Abstract
The evolutionarily conserved Smc5/6 complex is implicated in recombinational repair, but its function in this process has been elusive. Here we report that the budding yeast Smc5/6 complex directly binds to the DNA helicase Mph1. Mph1 and its helicase activity define a replication-associated recombination subpathway. We show that this pathway is toxic when the Smc5/6 complex is defective, because mph1Δ and its helicase mutations suppress multiple defects in mutants of the Smc5/6 complex, including their sensitivity to replication-blocking agents, growth defects, and inefficient chromatid separation, whereas MPH1 overexpression exacerbates some of these defects. We further demonstrate that Mph1 and its helicase activity are largely responsible for the accumulation of potentially deleterious recombination intermediates in mutants of the Smc5/6 complex. We also present evidence that mph1Δ does not alleviate sensitivity to DNA damage or the accumulation of recombination intermediates in cells lacking Sgs1, which is thought to function together with the Smc5/6 complex. Thus, our results reveal a function of the Smc5/6 complex in the Mph1-dependent recombinational subpathway that is distinct from Sgs1. We suggest that the Smc5/6 complex can counteract/modulate a pro-recombinogenic function of Mph1 or facilitate the resolution of recombination structures generated by Mph1.
The Smc5/6 complex is one of the three structural maintenance of chromosomes (SMC) complexes in eukaryotic cells and contains Smc5, Smc6, and six non-SMC elements, Nse1–6 (1–4). Similar to SMC proteins in the other two complexes, namely cohesin and condensin, Smc5 and Smc6 form the backbone of the complex upon which Nse subunits assemble (3, 5, 6). However, the Smc5/6 complex contains a unique small ubiquitin-like modifier (SUMO) ligase subunit, Nse2/Mms21 (hereafter Mms21), that facilitates the addition of SUMO to other proteins (1, 7, 8). The functions of the Smc5/6 complex are not as well understood as those of cohesin and condensin. Nevertheless, the importance of this complex is clear from the multiple defects manifested by its mutants, including sensitivity to DNA damage, telomere and rDNA defects, slow growth, and cell death (9, 10). Previous studies suggest that some of these defects indicate a role for the Smc5/6 complex in recombinational repair during impaired replication. In particular, it was found that, in budding yeast, when replicating in the presence of the DNA damaging agent methylmethane sulfonate (MMS), mutants of this complex accumulate recombination intermediates, which are detected as X-shaped DNA molecules by 2D agarose gel electrophoresis (2D gel) (11, 12). Because the absence of the DNA helicase Sgs1, which binds to Top3 and Rmi1 to dissolve double Holliday junctions (13), results in a similar defect, it was thought that the Smc5/6 complex might collaborate with Sgs1 to resolve such recombination intermediates (11, 14). However, a recent study suggests that these two protein entities do not associate with each other physically (12), raising the possibility that they perform different roles. Currently, the exact role of the Smc5/6 complex in recombinational repair is not known.
To understand the functions of the Smc5/6 complex, we sought to identify proteins that interact physically with this complex using yeast 2-hybrid (2H) screens coupled with co-immunoprecipitation. From this study, we recovered the DNA helicase Mph1 as the only interactor that has known functions in recombination. Mph1 is not a core recombination protein; rather, it defines a specialized recombination subpathway that operates when replication is impaired (15, 16). The nature of this subpathway remains to be elucidated but is expected to entail some of the biochemical activities exhibited by Mph1 and its orthologs, which include the Fanconi anemia M (FANCM) protein in humans, the Fml proteins in fission yeast, and the archaeal Hef protein (15, 17, 18). It is noteworthy that a FANCM mutation is implicated in Fanconi anemia, a genetic disease that affects development and causes cancer predisposition, suggesting the importance of Mph1-like proteins in cell physiology (19, 20).
An activity shared by Mph1 and its orthologs is the dissociation of DNA D-loop structures, a function important in limiting crossovers during mitotic recombination (21–23). In addition, Fml1, FANCM, and Hef can catalyze the regression or unwinding of replication forks, which can potentially lead to recombination (22–24). Although this process may subsequently restart the damaged replication fork, it also increases the risk of generating toxic recombination intermediates or other genetic alterations. Currently, it is unclear how this process is regulated in vivo. In addition to recombination, Mph1 also has possible roles in lagging-strand synthesis and in chromosomal rearrangement when overproduced (25, 26). Key residues for Mph1 helicase activity are required only for recombinational repair and not for its other functions (15, 21, 25, 26).
We show here that binding to the Smc5/6 complex does not lead to Mph1 sumoylation. Rather, several major defects in mutants of the Smc5/6 complex are suppressed by mph1Δ and mph1 helicase mutations. These observed suppressions correlate with a large reduction in the levels of recombination intermediates seen in these mutants. In contrast, mph1Δ does not suppress similar defects in sgs1Δ cells. Taken together, our results suggest that the Smc5/6 complex, but not Sgs1, modulates the Mph1-dependent recombination subpathway to prevent the accumulation of recombination intermediates and the associated deleterious effects.
Results
Mph1 Binds to the Smc5/6 Complex but Is Not Sumoylated.
To understand how the Smc5/6 complex is involved in recombinational repair, we examined whether it physically interacts with recombination factors. Using each subunit of the Smc5/6 complex as bait, we performed 2H screens against an arrayed library containing the majority of the yeast genes. The only interacting protein with a known function in recombination recovered from this screen was the Mph1 helicase, with Smc5 as the bait. The Smc5–Mph1 interaction was confirmed subsequently by pair-wise 2H tests (Fig. 1A).
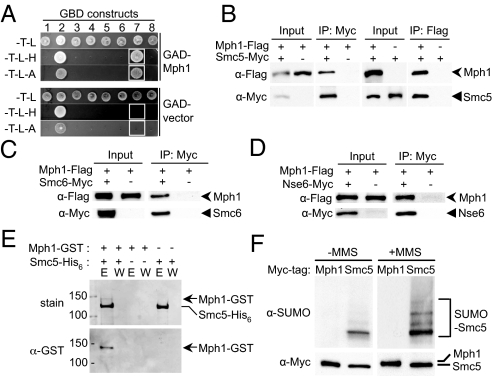
Fig. 1.
Mph1 interacts with the Smc5/6 complex but is not sumoylated. (A) Mph1 interacts with Smc5 in 2H. The 2H strain pJ69–4a containing either a GAD-Mph1 or a GAD vector was mated to pJ69–4alpha cells containing GBD-fused Nse1–6 (lanes 1–6), Smc5 (lane 7), and Smc6 (lane 8). The resulting diploids were selected on -TRP-LEU (-T-L) plates, and reporter activation was scored by replica plating to -TRP-LEU-HIS (-T-L-H) and -TRP-LEU-ADE (-T-L-A) media. Note that GBD-Nse2/Mms21 self-activates the HIS3 and ADE2 reporters. White boxes indicate the interactions between Mph1 and Smc5 and the corresponding controls. (B–D) Mph1 co-immunoprecipitates with Smc5, Smc6, and Nse6. Lysates from cells containing the indicated tagged constructs were immunoprecipitated with the indicated antibody. Cell lysates (input) and immunoprecipitated proteins (IP) were analyzed by protein blotting using anti-Flag (Upper) and anti-Myc (Lower) antibodies. In (B), Smc5 was precipitated by anti-Flag antibody only in the presence of Mph1-Flag, and vice versa. In (C and D), Mph1 was precipitated by anti-Myc antibody only in the presence of either Smc6-Myc (C) or Nse6-Myc (D). (E) Mph1 binds Smc5 in vitro. Recombinant Mph1-GST and Smc5-His6 were expressed in E. coli, and Mph1-GST was pulled down by the Ni-NTA resins only in the presence of Smc5-His6. E: eluate, W: wash. (Top) Coomassie stained gel; (Bottom) Western blot using anti-GST antibody. (F) Smc5, but not Mph1, is sumoylated. Cells containing Myc-tagged Mph1 or Smc5 were grown in the absence (−MMS) or presence (+MMS) of MMS. Mph1 and Smc5 were immunopurified by anti-Myc antibody and examined by protein blotting using anti-Myc (Bottom) and anti-SUMO antibodies (Top).
We then constructed epitope-tagged Smc5 and Mph1 at their chromosomal loci for co-immunoprecipitation experiments. As shown in Fig. 1B, Mph1 pulled down Smc5, and vice versa (Fig. 1B). In addition, Mph1 co-precipitated with Myc-tagged Smc6 and Nse6 (Fig. 1 C and D). Furthermore, we found that His-tagged Smc5 protein expressed in Escherichia coli pulled down recombinant Mph1 (Fig. 1E). These results demonstrate that Mph1 interacts with the Smc5/6 complex by directly binding to Smc5.
Because the Mms21 subunit of the Smc5/6 complex is a SUMO E3 (1), the interaction of Mph1 with the Smc5/6 complex may suggest that it is a SUMO target. However, we found that Mph1 was not sumoylated during normal growth or after MMS treatment, when the sumoylation of several other recombination proteins is induced (Fig. 1F). As a control, we can detect the sumoylation of Smc5 under both conditions (Fig. 1F). We thus conclude that the Mph1–Smc5/6 complex interaction does not lead to Mph1 sumoylation.
MPH1 Deletion Suppresses Several Defects of Mutants of the Smc5/6 Complex, Whereas Overexpression Exacerbates These Effects.
To understand the biological functions of the interaction between Mph1 and the Smc5/6 complex, we examined the effects of mph1Δ on three mutants of the Smc5/6 complex, including smc6–56 (27), smc6-P4 (or K239R), and mms21–11, which is defective in sumoylation (1). All three alleles exhibit strong sensitivity to replication-blocking agents and slow growth, with smc6–56 producing the most severe defects (Fig. 2 A and B). We found that mph1Δ suppressed the sensitivity to MMS, hydroxyurea (HU), and UV of all three alleles (Fig. 2 A and B). Moreover, mph1Δ suppressed the slow growth of these mutants, an effect most evident in smc6–56 cells (Fig. 2 A and B). In addition to slow growth, smc6–56 cells also exhibit defective centromere separation (28). Strikingly, we found that mph1Δ restored centromere separation in smc6–56 cells from approximately 65% to above 90% (Fig. 2 C and D). We conclude that the deletion of MPH1 suppresses three major defects associated with mutants of the Smc5/6 complex, namely sensitivity to DNA damage, slow growth, and defective centromere separation.
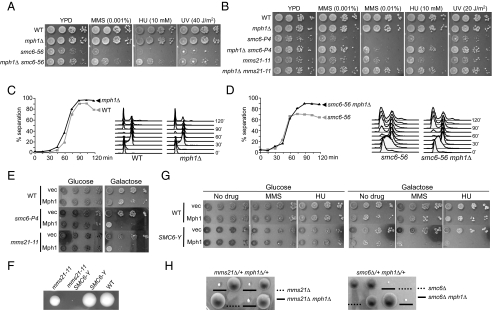
Fig. 2.
mph1Δ rescues several defects of smc6 and mms21 mutants, whereas Mph1 overexpression confers opposite effects. (A and B) mph1Δ rescues the growth defects and DNA damage sensitivity of smc6–56 (A), smc6-P4, and mms21–11 (B). WT and mutant cell cultures were diluted and spotted onto plates containing no drug or the indicated concentration of drugs or were treated with the indicated dose of UV light. (C and D) mph1Δ rescues centromere separation defects in smc6–56 cells. Cells were arrested in G1 at 23 °C and were shifted to 37 °C for 1 h to inactivate the smc6–56 allele before being released from G1. Samples were collected every 15 min to examine cell-cycle progression by FACS and chromatid separation by the appearance of 2 dots of GFP-LacI bound to tandem LacO repeats near the centromere on chromosome IV. To examine the events in the first cell cycle only, alpha-factor was added back to the cultures 45 min after release. (E and G) Mph1 overexpression sensitizes cells with defective Smc5/6 complexes. Cells containing smc6-P4 or mms21–11 (E) or SMC6-YFP (SMC6-Y, G) were transformed with pGAL-Mph1 or the control vector; cell cultures were diluted and spotted on plates lacking uracil with glucose or galactose and with and without MMS or HU. (F) Smc6-YFP confers normal growth but is synthetic sick with mms21–11. A representative tetrad from diploid strains heterozygous for SMC6-YFP and mms21–11 is shown. The genotype for each spore clone is indicated. (H) mph1Δ suppresses the lethality of mms21Δ and smc6Δ cells. Representative tetrads from diploid strains with the indicated genotypes are shown. The spore clones containing mms21Δ mph1Δ (Left) and smc6Δ mph1Δ (Right) are underlined; those containing mms21Δ (Left) and smc6Δ (Right) are marked with dotted lines, and their genotypes are deduced from sibling spore clones.
In a converse experiment, we found that MPH1 overexpression severely inhibited the growth of smc6-P4 and mms21–11 cells (Fig. 2E). To assess how MPH1 overexpression affects MMS and HU sensitivities, we used endogenously YFP-tagged Smc6, which leads to normal growth but has mild defects in the complex's functions as evidenced by its synthetic sick interaction with mms21–11 (Fig. 2F). We found that MPH1 overexpression rendered SMC6-YFP, but not WT cells, sensitive to MMS and HU (Fig. 2G). Taken together, the deletion and the overexpression experiments show that Mph1 causes toxicity during normal growth and in the presence of genotoxins when the Smc5/6 complex is defective.
These results prompted us to test whether Mph1 affects the essentiality of the Smc5/6 complex. Remarkably, we found that deletion of MPH1 suppressed the lethality of smc6Δ and mms21Δ spore clones (Fig. 2H). However, the double mutants mph1Δ smc6Δ and mph1Δ mms21Δ showed growth defects, indicating that the Smc5/6 complex possesses Mph1-independent functions. Nevertheless, the fact that cells can live without the Smc5/6 complex if Mph1 is removed suggests that an important function of the Smc5/6 complex pertains to Mph1 regulation during normal growth.
A Pro-Recombinogenic Function of Mph1 Is Toxic in Mutants of the Smc5/6 Complex.
Mph1 has been shown to function in a subpathway of recombinational repair to facilitate cell survival in MMS (15, 16). Consistent with previous reports, we found that mph1Δ exhibited moderate sensitivity to MMS, and very slight, if any, sensitivity to HU, UV, or X-rays and that mph1Δ did not increase the DNA damage sensitivity of the recombination mutants rad51Δ and rad52Δ (Fig. S1 A and B). In addition, we found that Mph1 formed nuclear foci during normal growth and that the percentage of cells containing Mph1 foci increased moderately after MMS treatment (Fig. 3A and Table S1). These Mph1 foci frequently co-localized with Rad52 and PCNA foci (Fig. 3A and Table S1), which are thought to represent recombination and replication centers, respectively (29, 30). This cell biological result is consistent with the genetic data and supports a role for Mph1 in replication-associated recombinational repair.
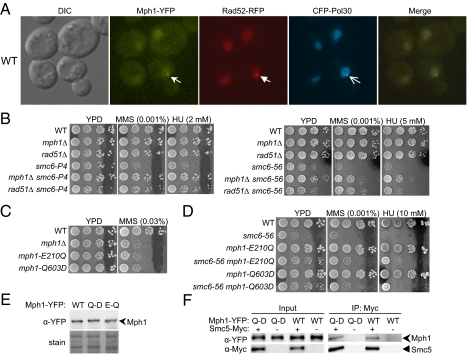
Fig. 3.
A pro-recombinogenic function of Mph1 is toxic in mutants of the Smc5/6 complex. (A) Mph1 foci co-localize with those of Rad52 and Pol30. Representative images of WT cells containing Mph1-YFP, Rad52-RFP, and CFP-Pol30 are shown. Foci formed by each protein are indicated by arrows. (B) rad51Δ, like mph1Δ, suppresses the sensitivity to MMS and HU of smc6-P4 cells (Left) and smc6–56 cells (Right). Experiments were performed as in Fig. 2A. (C and D) Helicase mutants mph1-E210Q and -Q603D exhibit sensitivity to MMS similar to that of mph1Δ (C) and suppress the sensitivity of smc6–56 cells to MMS and HU (D). Experiments were carried out as described in Fig. 2A. (E) Mph1-E210Q and -Q603D protein levels are similar to that of WT Mph1. WT and mutant Mph1 proteins were tagged with YFP at their chromosomal loci; their protein levels were examined by protein blotting using anti-YFP antibody (Top). Similar loading was verified by amido-black staining (Bottom). (F) Similar amounts of WT Mph1 and Mph1-Q603D are immunoprecipitated with Smc5. Experiments were carried out as in Fig. 1B.
Because Mph1 is also known to be involved in other processes, including lagging-strand synthesis and genomic rearrangements (25, 26), the question arises whether the toxicity of Mph1 in mutants of the Smc5/6 complex is related specifically to its recombination functions. We used two approaches to address this issue. First, we reasoned that if this hypothesis were true, then eliminating recombination by means other than deleting Mph1 should also suppress the defects in mutants of the Smc5/6 complex. We found that the removal of the recombinase Rad51 did indeed suppress the MMS and HU sensitivities of smc6–56 and smc6-P4 cells to a degree similar to that observed for mph1Δ (Fig. 3B). However, rad51Δ did not significantly suppress the slow growth of smc6–56, suggesting that other recombinational subpathways may be required for the proper growth of these cells.
In the second approach, we examined whether mutations of key residues of the Mph1 helicase domain can suppress defects of smc6 and mms21 mutants, because Mph1 helicase activity is required for its roles in recombination but not for its other functions (15, 21, 25, 26). To this end, we replaced WT MPH1 with mph1-E210Q or mph1-Q603D, which mutated the conserved glutamic acid in the DEAH motif or the key residue of the helicase motif, respectively (15, 17). Consistent with previous reports, mph1-E210Q and -Q603D behaved like mph1Δ for DNA damage sensitivity (Figs. 3C and S1A). We found that both alleles suppressed the sensitivity to MMS, HU, and UV in smc6 and mms21 mutant cells to the same degree as observed for mph1Δ (Figs. 3D and S2 A and B). In addition, they suppressed the slow growth of smc6–56 cells (Fig. 3D). Mph1-E210Q and -Q603D proteins were expressed at levels similar to WT Mph1 (Fig. 3E). Furthermore, Smc5 pulled down similar amounts of Mph1-Q603D and WT Mph1 (Fig. 3F), suggesting that Q603D does not affect the Smc5–Mph1 interaction. Therefore we conclude that the suppression conferred by these mutations is not caused by defects in protein levels or in the Mph1–Smc5 interaction but rather by defects in the Mph1 helicase function. The results concerning rad51Δ and mph1 helicase mutations suggest that a pro-recombinogenic role of Mph1 is toxic in cells with defective Smc5/6 complexes.
Absence of Mph1 or Its Helicase Function Suppresses the Accumulation of Recombination Intermediates in smc6 and mms21 Mutants.
To understand the molecular basis of the suppression conferred by mph1Δ and mph1 helicase mutations, we performed 2D gel analyses to examine the levels of recombination intermediates produced during impaired replication. Cells were synchronized in G2 phase and released into the cell cycle in the presence of sublethal concentrations of MMS. DNA from WT cells and relevant mutants was extracted at different time points and examined in 2D gels using a probe for the early firing replication origin ARS305 (Fig. 4A). It has been shown that smc6–56 and mms21–11 cells, but not WT cells, accumulate recombination intermediates at this DNA region and that rad51Δ suppresses these defects (11, 12). We confirmed these observations and found that smc6-P4 cells also accumulated X-shaped molecules for a prolonged period, whereas mph1Δ and mph1-Q603D behaved like WT cells (Fig. 4 A and B). Importantly, we found that both mph1Δ and mph1-Q603D greatly reduced the recombination intermediates detected in smc6–56, smc6-P4, and mms21–11 cells (Figs. 4 A and B, and S2C). These results strongly suggest that the helicase activity of Mph1 is largely responsible for the accumulation of X-shaped recombination structures in smc6 and mms21 mutant cells during impaired replication. This observation provides a likely explanation for the rescue of smc6 and mms21 cells' sensitivity to MMS by mph1 mutations.
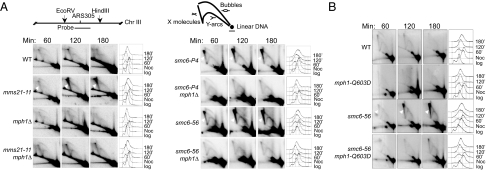
Fig. 4.
mph1Δ and mph1-Q603D decrease the levels of recombination intermediates in smc6 and mms21 mutants. (A and B) Cells were arrested at G2/M phase with nocodazole at 25 °C and then were released into YPD medium with 0.033% MMS at 30 °C. The replication and recombination intermediates at the ARS305 region 60, 120, and 180 min after release were analyzed by 2D gel electrophoresis followed by Southern blotting. Diagrams indicating the position of the probe and the replication structures are shown above the 2D gel images. The X-shaped DNA structures are indicated by arrowheads in smc6 and mms21 mutants. FACS analyses are presented to the right of each gel image.
The Genetic Interactions of Mph1 and the Smc5/6 Complex with the DNA Helicases Sgs1 and Srs2.
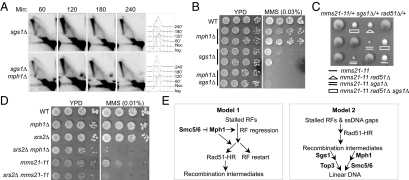
Several defects of smc6 and mms21 mutants are reminiscent of cells lacking the DNA helicase Sgs1. In particular, like smc6 and mms21 mutants, sgs1Δ cells accumulate recombination intermediates when cells replicate in the presence of MMS, with rad51Δ suppressing this defect (14). To understand whether mph1Δ, like rad51Δ, serves as a general suppressor for the accumulation of recombination structures, we examined its effect in sgs1Δ cells. We found that under the experimental conditions described above, the recombination intermediates in sgs1Δ were largely unchanged for a prolonged period when MPH1 was deleted (Fig. 5A). In addition, mph1Δ did not suppress the sensitivity of sgs1Δ cells to MMS but instead exacerbated it (Fig. 5B). Both observations are in contrast to the suppression conferred by mph1Δ in smc6 and mms21 mutants, suggesting different roles of Sgs1 and the Smc5/6 complex in preventing the accumulation of recombination intermediates. Consistent with this notion, sgs1Δ and mms21–11 are synthetic sick, and this defect is suppressed by the removal of Rad51 (Fig. 5C).
Fig. 5.
Genetic interactions of mph1Δ and mms21–11 with sgs1Δ and srs2Δ and models for the functions of the Smc5/6 complex and Mph1 in recombinational repair. (A) The amount of X-shaped DNA in sgs1Δ cells is largely unaffected by mph1Δ. Experiments were carried out as in Fig. 4. (B and D) mph1Δ enhances the MMS sensitivity of sgs1Δ (B) and srs2Δ (D) cells, and srs2Δ exacerbates MMS sensitivity of mms21–11 cells (D). Experiments were carried out as in Fig. 2A, except that plates were incubated for three days in B. (C) The synthetic sick interaction between sgs1Δ and mms21–11 is alleviated by rad51Δ. Shown are 3 tetrads from indicated diploids; the spore clones of the relevant genotype are indicated. (E) Models for the functions of the Smc5/6 complex and Mph1 in recombinational repair. See text for details. RFs: replication forks, HR: homologous recombination.
Another DNA helicase that plays a role in the suppression of recombinational events is Srs2. A shared function of Srs2 and Mph1 is preventing crossover events during mitotic recombination (31–33). However, we found that, unlike mph1Δ, srs2Δ increased the sensitivity of mms21–11 cells to MMS (Fig. 5D). In addition, srs2Δ and mph1Δ were synergistic for sensitivity to MMS, suggesting that they perform different roles in cellular resistance to MMS (Fig. 5D). Taken together, these genetic data indicate that the roles of the Smc5/6 complex in recombinational repair are at least partly different from those of Sgs1. Furthermore, Mph1 has functions distinct from the other two helicases, Sgs1 and Srs2, with regard to cell survival in MMS.
Discussion
Several lines of genetic evidence have indicated the involvement of the Smc5/6 complex in recombinational repair, but the mechanism underlying this function has been unclear. In this study, we have revealed an interaction between the Smc5/6 complex and the DNA helicase Mph1. We show that Mph1 is not sumoylated, although the Smc5/6 complex contains a SUMO ligase. Because mph1Δ alleviates several defects in smc6 and mms21 mutants, and MPH1 overexpression exacerbates some of these defects, Mph1 appears to be toxic in these cells. This toxicity is caused by a pro-recombinogenic function of Mph1, because eliminating recombination by rad51Δ or mutating key residues of Mph1 helicase motifs, required for recombination but not for other functions of Mph1, phenocopied mph1Δ in the suppression of smc6 and mms21 mutants. We further show that mph1Δ and its helicase mutations also reduced the levels of X-shaped recombination structures in smc6 and mms21 mutant cells. Collectively, these results suggest that the helicase activity of Mph1 leads to accumulation of recombination intermediates that is detrimental in cells containing defective Smc5/6 complexes.
Our genetic and physical evidence, when combined with the known biochemical activities of Mph1 and its orthologs, supports two models for the function of the Smc5/6 complex in recombinational repair (Fig. 5E). The first model proposes that the Smc5/6 complex binds Mph1 and regulates its action as a pro-recombinogenic factor. Mph1 orthologs can catalyze replication fork regression/migration (22–24). It is thus possible that the Smc5/6 complex modulates Mph1 to prevent fork regression or channels the regressed forks to direct restart instead of recombination. This action may prevent the formation of recombination intermediates and permit the use of safer pathways for rescuing damaged forks. The second model suggests that the Smc5/6 complex facilitates the resolution of the recombination structures generated by Mph1. Because Mph1 orthologs exhibit branch migration activities, it is formally possible that Mph1 can promote the formation of DNA joint molecules that need to be processed in an Smc5/6 complex-dependent manner. This relationship would be somewhat similar to that of Sgs1 and Top3. However, because its subunits do not contain sequences characteristic of topoisomerases, helicases, or nucleases, the Smc5/6 complex would play a structural role and/or recruit and assist additional enzymes. The first model has more experimental support at this stage. The Mph1 ortholog Fml1 has recently been shown to promote recombination at stalled replication forks in vivo, whereas it is unclear whether a potential role for Mph1 in branch migration could lead to specific types of joint molecules in cells. In fact, Mph1 was recently shown to dissociate D-loop structures and to disfavor joint molecule formation in mitotic recombination (21). Future work will be required to address the biochemical activities of Mph1 and the effect of the Smc5/6 complex on such activities and on other aspects of Mph1 functions.
In summary, the results presented here provide important insights into the role of the Smc5/6 complex in the Mph1-dependent recombination pathway. Because cells can live without the Smc5/6 complex when MPH1 is deleted, this role is important for normal growth. As defects in sgs1Δ cells are not suppressed by mph1Δ, Sgs1 probably has limited roles in this pathway. rad51Δ suppression of the synthetic interaction of sgs1Δ and mms21–11 also supports the notion that Sgs1 and the Smc5/6 complex have different roles in recombination. However, the Smc5/6 complex may work with proteins other than Sgs1 in the Mph1 pathway, because mph1Δ was recently shown to suppress the accumulation of X-shaped molecules in cells lacking Esc2, a protein containing SUMO-like domains (34). Since the Smc5/6 complex, Mph1, and Esc2 are conserved in humans, it will be interesting to examine whether a similar recombination subpathway exists in human cells. In this context, it is noteworthy that a mutation in Mms21 leads to sensitivity to inter-strand cross-linking agents (35), a hallmark of Fanconi anemia cells. Because improper recombination underlies the pathology of Fanconi anemia and several other human diseases, an in-depth investigation of the mechanisms involved is likely to contribute to the understanding and the development of potential treatments for these conditions.
Materials and Methods
Yeast Strains, Plasmids, Primers and Genetic Manipulations.
Yeast strains and plasmids are listed in Table S2. Primer sequences are available upon request. We carried out 2H screens and pair-wise testing as described (6). Standard yeast protocols were used for strain construction, growth, and medium preparation. Spot assay plates were incubated at 30 °C and photographed after two days, unless otherwise indicated.
Other Procedures.
For live-cell imaging, cells were processed for microscopy as described (29), except for the exposure times used for fusion proteins: CFP-Pol30, 0.3 s; Mph1-YFP, 3 s; and Rad52-RFP, 0.5 s. Spot assays for detecting sensitivity to damage and protein analyses were carried out as described (1). The primary antibodies used were anti-Myc (9E10), anti-Flag (Sigma), anti-GFP (Roche), and anti-SUMO (1). Chromatid separation assays (36), 2D gel analyses (11), and an in vitro protein pull-down assay (6) were performed as described.
Supplementary Material
Acknowledgments.
We thank S. Fields (University of Washington, Seattle, WA) for providing the 2H library; R. Rothstein (Columbia University, New York) and H. Klein (New York University, New York) for yeast strains; X. Liu for constructing the smc6-P4 strain; Y. Yan for helping in sumoylation detection; and S. Keeney, M. Jasin, P. Thorpe, J. Petrini, and members of the Zhao laboratory for comments on the manuscript. This work was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro to D.B. and by National Institute of Health Grants R01GM079516 to H.Y. and R01GM080670 to X.Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908258106/DCSupplemental.
References
- 1.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sergeant J, et al. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol Cell Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, III, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. Identification of the proteins, including MAGEG1, that make up the human SMC5–6 protein complex. Mol Cell Biol. 2008;28:1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR. The Smc5-Smc6 DNA repair complex. Bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J Biol Chem. 2006;281:36952–36959. doi: 10.1074/jbc.M608004200. [DOI] [PubMed] [Google Scholar]
- 6.Duan X, et al. The architecture of the Smc5/6 complex of S. cerevisiae reveals a unique interaction between the Nse5–6 subcomplex and the hinge regions of Smc5 and Smc6. J Biol Chem. 2009;284:8507–8515. doi: 10.1074/jbc.M809139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews EA, et al. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray JM, Carr AM. Smc5/6: A link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- 10.De Piccoli G, Torres-Rosell J, Aragon L. The unnamed complex: What do we know about Smc5-Smc6? Chromosome Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- 11.Branzei D, et al. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 12.Sollier J, et al. The S. cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction mediated intra-S repair. Mol Biol Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: ADNA structure-specific ‘dissolvasome’? Trends Biochem Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Liberi G, et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schurer KA, Rudolph C, Ulrich HD, Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash R, et al. S. cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- 18.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure (London) 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosedale G, et al. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nature Structural and Molecular Biology. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 21.Prakash R, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun W, et al. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Komori K, et al. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of archaeal Hef protein in processing stalled replication forks. J Biol Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee S, et al. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J Cell Biol. 2008;181:1083–1093. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang YH, et al. The MPH1 gene of S. cerevisiae functions in Okazaki fragments processing. J Biol Chem. 2009;284:10376–10386. doi: 10.1074/jbc.M808894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onoda F, et al. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in S. cerevisiae. DNA Repair. 2004;3:429–439. doi: 10.1016/j.dnarep.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Lindroos HB, et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo YC, et al. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mankouri HW, Ngo HP, Hickson ID. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in S. cerevisiae. Mol Biol Cell. 2009;20:1683–1694. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoch NC, et al. Allelism of Saccharomyces cerevisiae gene PSO10, involved in error-prone repair of psoralen-induced DNA damage, with SUMO ligase-encoding MMS21. Curr Genet. 2008;53:361–371. doi: 10.1007/s00294-008-0192-z. [DOI] [PubMed] [Google Scholar]
- 36.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.