronizing these to the solar day. The circadian oscillator in mammals is based on an interlocked feedback loop. Transcription factor BMAL1 heterodimerizes with either CLOCK or NPAS2 to drive the expression of Cryptochrome (Cry1 and -2), and Period (Per 1, -2, and -3) genes. In turn, CRY and PER proteins inhibit CLOCK/NPAS2:BMAL1 transcriptional activity, creating approximately 24-h oscillations in mRNA and protein levels of Per, Cry (1, 2), and various output genes. The cell autonomous molecular clock sustains these oscillations in the absence of rhythmic environmental cues, such as light:dark or temperature cycles.
The hypothalamic suprachiasmatic nucleus (SCN) functions as the master circadian pacemaker, determining the overall diurnal/nocturnal activity preference of the animal. While the SCN oscillator is sensitive to light signals, it is hardly perturbed by feeding pattern. On the other hand, the oscillator in peripheral organs, as in the liver, is largely insensitive to acute changes in light regime, but its phase and amplitude are influenced by feeding activity (3, 4). The liver oscillator is thought to help the organism adapt to a daily pattern of food availability by temporally tuning expression of a large number of genes regulating metabolism and physiology. Such temporal regulation of metabolism is important, since the absence of a robust circadian clock predisposes the organism to various metabolic dysfunction and diseases (5–7).
To understand the molecular mechanisms of rhythmic gene expression sustained in the absence of environmental cues, specifically light:dark input, several studies have examined temporal gene expression patterns in ad lib fed mice held under constant darkness (DD). These studies revealed that up to 15% of the expressed transcripts in the liver exhibit circadian rhythms (8). These transcriptional rhythms were mainly attributed to the circadian clock. However, rhythmic feeding is sustained under DD. Interestingly, only a small number of rhythmic transcripts in the liver are direct targets of oscillator components (9–11). Additionally, in liver-specific clock-deficient mice, a subset of hepatic circadian transcripts was shown to oscillate in the presence of an intact SCN clock (12), which drives rhythmic feeding behavior (7). Therefore, it is unclear whether the circadian rhythms of the hepatic transcripts are under control of the liver oscillator, driven by a rhythmic feeding pattern, or a combination of both.
To systematically dissect the role that food, feeding pattern, and the circadian oscillator play in determining rhythmic gene expression, we assessed genome-wide transcriptional changes in response to fasting and refeeding in wild-type (WT) C57B6 mice. Circadian gene expression profiles of these mice were examined under three different conditions: ad libitum (ad lib), daytime-restricted feeding (tRF), and prolonged fasting. We found that food intake and time of feeding had profound effect on rhythmic gene expression. Temporal consolidation of feeding could restore approximately 24-h rhythms in gene expression in hundreds of transcripts in oscillator-deficient cry1−/−;cry2−/− mice. Our results indicate that rhythmic gene expression in mammalian liver results from synergistic interaction between the circadian clock and feeding pattern.
Results
Temporally Consolidated Feeding Generates Rhythm in Respiratory Exchange Ratio Independent of the Circadian Clock.
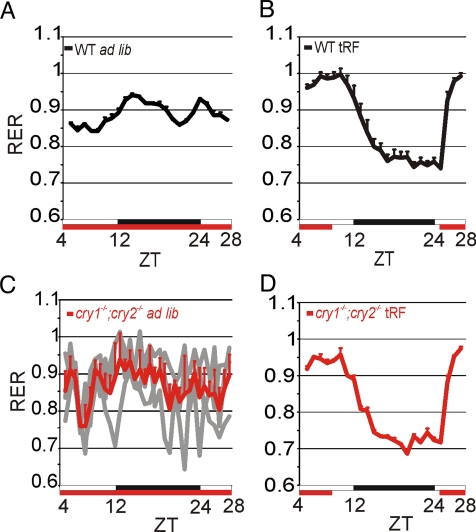
Under steady-state conditions, the feeding behavior of mice fed with a standard lab diet is reflected in the respiratory exchange ratio (RER), which exhibits daily oscillations. In WT mice fed ad lib, the RER show a bimodal rhythm with a major peak around Zeitgeber time (ZT)12 and a minor peak around ZT24, mirroring feeding behavior (Fig. 1A and Fig. S1) (13). If food availability is temporally restricted (tRF) to 8 h during the daytime, variations of the RER are consolidated and shifted to a single peak in the daytime (Fig. 1B).
Fig. 1.
Restricted feeding restores feeding rhythms in cry1−/−;cry2−/− mice. The daily variation of the RER in mice is shown as an indicator of food intake. (A–D) Average (+SEM, n = 8 for WT, n = 4 for cry1−/−;cry2−/− mice under tRF, and n = 3 under ad lib conditions). (C) In addition to the average (red), the RER of the single mice is shown (gray). x-axis, white and black bars indicate day and night, respectively; red bars indicate food availability. ZT0 coincides with the onset of light, while ZT12 coincides with the onset of darkness.
Next, we tested the RER of Cry deficient (cry1−/−;cry2−/−) mice. As expected, these mice, lacking a functional oscillator, showed no approximately 24-h rhythm in activity or RER when food was available ad lib (Fig. 1C and Fig. S1C). However, under tRF conditions, these mice showed food anticipatory activity and a robust rhythm in RER indistinguishable from those of the WT mice (Fig. 1D and Fig. S1C). Therefore, despite the lack of a functional oscillator, these mice can adapt to schedule feeding. As such, cry1−/−;cry2−/− mice under tRF conditions can serve as a model to test whether daily feeding rhythms can drive approximately 24-h rhythms in transcription.
Daily Rhythms in Metabolic Regulators Are Primarily Driven by Temporal Feeding Pattern.
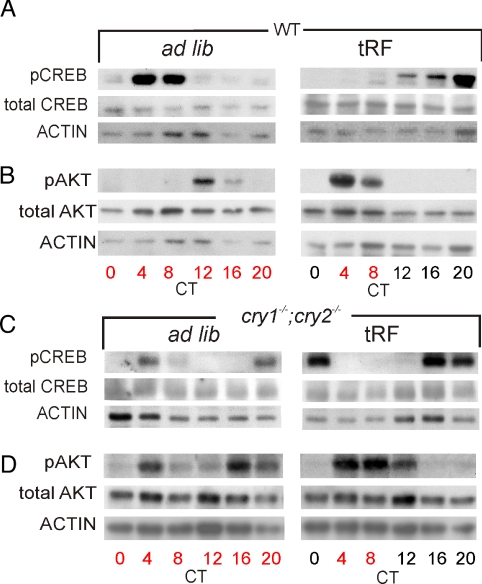
Subsequently, we assessed whether the overt rhythm in food consumption accompanies temporal changes in the activity of major metabolic regulators of the liver. We focused on CREB and AKT. CREB is phosphorylated and activated by extended periods of fasting, while AKT is phosphorylated upon insulin receptor activation. Both in turn regulate cellular energy metabolism. We found that levels of the transcriptionally active phospho-Serine 133-CREB (pCREB) (14) and phospho-Serine 473-AKT (pAKT) (15) levels oscillated in all conditions except in cry1−/−;cry2−/− mice subjected to ad lib conditions, where feeding rhythms were absent (Fig. 2). While pCREB peak levels coincided with periods of fasting, pAKT levels peaked after the onset of feeding (Fig. 2 A and B). Although the rhythms in pAKT and pCREB in both WT and cry1−/−;cry2−/− mice under tRF were comparable, the peak levels in cry1−/−;cry2−/− mice were delayed. Thus, these rhythms can be driven by feeding schedule in the absence of a clock, while the circadian oscillator fine-tunes their phases.
Fig. 2.
Daily rhythms in AKT and CREB phosphorylation are driven by temporal feeding pattern. Total and phosphorylated protein levels of CREB (A and C) and AKT (B and D) in livers of WT (A and B), and cry1−/−;cry2−/− mice (C and D) under ad lib and tRF feeding conditions. Food availability is indicated in red.
Targets of Metabolism-Driven Transcription Factors Show Clock-Independent Rhythmic Transcription.
We used high-density arrays (HDAs) to measure gene expression changes in the livers of WT and cry1−/−;cry2−/− mice under ad lib, tRF, fasting, and refeeding conditions (Fig. S2) to test whether metabolic and stress responsive regulators cause rhythmic transcription under physiological feeding conditions. We focused on transcriptional changes of genes known to be regulated by CREB, AKT (FoxO1-mediated), SREBP1/2 (feeding-activated), and ATF6 (a mediator of cellular stress).
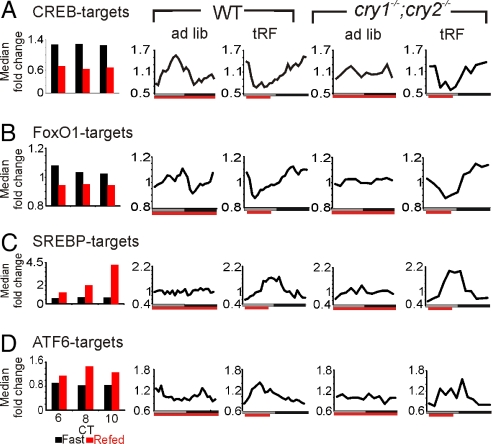
The mRNA levels of CREB and FoxO1 targets showed similar expression patterns under different feeding conditions and genotypes. Fifty targets of CREB (16) and 30 targets of FoxO1 (17) were repressed by food intake following prolonged fasting. In WT mice under both ad lib and tRF conditions, transcript levels oscillated, peaking during the respective periods of fasting (Fig. 3 A and B).
Fig. 3.
Circadian and ultradian transcription is driven by feeding state controlled transcription factors CREB, FoxO1, SREBP-1/2, and ATF6. Median temporal expression pattern of transcriptional targets of CREB (A), FoxO1 (B), the SREBPs (C), and ATF6 (D) in the liver of WT and cry1−/−;cry2−/−.
Feeding after 24 h fasting induced the expression of 31 ATF6 targets and 32 SREBP1/2 targets (18) with distinct kinetics. ATF6 targets were rapidly induced to peak levels 4 h after feeding onset and declined to baseline levels quickly afterward. Such rapid response kinetics of ATF6 targets resulted in weak 12-h transcriptional rhythms under ad lib conditions in WT mice reflecting their bimodal feeding pattern, and they assumed 24-h periodicity under tRF conditions. In contrast to ATF6 targets, SREBP targets displayed a slow induction to about 4.5-fold above fasting levels after 6 h of food availability. Such slow response to feeding and the lack of robust feeding consolidation under ad lib feeding blunts transcriptional rhythms in SREBP targets to almost undetectable levels. Under tRF conditions, however, SREBP targets showed rhythmic expression patterns with peak phases attained at 6–8 h after feeding onset (Fig. 3 C and D).
In cry1−/−;cry2−/− mice under ad lib conditions, the targets of the above described transcription factors were arrhythmic. Under tRF conditions, these transcripts showed clear 24-h expression rhythms similar to expression in WT mice (Fig. 3). However, the peak phases of transcript expression occur slightly earlier in WT mice than in their cry1−/−;cry2−/− mice counterparts, suggesting that the oscillator may potentiate transcriptional responses of these metabolic regulators to feeding and fasting.
The observation that four different classes of metabolic/stress responsive transcription factors drive liver-clock-independent rhythmic transcription raised the question, whether all food-responsive transcripts show clock-independent rhythmic expression reflecting feeding behavior.
Feeding-Responsive Transcripts Show Ultradian and Circadian Transcription Following the Pattern of Food Intake.
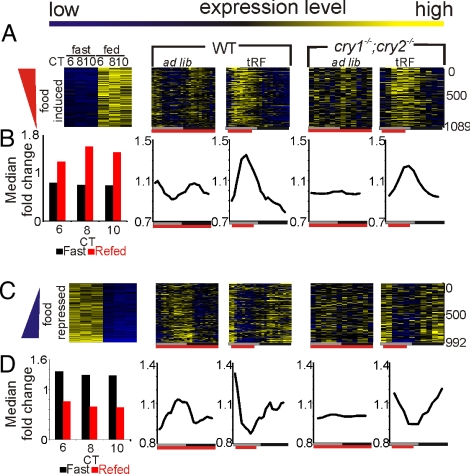
To identify transcripts that are acutely affected by feeding and to minimize any confounding effect of light and the circadian clock on the fasting/feeding modulated transcription, we subjected WT mice held under DD to a full circadian cycle (24 h) of fasting starting from CT4 before refeeding. This length of fasting has moderate effect on hepatic gene expression (19) and does not arrest the intrinsic clock (3). Since transcriptional changes in response to refeeding can have different kinetics (20), we monitored expression every 2 h for three time points after feeding. As previously defined (15), we set a 1.6-fold change threshold criteria and identified 1,089 food-induced and 992 food-repressed probe sets. Heat map rendering of the 24-h expression profile of these probe sets in WT or cry1−/−;cry2−/− mice showed a clear rhythmic expression pattern reflecting feeding behavior (Fig. 4). The rhythmic transcription of feeding-modulated genes is well pronounced in the median of their normalized expression profiles (Fig. 4 B and D). In WT mice under ad lib feeding conditions, many food-induced probe sets showed synchronous, low-amplitude rhythms with elevated levels during the nighttime and two peaks, coinciding with the two nocturnal feeding bouts (Fig. 4 A and B). Under tRF conditions, as feeding was consolidated to one bout, feeding regulated transcripts displayed distinct 24-h rhythms in WT and cry1−/−;cry2−/− mice, with peak levels of food-induced transcripts following food intake and food-repressed transcripts preceding food intake (Fig. 4).
Fig. 4.
Rhythmic food intake drives rhythmic gene expression. Heatmap rendering (A and C) and median expression changes (B and D) of food-induced and food-repressed transcripts in the indicated genotypes and feeding conditions. (A and B) Columns indicate time points, and rows indicate single transcripts. High levels are shown in yellow, whereas low levels are shown in blue. Data for WT are at 1-h resolution, while data for cry1−/−;cry2−/− mice are at 2-h resolution. Median expression values were smoothed by a 3-h moving average.
Global Transcriptional Rhythms in tRF fed cry1−/−;cry2−/− Mice Can Be Partly Restored by Food Intake.
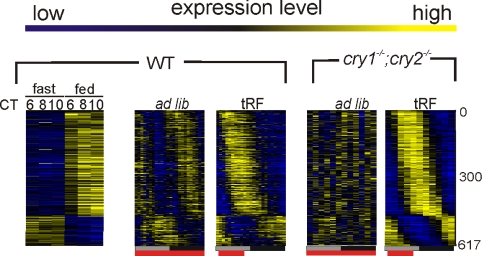
The heatmap rendering of feeding-modulated probe sets showed that feeding consolidation can drive rhythmic gene expression in cry1−/−;cry2−/− mice into two principal phases coinciding with time of feeding and fasting (Fig. 4). This prompted us to assess the extent of global rhythms in gene expression driven by an imposed feeding rhythm in the oscillator deficient cry1−/−;cry2−/− mice. Hepatic transcription profiles of cry1−/−;cry2−/− mice under ad lib or tRF feeding conditions were analyzed to find transcripts with robust approximately 24-h periodicity (21). As shown earlier (10), no significant rhythm in gene expression was found in ad lib fed cry1−/−;cry2−/− mice (Table S1). However, feeding consolidation restored rhythmicity for 617 probe sets. These probe sets were clearly split into two groups, one consisting of food-induced transcripts and the other consisting of food-repressed transcripts (Fig. 5). The probe sets also showed 24-h rhythms in WT mice under tRF conditions with comparable, but not identical phases of expression (Fig. 5). Additionally, we found that nearly all transcripts previously shown to be rhythmic in the liver of liver-specific Bmal1 knockdown mice (12) gained rhythmic expression in cry1−/−;cry2−/− mice liver under tRF (Fig. S3).
Fig. 5.
Temporally restricted feeding can partially restore approximately 24-h rhythm in gene expression in the livers of cry1−/−;cry2−/− mice. Heatmap rendering of temporal gene expression pattern of 617 probe sets scored rhythmic in cry1−/−;cry2−/− mouse liver under tRF.
Plasticity of Circadian Transcriptome Under Different Feeding Conditions.
The effect of feeding and fasting on temporal gene expression suggested that the repertoire, phase, and amplitude of rhythmic transcripts in the WT liver are products of both feeding pattern and the intrinsic circadian clock.
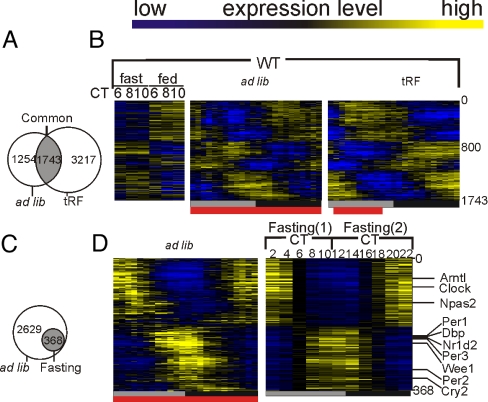
Feeding consolidation under tRF condition improved rhythms in a set of 3,217 probe sets, while dampening the rhythms of a set of 1,254 (Fig. 6A and Fig. S4 A and B). Many oscillator components continued rhythmic expression under both conditions.
Fig. 6.
Feeding schedule affects rhythmic transcription. (A) Transcripts that were scored as rhythmic in WT mice under both ad lib and tRF or fasting condition are shown in a Venn-Diagram. (B) Heatmap rendering of temporal gene expression pattern of the 1,743 “common cycler” transcripts is shown. (C) Venn-Diagram depicting only 368 of 2,997 transcripts rhythmic in ad lib-fed WT mice maintained a robust transcriptional rhythm under fasting conditions. (D) Heatmap rendering of these 368 transcripts, including most core clock components and known clock-controlled genes under ad lib and fasting conditions.
Also, nearly 28% of transcripts rhythmic in both conditions (common cyclers) were induced or repressed by a factor of 1.6-fold upon refeeding. Heatmap rendering of their expression patterns (Fig. 6B) revealed effect of feeding on the expression of many of the remaining common cyclers. Accordingly, feeding pattern also influenced the phase and amplitude of oscillation of most of the common cyclers. When feeding was changed from the predominantly nocturnal ad lib to daytime tRF conditions, the vast majority (84%) of common cyclers showed >6-h shift in their phase of expression (Fig. 6A and Fig. S5). Furthermore, >73% of the common cyclers, including the core oscillator components and their immediate targets, showed an increase in amplitude of oscillation (Fig. S5 A–D).
Most of the “common cyclers” group of transcripts were not scored rhythmic by the applied filters in cry1−/−;cry2−/− mice under tRF conditions. However, many showed a low amplitude rhythmic transcription pattern with phases similar to those in the WT mice (Fig. S3B). Therefore, feeding consolidation may generate low amplitude rhythms, however a functional hepatic clock may fine tune their phases (Fig. S6) and make them more robust (example in Fig. S7). Interestingly, we identified only a small group of nine transcripts displaying similar expression patterns, which did not change their phase of peak-expression between the two feeding conditions (Fig. S5 F and G). We hypothesized that these transcripts might be driven by systemic cues originating in the SCN, which is unperturbed by feeding pattern (3, 4).
Food Driven Transcripts Fail to Sustain Their Rhythms in the Absence of Food Intake.
The circadian oscillations of the core clock mechanism are sustained during fasting of up to 48 h (3, 22). Output transcripts whose expression is dominantly driven by the hepatic circadian clock should also sustain their oscillation under fasting conditions.
In separate experiments, we subjected WT mice to approximately 24-h fasting periods starting from CT4 or CT14 and performed gene expression profiling at 2-h intervals for up to 10 h starting at CT2 or CT12, respectively (see Fig. S2 for experimental outline). Covering only 22 h, the sampling protocol may slightly under-report the number of rhythmic transcripts.
The analyses revealed that >80% of transcripts rhythmic under ad lib condition cease to oscillate when food is withheld, while only 368 of the 2,997 rhythmic transcripts including the core oscillator components continue self-sustained rhythms in the absence of feeding (Fig. 6 C and D). This suggested that the perturbed metabolic state under prolonged fasting does not stop the circadian oscillator, but blunts rhythmic expression of majority of hepatic transcripts. Hence, food intake is a major cause of rhythmic transcription even in the presence of a functional clock.
Per1 and Per2 Genes Respond Differently to Feeding Signal.
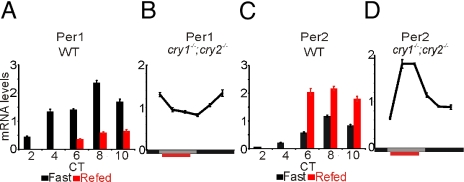
Although the core oscillator functions under fasting conditions (22), feeding time ultimately sets the phase of the peripheral clock (4). Among all of the known oscillator components, the mRNA levels of the Period genes were acutely affected by refeeding after 24 h of fasting. RT-qPCR confirmed that levels of Per1 mRNA declined following food intake, while the levels of Per2 transcripts increased (Fig. 7 A and C). This acute effect of feeding on Per gene expression is independent of the oscillator itself. In the liver of cry1−/−;cry2−/− mice under daytime feeding condition, the Per1 mRNA level also declined immediately after food intake, while Per2 level increased (Fig. 7 B and D). Both genes showed rhythmic expression, albeit with low amplitude, under tRF condition in these mice.
Fig. 7.
Feeding cue is essential for rhythmic transcription of a subset of transcripts and is integrated into the circadian clock through Per1 and Per2. (A and C) Mice were fasted for 24 h and re-fed at CT4 or left under fasted condition. mRNA levels of Per1 and Per2 in the livers of fasted (black) or re-fed (red) mice were determined by RT-qPCR at 2-h resolution. (B and D) mRNA levels of Per1 and Per2 in the liver of cry1−/−;cry2−/− mice under tRF conditions were determined by RT-qPCR at 4-h resolution.
Discussion
Circadian expression of several oscillator components in the mouse liver persist under prolonged fasting, daily restricted feeding (3), frequent meal (23), and high fat diet conditions (24). Rhythmic expression of these components is even conserved in cultured cells growing under constant temperature and nutrient availability (25). Despite the sustenance of rhythms in the core clock, rhythms in output transcripts are highly variable. For example, in cultured mouse cells with saturating nutrient availability, only a small set of <50 transcripts show robust oscillation, while in the liver of predominantly nocturnal ad lib fed WT mice, thousands of transcripts show circadian oscillation (26). Blunting the day:night differences in food intake also affects many output transcripts by reducing the amplitude of their rhythmic expression (24). Since feeding is a daily event that can trigger changes in gene expression through systemic thermogenic, endocrine, and metabolic cues, we tested the contribution of circadian oscillator and temporal feeding pattern in determining the circadian transcriptome in mouse liver.
Food Modulated Transcripts Constitute a Metabolic Sand Timer.
A large number of hepatic transcripts show increased expression upon feeding or fasting. Food-induced transcripts rise following daily physiological feeding bouts, while the fasting-induced transcripts accumulate with increasing length of fasting. Many of these feeding/fasting-regulated transcripts are direct targets of metabolic and stress regulators CREB, SREBP, ATF6, and FoxO1. The cyclic activities of these metabolic regulators loosely coupled to the circadian clock, but a daily rhythm in feeding can drive their rhythmic activities even in the absence of a clock. Conversely, in the absence of feeding, a functional clock cannot impose rhythmic expression of their targets. Hence, the food-driven transcripts are analogous to a metabolic sand timer as opposed to the self-sustaining rhythms of a circadian oscillator (Fig. S8).
Prolonged Fasting Suppresses Rhythmic Transcription of a Large Number of Transcripts.
Under fasting conditions, only 368 probe sets continued to oscillate robustly (Fig. 6 C and D and Fig. S4C). This small subset includes the known core oscillator components and their well-characterized immediate targets. In the absence of food intake, most food-driven transcripts fail to sustain circadian variations, highlighting their nature as a response to and not in anticipation of feeding. The cellular levels of various products of energy metabolism, such as NADH and ATP, which are directly used in transcription or are necessary cofactors for various chromatin modifying enzymes, drop during low nutrient states (27–29). Therefore, prolonged fasting may globally suppress dynamic changes in transcription. However, resistance of the circadian oscillator to severe perturbations of general transcription (30) may allow sustenance of the core clock under prolonged fasting.
Down-regulation of circadian transcriptional outputs under low nutrient state may be a universal phenomenon across kingdoms. In plants, up to 30% of the transcriptome, including several enzymes and regulators for nutrient production and metabolism, shows rhythmic expression under constant light when photosynthesis persists (31, 32). However, under DD only a handful of transcripts including the components of the oscillator and their immediate outputs show sustained rhythms (33). This may be a mechanism to conserve energy by preventing wasteful synthesis and degradation of a large number of transcripts.
Circadian Oscillator Modulates Expression of Food-Driven Transcripts.
In the absence of a circadian oscillator in cry1−/−;cry2−/− mice, scheduled feeding can rescue rhythmic expression of only 617 food-driven transcripts. A large number of food-driven transcripts do not attain the same level of amplitude as they do under similar feeding conditions in the WT mice, thus are not detected as rhythmic in cry1−/−;cry2−/− mice. This highlights the role of an oscillator in enhancing the amplitude of oscillation in food-driven transcripts.
Under identical tRF conditions, the peak phases of expression of feeding driven transcripts in WT mice occur 1–2 h earlier than in cry1−/−;cry2−/− mice (Fig. S6). This delay of peak expression in cry1−/−;cry2−/− mice does not arise from changes in the feeding pattern as reflected in their RER (Fig. 1). A possible explanation is that the circadian oscillator anticipates changes in the feeding state and accelerates the transcriptional response to an acute activation or repression by feeding cues in food-dependent genes. Similar oscillator-mediated potentiation of rhythmic posttranslational modifications is also observed for CREB and AKT phosphorylation. In summary, although an anticipatory role of the circadian clock has long been hypothesized, here it has been demonstrated in genome-wide transcriptional regulation.
Direct Effect of Feeding on the Circadian Clock.
Among various circadian clock components, the mRNA level of Per1 is down-regulated, while Per2 is up-regulated by food input, causing their peak expression to occur before or after the feeding onset, respectively (Fig. S8A). It is well known that feeding-induced changes in CREB and heat shock factor (HSF) activities can regulate Per1 and Per2 expression, respectively, from their target binding sites present in Per1 and Per2 promoters (34, 35). The acute dephosphorylation and consequently inactivation of CREB (Fig. 2A) following feeding may down-regulate Per1. Postprandial rise in body temperature, on the other hand, may activate Per2 transcription via HSF binding to the cis-acting element in the Per2 promoter (12). These mechanisms are independent of a functional clock, as the differential effects of refeeding on Per1 and Per2 mRNA levels are preserved in the cry1−/−;cry2−/− mice.
Interaction between Clock and Temporal Feeding Pattern Determines the Circadian Transcriptome.
Rhythmic feeding behavior increases the amplitude in the oscillations in core clock components and their immediate target genes, thereby causing robust rhythmic transcription (Fig. S5 A–D). In turn, the circadian clock modifies and enhances the transcriptional response to feeding cues. As a result, 4,960 transcripts display rhythmic expression in WT mice under consolidated feeding (tRF). This largely surpasses the combined numbers of food-only (617 transcripts are rhythmic in cry1−/−;cry2−/− mice under tRF conditions) and circadian clock-only (368 rhythmic transcripts under fasting condition in WT mice) driven transcripts, even when lower sampling frequency is taken into account (26). This synergistic interaction between clock and metabolism is exemplified in the subset of rhythmic transcripts that encode the mitochondrial components (Fig. S7). In WT mice under tRF conditions, these transcripts exhibit more coherent phases and robust rhythms in expression than WT mice under ad lib conditions or cry1−/−;cry2−/− mice under tRF conditions.
Significance of Oscillator-Feeding Time Interaction.
Altogether, the independent but interlinked organization of metabolic sand timer and circadian clock allows an organism to adapt faster to predictable changes in the energy state. This organization also enables the uncoupling of metabolic sand timer and circadian clock to facilitate the acute induction or repression of thousands of transcripts in response to unpredictable changes in the energy state. Finally, differences in the coherence and robustness of rhythmic transcripts between ad lib and tRF conditions in WT mice raises the question on whether tRF conditions offer a better overall metabolic state for the organism than ad lib conditions. Although these two groups had equal caloric intake (Fig. S1A), ad lib fed mice had lower amplitude rhythms in clock gene expression. Reduction in amplitude has also been reported in mice fed a high fat diet ad lib, and some changes in expression pattern were seen when mice were put under a frequent meal paradigm (36). Interestingly, the extra calories consumed by mice on a high fat diet were mostly consumed during the daytime (24). Hence, temporal spreading of the feeding period itself can lead to a dampening of rhythmic transcription. As one major role of a robust circadian clock is to temporally separate incompatible processes, a low amplitude rhythm may favor biochemically incompatible processes to temporally overlap, leading to adverse long-term effects on general health and longevity. As such, it remains to be determined if temporally restricting feeding to daytime or nighttime can have beneficial effect on healthy living.
Materials and Methods
Detailed experimental procedures are outlined in the SI Text.
Animals.
All animal experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Salk Institute. Male wild-type C57BL/6J mice (8- to 12-week-old) and 8- to 16-week-old cry1−/−;cry2−/− mice were used. Metabolic data were measured by indirect calorimetry.
Real-Time Quantitative PCR.
Total RNA was pooled from at least three livers and used for cDNA synthesis. Transcript levels were then quantified by qPCR using the SYBR Green PCR Master Mix (Applied Biosystems). Results for the respective gene of interest were normalized to Gapdh and subsequently median-normalized to 1.
Western Blot Analysis.
Proteins were detected using antibodies against pCREB (37), CREB (38), pAKT (#7100; Cell Signaling), AKT (#9272; Cell Signaling), and βACTIN (A2066; Sigma–Aldrich), which were used at the dilution proposed by the manufacturer.
Probe Hybridization and Data Analysis.
Total RNA was pooled from three liver samples and amplified, labeled, and hybridized on Affymetrix MOE 430_2 HDAs using the One-Cycle Amplification kit (Affymetrix) following the manufacturer's protocol. Raw data were analyzed in R. Rhythmic transcripts were identified using COSOPT and Fisher G test as described in refs. 11 and 39. Heatmaps were generated with the Cluster and Treeview (40) program.
Data Access.
The data discussed in this publication have been deposited in NCBI-GEO with accession number GSE13093 and is publicly accessible in a database at http://circadian.salk.edu. Gene expression of WT mice under ad lib conditions (26) is also accessible from http://wasabi.itmat.upenn.edu/circa/mouse.
Supplementary Material
Acknowledgments.
The authors thank Aziz Sancar for the cry1−/−;cry2−/− mice. This work was supported by the National Institutes of Health (NIH) Grant NRSA 1F32GM082083–01 (to L.D.) and the Pew Scholars Program in Biomedical Sciences and by NIH Grant EY016807 (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE13093).
This article contains supporting information online at www.pnas.org/cgi/content/full/0909591106/DCSupplemental.
References
- 1.Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: Photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu Rev Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 5.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turek FW, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oishi K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 11.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 12.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheward WJ, et al. Entrainment to feeding but not to light: Circadian phenotype of VPAC2 receptor-null mice. J Neurosci. 2007;27:4351–4358. doi: 10.1523/JNEUROSCI.4843-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 15.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 16.Conkright MD, et al. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, et al. FoxO1 regulates multiple metabolic pathways in the liver: Effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 18.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokolovic M, et al. The transcriptomic signature of fasting murine liver. BMC Genomics. 2008;9:528. doi: 10.1186/1471-2164-9-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straume M. DNA microarray time series analysis: Automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol. 2004;383:149–166. doi: 10.1016/S0076-6879(04)83007-6. [DOI] [PubMed] [Google Scholar]
- 22.Kawamoto T, et al. Effects of fasting and re-feeding on the expression of Dec1, Per1, and other clock-related genes. J Biochem. 2006;140:401–408. doi: 10.1093/jb/mvj165. [DOI] [PubMed] [Google Scholar]
- 23.van der Veen DR, et al. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc Natl Acad Sci USA. 2006;103:3393–3398. doi: 10.1073/pnas.0507825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 26.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 28.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 29.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dibner C, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmer SL, et al. Orchestrated transcription of key pathways in arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 33.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 34.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinke H, et al. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cailotto C, et al. The suprachiasmatic nucleus controls the daily variation of plasma glucose via the autonomic output to the liver: Are the clock genes involved? Eur J Neurosci. 2005;22:2531–2540. doi: 10.1111/j.1460-9568.2005.04439.x. [DOI] [PubMed] [Google Scholar]
- 37.Hagiwara M, et al. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez GA, Menzel P, Leonard J, Fischer WH, Montminy MR. Characterization of motifs which are critical for activity of the cyclic AMP-responsive transcription factor CREB. Mol Cell Biol. 1991;11:1306–1312. doi: 10.1128/mcb.11.3.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.