Abstract
In response to DNA damage, checkpoint proteins halt cell cycle progression and promote repair or apoptosis, thereby preventing mutation accumulation and suppressing tumor development. The DNA damage checkpoint protein Hus1 associates with Rad9 and Rad1 to form the 9-1-1 complex, which localizes to DNA lesions and promotes DNA damage signaling and repair. Because complete inactivation of mouse Hus1 results in embryonic lethality, we developed a system for regulated Hus1 inactivation in the mammary gland to examine roles for Hus1 in tissue homeostasis and tumor suppression. Hus1 inactivation in the mammary epithelium resulted in genome damage that induced apoptosis and led to depletion of Hus1-null cells from the mammary gland. Conditional Hus1 knockout females retained grossly normal mammary gland morphology, suggesting compensation by cells that failed to undergo Cre-mediated Hus1 deletion. p53-deficiency delayed the clearance of Hus1-null cells from conditional Hus1 knockout mice and caused the accumulation of damaged, dying cells in the mammary gland. Notably, compensatory responses were impaired following combined Hus1 and p53 loss, resulting in aberrant mammary gland morphology and lactation defects. Overall, these results establish a requirement for Hus1 in the survival and proliferation of mammary epithelium and identify a role for p53 in mammary gland tissue regeneration and homeostasis.
Keywords: breast cancer, DNA damage checkpoint
DNA is constantly subjected to intrinsic and extrinsic genotoxins that may lead to mutations in growth regulatory genes, resulting in cancer formation (1). To prevent mutation accumulation, evolutionarily conserved DNA damage checkpoint pathways survey the genome and respond to DNA lesions by halting the cell cycle, stabilizing replication forks, and inducing repair. In cases of extensive or unrepairable damage, checkpoints instead trigger apoptosis, which eliminates defective cells that are at risk for malignant transformation and stimulates regenerative proliferation that ensures tissue homeostasis.
Two primary checkpoint pathways, the Atm and Atr pathways, function in parallel to respond to DNA damage in mammals (2). The Atm pathway, which includes Atm, Chk2, p53, and additional components, responds to double-stranded DNA breaks (DSBs). Inactivating mutations in genes of the Atm pathway cause increased risk for a variety of malignancies, including breast cancers (3). Notably, over half of all solid human tumors are estimated to have p53 mutations (4). In addition to its well-established functions in apoptosis induction, cell cycle control, and safeguarding of genome stability, p53 also plays a complex and incompletely understood role in tissue homeostasis. In some settings, p53 appears to impede regenerative processes (5–7), while in others it facilitates tissue renewal and recovery from cell loss (8–10).
The Atr pathway, which includes Atr, Chk1, and the Rad9-Rad1-Hus1 (9-1-1) complex, responds to stalled replication forks and a broad array of DNA lesions (2). Because the Atr pathway is essential for embryonic development in mammals, many of its physiological roles have not been fully characterized. Although Chk1 activation is known to occur in response to oncogenic signals (11), there is limited direct evidence indicating that Atr pathway defects are strongly tumor predisposing. Monoallelic ATR and CHK1 mutations, as well as misexpression of CHK1 and 9-1-1 complex components, have been reported in human cancers (12–19). In mouse models, Atr heterozygosity causes a slight increase in tumor incidence (20). Chk1 haploinsufficiency is associated with defects suggestive of cell transformation in the mammary gland (21) and modestly accelerates mammary tumorigenesis in Wnt-1 transgenic mice (22). These data suggest a possible role for the Atr pathway in tumor suppression; however, heterozygosity for these genes in mice results in relatively mild phenotypes.
As a member of the 9-1-1 complex, Hus1 shows predicted structural homology to proliferating cell nuclear antigen (23), the sliding clamp that serves as a processivity factor during DNA replication. 9-1-1 is loaded onto DNA in response to damage and functions as a scaffold that facilitates the formation of checkpoint signaling complexes, leading to efficient phosphorylation and activation of downstream effectors in response to DNA damage. Notably, 9-1-1 promotes the Atr-dependent phosphorylation of Chk1 through the recruitment of the checkpoint mediator TopBP1, which stimulates Atr kinase activity (24). Targeted deletion of Hus1 (25) or Rad9 (26) results in severe genomic instability, profound hypersensitivity to genotoxic stress, and mid-gestational embryonic lethality.
To study the physiological functions of the Atr checkpoint pathway in tumor suppression and tissue homeostasis while bypassing the embryonic lethality of conventional knockout models, we previously developed a conditional loxP-flanked Hus1 allele (Hus1Flox) (27). Hus1Flox is a fully functional allele that expresses wild-type Hus1, but is converted to the null allele Hus1Δ2,3 upon Cre-mediated recombination. In this study, we generated conditional Hus1 knockout mice that express Cre from the beta-lactoglobulin (Blg) promoter to inactivate Hus1 selectively in the mammary glands of pregnant and lactating mice (28). The mammary gland was targeted because this tissue expresses Hus1 (29), has a well-characterized developmental pattern (30), and is susceptible to tumor development due to defects in DNA damage response factors (3). Complete Hus1 inactivation was not associated with increased mammary tumor predisposition; instead Hus1 was found to be essential for genome maintenance and cell survival in mammary epithelium. Unexpectedly, p53 loss exacerbated the deleterious effects of Hus1 inactivation and impaired a compensatory response that in p53-proficient animals promoted tissue regeneration by cells that failed to delete Hus1. These data identify a role for p53 in tissue homeostasis in the mammary gland and additionally suggest that inhibition of Atr signaling may be an effective tool for the treatment of p53-deficient cancers.
Results
Depletion of Hus1-Null Cells from the Mouse Mammary Gland Following Conditional Hus1 Inactivation.
The conditional allele Hus1Flox (27) was used in combination with Blg-Cre transgenic mice (28) to produce mice in which Hus1 was deleted selectively in mammary epithelium. Hus1Flox/Δ1 Cre+ conditional Hus1 knockout mice contained the conditional allele and one copy of the null allele Hus1Δ1 such that Cre-mediated recombination in the mammary gland upon pregnancy would generate Hus1-deficient cells. Control Hus1+/Flox Cre+ mice served as an indicator of the maximum possible Hus1 deletion in the absence of selection against Hus1 loss, as the mammary glands of these mice retain one wild-type Hus1 copy following recombination. Hus1+/+ and Hus1Flox/Flox Cre− mice express wild-type levels of Hus1 and were used to establish baseline values in all assays.
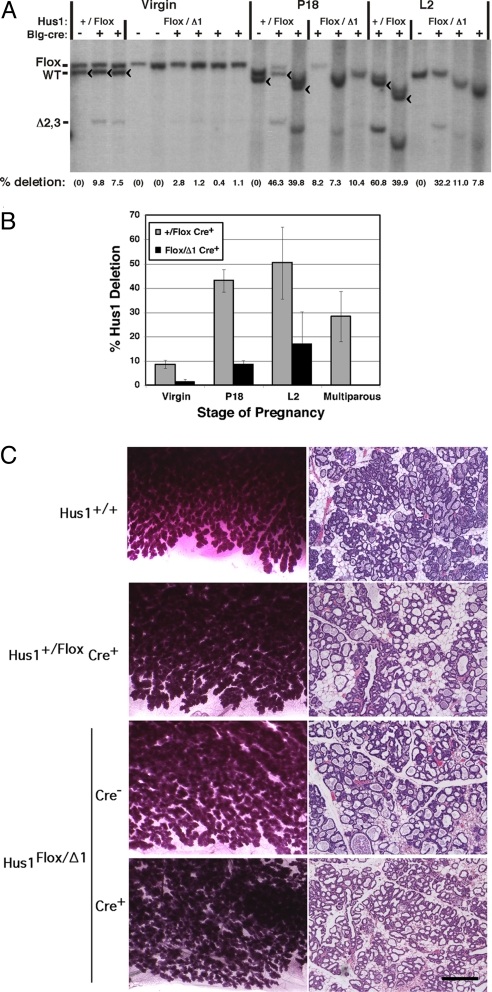
Conditional Hus1 knockout and control mice were born at expected frequencies and appeared grossly normal (Table S1). To determine the extent of Cre-mediated Hus1 inactivation, we performed Southern blotting on DNA extracted from mammary glands from mice as virgins, at the 18th day of pregnancy (P18), or at the second day of lactation (L2) using a probe that can distinguish the unrecombined Hus1Flox, wild-type Hus1+, and inactivated Hus1Δ2,3 alleles (Fig. 1 A and B). No Hus1 deletion was seen in the Cre− control animals as expected, and only limited deletion was seen in virgin Cre+ control mice, likely due to basal Blg promoter activity as reported previously (28). Blg-Cre expression increases during pregnancy and lactation, and accordingly, control Hus1+/Flox Cre+ mammary glands showed an average of 43% Hus1 deletion at P18 and 50% at L2. Notably, the extent of Hus1Flox deletion was substantially reduced in Hus1Flox/Δ1 Cre+ mice relative to Hus1+/Flox Cre+ mice at every developmental stage. Hus1Flox/Δ1 Cre+ mammary glands showed an average of 1.4% (virgin), 9% (P18), and 17% (L2) Hus1 deletion. Relative to Hus1+/Flox Cre+ controls, these values represent decreases in Hus1 deletion of 84, 80, and 66% in virgin, P18, and L2 mice, respectively. These data suggest that Hus1 loss puts cells at a selective disadvantage and results in their depletion from the mammary gland.
Fig. 1.
Depletion of Hus1-deficient cells from mammary glands of conditional Hus1 knockout mice. (A) DNA was isolated from mammary glands of virgin mice, mice at the 18th day of pregnancy (P18) or mice at the second day of lactation (L2), and analyzed by Southern blotting. The position of the bands for Hus1Flox, wild-type Hus1, and the recombined inactivated Hus1 allele, Hus1Δ2,3, are indicated. The Hus1Δ1 allele is not detected in this assay. Because there is variation in gel mobility between samples, the position of the band for wild-type Hus1 is additionally marked with a “<” symbol. “% deletion” refers to the extent of Cre-mediated conversion of Hus1Flox to Hus1Δ2,3, as determined by quantification of Southern blot signal by PhosphorImager. (B) Bar graph shows the average percentage of Hus1 deletion at each developmental stage for mice of the indicated genotypes. Values are means derived from the Southern blots in Fig. 1A and Fig. S1A; error bars, standard deviation. Hus1 deletion was significantly lower in Hus1Flox/Δ1 Cre+ mice relative to Hus1+/Flox Cre+ controls at all stages except in virgins (virgin, P = 0.856; P18, P < 0.001; L2, P < 0.001; Multiparous, P < 0.001) as determined by one-way ANOVA. (C) Representative images of whole mount preparations and histological sections of mammary glands from conditional Hus1 knockout and control females are shown. (Scale bar, 400 μm.)
To determine whether Hus1-deficient cells would accumulate in the mammary glands of multiparous females, we continuously bred conditional Hus1 knockout females to wild-type males. On average, Hus1+/Flox Cre+ females gave birth to 10 litters, while Hus1Flox/Δ1 Cre− and Hus1Flox/Δ1 Cre+ females gave birth to 11 litters. Mammary glands were harvested from these females several months after they gave birth to their final litter, and DNA was isolated and analyzed by Southern blot for Hus1 deletion (Fig. 1B and Fig. S1). Strikingly, there was a complete absence of Hus1-deficient cells in Hus1Flox/Δ1 Cre+ multiparous mammary glands. By contrast, an average of 28% Hus1 deletion was observed in Hus1+/Flox Cre+ multiparous mammary glands; this Hus1 deletion level is lower than that seen in control females at L2 of their first pregnancy because of reduced epithelial cell content following involution. Together, these data indicate that Hus1-deficient cells are selected against and, following multiple pregnancies, completely eliminated from mammary glands of conditional Hus1 knockout mice.
Grossly Normal Mammary Gland Morphology Following Cre-Mediated Hus1 Inactivation.
To determine the effects of Hus1 deletion on mammary gland development and morphology, we performed histopathological analysis of mammary glands from conditional Hus1 knockout and control mice at P18 (Fig. S2), L2 (Fig. 1C), or following multiple rounds of pregnancy (Fig. S1C). Whole mounts and histological sections of Hus1Flox/Δ1 Cre+ mammary glands were similar to those prepared from control Hus1+/Flox Cre+, Hus1Flox/Δ1 Cre−, and wild-type females at all developmental stages. Normal mammary gland development was observed during pregnancy and lactation in mammary glands of all genotypes (Fig. 1C), including similar densities of epithelial and adipose cells as well as typical patterns of side branching, ductal epithelial expansion, and terminal differentiation of milk-producing alveoli (30). These findings suggest that cells that failed to undergo Cre-mediated recombination were able to compensate for the loss of Hus1-deficient cells from the mammary gland and promote normal development.
Consistent with these observations of normal mammary gland architecture, Hus1Flox/Δ1 Cre+ mice were capable of lactating and nursing offspring. Both Hus1Flox/Δ1 Cre+ and control multiparous females had an average of seven pups per litter at weaning, suggesting that multiparous Hus1Flox/Δ1 Cre+ females were capable of nourishing their offspring. In addition, conditional Hus1 inactivation had no significant effect on mammary tumor incidence. Mammary epithelial tumors were found in two of 22 (9.1%) multiparous Hus1Flox/Δ1 Cre+ females, and one of 16 (6.25%) multiparous Hus1+/Flox Cre+ and Hus1Flox/Δ1 Cre− control females. The mammary tumors were composed of cuboidal cells arranged in solid to cystic lobules with variable amounts of glandular formation and scant supporting stroma (Fig. S1D). The cells had small to moderate amounts of eosinophilic cytoplasm and round to oval nuclei with few (<5) mitotic figures in ten 400× fields. A third Hus1Flox/Δ1 Cre+ mouse developed a palpable mammary mass that fully regressed before euthanasia. Southern blot analysis further indicated that Hus1 loss did not directly contribute to tumor formation in multiparous Hus1Flox/Δ1 Cre+ mice, as these tumors showed no Hus1 deletion, whereas complete recombination of the Hus1Flox allele was observed in the neoplasm from the multiparous Hus1+/Flox Cre+ control female (Fig. S1B). The tumors arose in mice that were on average 14 months old and had delivered multiple litters each, suggesting that these were spontaneous background neoplasms in aged mice. Overall, the analysis of conditional Hus1 knockout mice indicates that Hus1-deleted cells are cleared from the mammary gland and do not cause increased cancer risk. Moreover, remaining Hus1-expressing cells compensate for the loss of Hus1-deficient cells and regenerate a morphologically normal, functional mammary gland.
Increased Genome Damage and Apoptosis in Hus1-Deficient Mammary Epithelium.
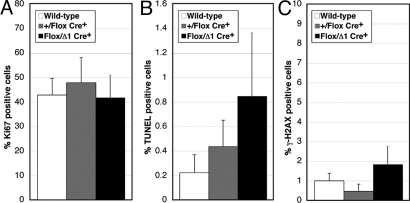
Checkpoint dysfunction can cause increased cell death or impaired cell cycle progression (2). To determine if the depletion of Hus1-deficient cells from the developing mammary gland was due to apoptosis or defective proliferation, we performed terminal uridine deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) and Ki67 antigen staining. TUNEL staining detects extensive DNA fragmentation, a characteristic feature of apoptotic cells (31), while Ki67 antigen is expressed in actively dividing but not quiescent cells and therefore is commonly used as a proliferation marker (32). At L2, control Hus1+/Flox Cre+ mammary glands contained an average of 0.44% apoptotic cells, whereas Hus1Flox/Δ1 Cre+ glands contained an average of 0.85% apoptotic cells, indicating that Hus1 deficiency led to a moderate but significant increase in apoptosis in the developing mammary gland (P = 0.049) (Fig. 2B). The increased apoptosis was not associated with a significant change in cellular proliferation (P = 0.265) (Fig. 2A).
Fig. 2.
Significantly increased apoptosis and genome damage in mammary glands from conditional Hus1 knockout mice. Sections from the fourth mammary gland of conditional Hus1 knockout mice at L2 were stained for Ki67 to assess proliferation, by TUNEL assay to detect apoptosis, or for γ-H2AX to detect DNA damage. The percentage of (A) Ki67, (B) TUNEL, or (C) γ-H2AX positive cells was quantified. Values are means of at least six fields per genotype; error bars, standard deviation. The respective number of wild-type (Hus1+/+ or Hus1Flox/Flox Cre−), Hus1+/Flox Cre+, and Hus1Flox/Δ1 Cre+ mice analyzed was 4, 2, and 3 in A; 4, 4, and 5 in B; and 4, 2, and 2 in C.
To determine the basis for the increased apoptosis, we performed immunohistochemistry against γ-H2AX, the phosphorylated histone variant that accumulates at DSB sites and serves as a robust marker for genome damage (33). Hus1Flox/Δ1 Cre+ mammary glands showed significantly elevated levels of γ-H2AX staining as compared to control Hus1+/Flox Cre+ mammary glands (P = 0.016) (Fig. 2C). A trend toward increased genome damage in Hus1-deficient mammary epithelium also was observed when comparing Hus1Flox/Δ1 Cre+ and wild-type samples, although in this case the difference was not statistically significant (P = 0.088). Overall, these histological analyses suggest that Hus1 deficiency in the mammary gland results in genome damage that impairs cell viability, while cells that escape recombination and retain wild-type Hus1 compensate to maintain the structure of the developing gland.
p53 Loss Delays the Clearance of Hus1-Deficient Cells from the Mammary Gland but Impairs Mammary Gland Structure and Function.
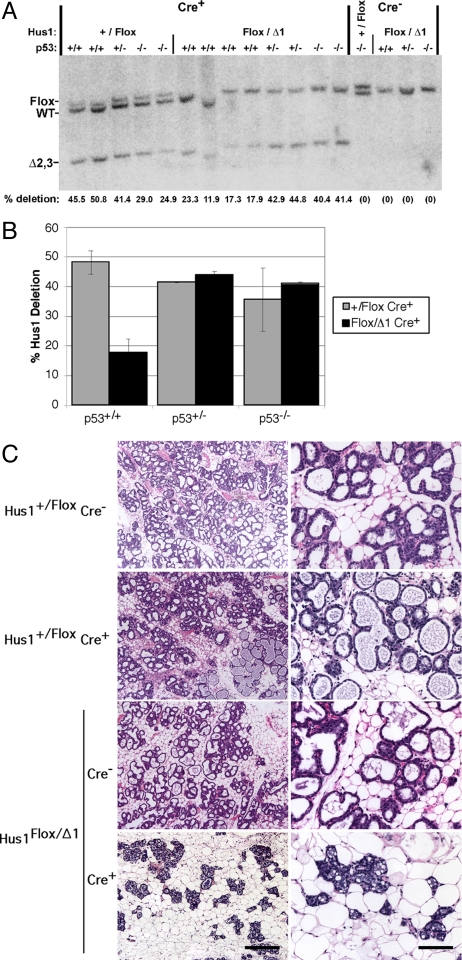
Hus1 loss results in genome damage that can trigger Hus1-independent DNA damage responses (25, 34). Because p53 induction is observed following Hus1 inactivation in embryos and cultured fibroblasts (35), we hypothesized that the increased apoptosis in Hus1-deficient mammary tissue may be due to a p53-dependent checkpoint response to genome damage that occurs following Hus1 loss. We therefore tested whether Hus1-deficient cells would be retained in the mammary gland in the absence of p53 by crossing conditional Hus1 knockout mice onto a p53-deficient background. Southern blot analysis of mammary glands from control Hus1+/Flox Cre+ mice on p53−/− and p53+/− backgrounds revealed levels of Hus1 deletion similar to those in Hus1+/Flox Cre+ p53+/+ controls (Fig. 3 A and B and Fig. S3). Notably, Hus1Flox/Δ1 Cre+ mice on p53−/− and p53+/− backgrounds showed deletion levels similar to those of Hus1+/Flox Cre+ controls, suggesting that inactivation of even one p53 allele reduced the loss of Hus1-deficient cells from the mammary gland. At L2, p53−/− conditional Hus1 knockout mammary glands showed an average of 41% deletion, a 2.3-fold increase in Hus1 deletion as compared to that in p53+/+ conditional Hus1 knockouts. Likewise, p53-deficiency caused a 2.2-fold increase in Hus1 deletion in the p53-deficient conditional knockout glands at P18 (Fig. S3), and similar results also were observed at L4 (Fig. S4A).
Fig. 3.
Increased retention of Hus1-null cells in mammary glands from p53-deficient conditional Hus1 knockout mice. (A) Southern blot analysis was performed to determine the extent of Hus1 deletion in mammary glands of conditional Hus1 knockout mice that differ in p53 status. DNA was extracted from mammary glands of mice of the indicated genotypes at L2 and probed by Southern blot as described in Fig. 1. (B) The percentage of Hus1 deletion for each genotype was calculated as the average from the following number of mice: Hus1+/Flox Cre+ p53+/+ (n = 2), Hus1Flox/Δ1 Cre+ p53+/+ (n = 4), Hus1+/Flox Cre+ p53+/− (n = 1), Hus1Flox/Δ1 Cre+ p53+/− (n = 2), Hus1+/Flox Cre+ p53−/− (n = 4), and Hus1Flox/Δ1 Cre+ p53−/− (n = 2). Error bars, standard deviation. Hus1 deletion was significantly lower in Hus1Flox/Δ1 Cre+ mice relative to Hus1+/Flox Cre+ controls in the p53+/+ background (P = 0.006) but not in the p53−/− background (P = 0.839) as determined by one-way ANOVA. (C) Representative histological sections of mammary glands from p53-deficient conditional Hus1 knockout and control mice are shown. [Scale bars, 400 μm for low-magnification images (Left); 100 μm for high-magnification images (Right).]
To determine the effects of combined Hus1 and p53 loss on mammary gland development and morphology, we prepared whole mounts and histological sections from the mammary glands of these mice at P18 and L2. Germline p53−/− mice exhibit normal mammary gland growth and morphogenesis during pregnancy and lactation (36). These mice typically do not develop mammary neoplasms, but succumb to other malignancies. In agreement with these previous findings, grossly normal mammary gland growth and morphogenesis was observed at P18 and L2 in Hus1+/Flox Cre− p53−/−, Hus1+/Flox Cre+ p53−/−, and Hus1Flox/Δ1 Cre− p53−/− control mice (Fig. 3C and Fig. S3 B and C). However, histological analysis of Hus1Flox/Δ1 Cre+ p53−/− mammary glands at P18 unexpectedly showed delayed lobuloalveolar development and fewer lipid vacuoles (Fig. S3B). These defects were even more striking at L2 (Fig. 3C and Fig. S3C) and L4 (Fig. S4B), at which point the mammary epithelium appeared sparser and the formation of milk-filled alveoli was impaired. Quantification of epithelial content revealed that Hus1Flox/Δ1 Cre+ p53−/− mammary glands contained 2.2-fold less epithelium than those from Hus1+/Flox Cre+ p53−/− control mice (P < 0.001) (Fig. S5). Consistent with these findings, offspring from four Hus1Flox/Δ1 Cre+ p53−/− females showed delayed growth relative to cross-fostered littermates and in three of four cases failed to survive to weaning age, suggesting that these females were incapable of normal lactation. Subsequent litters from these females showed less severe phenotypes, and in one case, apparently normal pups were obtained, presumably because the involution defects of p53-deficient mice (36) resulted in retention of mammary epithelium into subsequent pregnancies.
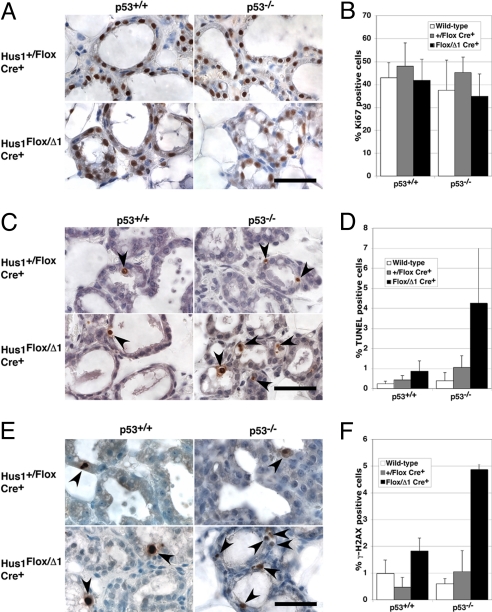
To determine how p53 loss affected the proliferation and survival of mammary epithelium, we performed TUNEL and Ki67 staining on histological sections from p53-deficient conditional Hus1 knockout mice at L2 (Fig. 4). There was no significant difference in proliferation between conditional Hus1 knockout and control mice on either p53+/+ or p53−/− backgrounds (Fig. 4 A and B). Interestingly, the 2-fold increase in apoptosis observed in p53+/+ conditional Hus1 knockout mammary glands as seen in Fig. 2 was elevated to a 4-fold increase in the absence of p53 (Fig. 4 C and D). The significantly increased apoptosis in the p53-deficient conditional Hus1 knockout as compared to p53-deficient Hus1+/Flox Cre+ control mice (P = 0.021) also was associated with a significantly greater level of H2AX phosphorylation (P < 0.001) (Fig. 4 E and F). Similar results were observed in mammary glands at L4 (Fig. S4 C–E). Together, the data suggest that while p53 loss delays the clearance of Hus1-deficient cells from the mammary gland, these cells do undergo apoptosis through a p53-independent pathway that responds to unrepaired DNA damage. In contrast to what happens in p53+/+ conditional Hus1 knockout mammary glands, tissue regeneration by cells that have not undergone Hus1 deletion is limited in the absence of p53, resulting in impaired mammary gland development and function.
Fig. 4.
Increased genome damage and apoptosis in mammary glands from p53-deficient conditional Hus1 knockout mice. Sections from the fourth mammary gland of p53-deficient conditional Hus1 knockout mice at L2 were stained for Ki67 to assess proliferation, by TUNEL assay to detect apoptosis, or for γ-H2AX to detect DNA damage. Representative images of mammary glands stained for (A) Ki67, (C) TUNEL, or (E) γ-H2AX are shown. Arrows highlight positively-stained cells. (Scale bars, 40 μm.) Bar graphs show the average percentage of cells positive for (B) Ki67, (D) TUNEL, or (F) γ-H2AX staining, with error bars denoting standard deviation. Wild-type refers to Hus1+/+ Cre− p53+/+ and Hus1Flox/Flox Cre− p53+/+ or Hus1+/Flox Cre− p53−/−. Values for p53−/− mice are the mean of at least six fields from two animals. Data for Ki67, TUNEL, and γ-H2AX staining on sections from p53+/+ mice are the same as those shown in Fig. 2. The difference between the frequency of positively stained cells in Hus1Flox/Δ1 Cre+ p53−/− and Hus1+/Flox Cre+ p53−/− mice was significant for TUNEL (P = 0.021) and H2AX (P < 0.001) but not Ki67 (P = 0.059) assays as determined by Student's t-test.
Discussion
Hus1 is an essential checkpoint gene, inactivation of which results in spontaneous chromosomal aberrations and embryonic lethality in mice. The ability to conditionally delete Hus1 in adult mice using Cre-loxP recombination allowed us to circumvent this lethality and investigate the physiological consequences of checkpoint dysfunction specifically in the mammary gland. Southern blot analysis revealed that Hus1-deficient cells were under-represented in mammary glands from Hus1Flox/Δ1 Cre+ mice at all developmental stages and, instead of accumulating over multiple rounds of pregnancy and repeated Cre induction, were entirely absent from multiparous females. Mammary glands from conditional Hus1 knockout mice exhibited increased DNA damage and apoptosis, suggesting that Hus1-null cells were selected against due to impaired genome maintenance. Together with previous observations that Hus1 inactivation has severe consequences on genomic integrity in embryos and embryonic fibroblasts (25, 34), these data further establish a fundamental requirement for Hus1 in responding to spontaneous genome damage, even in the absence of extrinsic genotoxic stresses.
Importantly, conditional Hus1 knockout females were not predisposed to mammary tumor formation. This finding is consistent with the fact that complete inactivation of Atr pathway components has not been observed in human cancers. In mice, nullizygous mutations in Atr, Chk1, Hus1, or Rad9 cause embryonic lethality (20, 22, 25, 26). Conditional inactivation of these genes also has not been reported to result in tumor predisposition. Chk1 deletion in the mammary glands of Chk1Flox/Flox Wap-Cre+ mice leads to loss of proliferative potential and clearance of Chk1-deficient epithelium by apoptosis (21), similar to the effects of Hus1 deficiency we observed. In thymocytes, Chk1 deletion causes apoptosis and cell loss, but is not associated with spontaneous tumorigenesis or leukemogenesis (37). Atr adult mosaic knockout mice similarly are not tumor prone (38). Partial inactivation of the Atr pathway, however, may have tumor-promoting effects. Deletion of one Chk1 allele causes inappropriate S-phase entry and other features common to cancer cells (21), and also moderately enhances mammary tumor predisposition in WNT-1 transgenic mice (22). In addition, Atr+/− mice develop various neoplasms at a low frequency (20). On the other hand, mice with a partial reduction in Hus1 expression are not prone to spontaneous tumor development, despite increased genomic instability (39). Clearly, additional studies are required to fully resolve the importance of Hus1 and the Atr checkpoint pathway in tumorigenesis.
Fifty to sixty percent of cells in the mammary glands of control Hus1+/Flox Cre+ mice underwent Cre-mediated Hus1 deletion. Depletion of an equivalent number of cells from the mammary glands of conditional Hus1 knockout mice would be predicted to result in significant morphological defects. Yet, Hus1Flox/Δ1 Cre+ mammary glands were morphologically indistinguishable from controls, suggesting that cells that failed to undergo Hus1 deletion compensated for the loss of Hus1-deficient cells and promoted normal mammary gland development. Similarly, Atr deletion in adult mice by Cre-loxP recombination leads to widespread cell loss, followed by the recovery of cellularity in most tissues due to expansion of cells retaining Atr expression (38). Apoptosis-induced compensatory proliferation is a well-established phenomenon known to occur in Drosophila and mammals (8, 10, 40, 41). Dying cells, as well as the phagocytes that clear them, signal to surrounding cells via mitogens and immune modulators (40–42). The relatively normal mammary gland architecture in conditional Hus1 knockout mice reflects the action of powerful homeostatic mechanisms that are capable of regenerating a functional mammary gland following extensive cell loss.
Based on the central role of p53 in the apoptotic response to DNA damage (5), we hypothesized that p53 was responsible for the elimination of Hus1-deficient cells from the mammary gland. In mouse embryos, Hus1 deficiency triggers p53-dependent induction of p21 and Perp, indicating that p53 is activated by genome damage induced by Hus1 loss (35). In this study, Hus1-deficient cells were retained in the mammary gland longer in the absence of p53, suggesting that p53 normally contributes to the death and/or clearance of Hus1-deficient cells. Although p53 loss did reduce the clearance of Hus1-deficient cells from the mammary gland at P18, L2, and L4, the combined loss of p53 and Hus1 ultimately resulted in significantly increased DNA damage and apoptosis in the mammary gland. The increased TUNEL staining observed in p53-deficient conditional Hus1 knockout mice may occur because cell death becomes restricted to a narrower time frame, or because dying cells accumulate due to inefficient clearance. Alternatively, p53 deficiency may cause an even greater amount of genome damage in Hus1-null mammary epithelial cells, resulting in elevated apoptosis. The latter possibility is supported by the prior observation that p53 loss increases genomic instability in primary conditional Hus1 knockout fibroblasts (34).
These results add to the emerging picture that dysfunction of the Atr checkpoint pathway triggers p53-independent apoptotic responses. For instance, Chk1-deficient embryos undergo p53-independent apoptosis (22). Similarly, IR or replication stress in the absence of Chk1 trigger apoptosis through p53-independent pathways that are dependent on caspase-2 or caspase-3, respectively (43, 44). Here we show that Hus1 loss sensitizes p53-deficient cells to apoptosis in vivo, without additional exogenous stresses. Altogether, the data presented here suggest that Hus1-deficient cells in the developing mammary gland are targeted for apoptosis even when p53 is absent, a process that eliminates cells that are at risk for malignant transformation due to increased genomic instability. While this manuscript was in preparation, Greenow and colleagues published similar results indicating that conditional Chk1 inactivation in the small intestine causes p53-independent apoptosis followed by compensatory proliferation by remaining Chk1-proficient cells (45).
It follows that Hus1 impairment also may reduce cancer cell viability. Interestingly, tumor-initiating cells from p53-null mammary tumors show up-regulation of Hus1 and other DNA damage response genes, suggesting that these cancer stem cells may be highly dependent upon checkpoint functions (46). Inactivation of Hus1 or other Atr checkpoint pathway components therefore represents a potential strategy to sensitize tumors, particularly those harboring p53 mutations, to DNA-damaging chemotherapeutics. The Chk1 inhibitors currently undergoing clinical testing represent one promising means to accomplish this, and evidence from cell culture models suggests that these agents are indeed effective in cells with mutant p53 (47, 48).
Among the most striking findings from this study was that p53 deficiency greatly limited compensatory tissue regeneration by cells that escaped recombination of the conditional Hus1 allele, resulting in substantial morphological defects in the mammary glands of p53−/− conditional Hus1 knockout mice. In p53+/+ conditional Hus1 knockout mice, p53-induced apoptosis and clearance of Hus1-deleted cells during mammary development may signal surrounding cells to regenerate a functional mammary gland. This could be a physical cue, such as the creation of space for the expansion of neighboring cells, or a biochemical signal, such as the production of a secreted mitogen, that is lacking in the p53-deficient genetic background. Such a mechanism has been elucidated in Drosophila, where p53 is required for compensatory proliferation in damaged tissues containing dying cells (10). An alternative model is that the damaged Hus1-deficient cells that accumulate in the absence of p53 secrete a dominant inhibitory signal that acts on surrounding cells. Cells that are induced to senesce by genotoxic stress are known to produce various secreted factors, some of which are growth inhibitory, and a recent study indicates that the senescence-associated secretory phenotype is more vigorous in the absence of p53 (49). These mechanisms are not mutually exclusive; in p53-deficient conditional Hus1 knockout mammary glands, compensatory growth may be prevented through a combination of the absence of a regenerative signal and the presence of a dominant inhibitory signal. The conditional Hus1 knockout mouse model described here will be a valuable tool for deciphering the precise molecular mechanism underlying the role for p53 in mammary gland tissue regeneration and homeostasis.
Materials and Methods
Mice.
Details of the mouse strains used are provided in the SI Text. Mice were housed in accordance with institutional animal care and use guidelines.
Southern Blot.
Deletion of the conditional Hus1 allele was quantified by Southern blotting as described in SI Text.
Whole Mounts and Histology.
Mammary glands from conditional Hus1 knockout and control mice were subjected to whole mount and histological analysis as described in SI Text.
Immunohistochemistry and TUNEL Staining.
Mammary gland sections were stained by TUNEL assay or with antibodies specific for Ki67 or γ-H2AX as described in SI Text.
Supplementary Material
Acknowledgments.
We thank Phil Leder for support during the early stages of this project; Eric Brown for helpful discussions and sharing unpublished results; Alex Nikitin, Yves Boisclair, Joe Wakshlag, and members of the Weiss laboratory for valuable suggestions and comments on the manuscript; and the staff of the Cornell Center for Animal Resources and Education and Laboratory Animal Services for excellent animal care. This work was supported by National Institutes of Health Grant R01 CA108773 and an American Cancer Society Institutional Exploratory Research Grant (to R.S.W.). S.A.Y. was supported by a Department of Defense Breast Cancer Research Program predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904965106/DCSupplemental.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Bartek J, Lukas J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Soussi T. p53 alterations in human cancer: More questions than answers. Oncogene. 2007;26:2145–2156. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- 5.Rodier F, Campisi J, Bhaumik D. Two faces of p53: Aging and tumor suppression. Nucleic Acids Res. 2007;35:7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 8.Valentin-Vega YA, Okano H, Lozano G. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 2008;15:1772–1781. doi: 10.1038/cdd.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 10.Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol. 2006;16:1606–1615. doi: 10.1016/j.cub2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartek J, Bartkova J, Lukas J. DNA damage signaling guards against activated oncogenes and tumor progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 12.Vassileva V, Millar A, Briollais L, Chapman W, Bapat B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002;62:4095–4099. [PubMed] [Google Scholar]
- 13.Menoyo A, et al. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 14.Bertoni F, et al. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer. 1999;26:176–180. [PubMed] [Google Scholar]
- 15.de la Torre J, et al. Expression of DNA damage checkpoint protein Hus1 in epithelial ovarian tumors correlates with prognostic markers. Int J Gynecol Pathol. 2008;27:24–32. doi: 10.1097/pgp.0b013e31812dfaef. [DOI] [PubMed] [Google Scholar]
- 16.Zhu A, Zhang CX, Lieberman HB. Rad9 has a functional role in human prostate carcinogenesis. Cancer Res. 2008;68:1267–1274. doi: 10.1158/0008-5472.CAN-07-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verlinden L, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res. 2007;67:6574–6581. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 18.Cheng CK, Chow LW, Loo WT, Chan TK, Chan V. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res. 2005;65:8646–8654. doi: 10.1158/0008-5472.CAN-04-4243. [DOI] [PubMed] [Google Scholar]
- 19.Maniwa Y, et al. Accumulation of hRad9 protein in the nuclei of nonsmall cell lung carcinoma cells. Cancer. 2005;103:126–132. doi: 10.1002/cncr.20740. [DOI] [PubMed] [Google Scholar]
- 20.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 21.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 23.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9–1-1 for DNA damage: The Rad9-Hus1-Rad1 (9–1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 24.Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 26.Hopkins KM, et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitt PS, Liu H, Manning C, Weiss RS. Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics. 2005;86:212–224. doi: 10.1016/j.ygeno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Selbert S, et al. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 1998;7:387–396. doi: 10.1023/a:1008848304391. [DOI] [PubMed] [Google Scholar]
- 29.Weiss RS, Kostrub CF, Enoch T, Leder P. Mouse Hus1, a homolog of the Schizosaccharomyces pombe hus1+ cell cycle checkpoint gene. Genomics. 1999;59:32–39. doi: 10.1006/geno.1999.5865. [DOI] [PubMed] [Google Scholar]
- 30.Silberstein GB. Postnatal mammary gland morphogenesis. Microsc Res Tech. 2001;52:155–162. doi: 10.1002/1097-0029(20010115)52:2<155::AID-JEMT1001>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 31.Heatwole VM. TUNEL assay for apoptotic cells. Methods Mol Biol. 1999;115:141–148. doi: 10.1385/1-59259-213-9:141. [DOI] [PubMed] [Google Scholar]
- 32.Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: Histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Zhu M, Weiss RS. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol Biol Cell. 2007;18:1044–1055. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss RS, Matsuoka S, Elledge SJ, Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Curr Biol. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- 36.Jerry DJ, et al. Delayed involution of the mammary epithelium in BALB/c-p53null mice. Oncogene. 1998;17:2305–2312. doi: 10.1038/sj.onc.1202157. [DOI] [PubMed] [Google Scholar]
- 37.Zaugg K, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci USA. 2007;104:3805–3810. doi: 10.1073/pnas.0611584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levitt PS, et al. Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol Cell Biol. 2007;27:2189–2201. doi: 10.1128/MCB.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandivier RW, Henson PM, Douglas IS. Burying the dead: The impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest. 2006;129:1673–1682. doi: 10.1378/chest.129.6.1673. [DOI] [PubMed] [Google Scholar]
- 41.Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–521. doi: 10.1038/nature01991. [DOI] [PubMed] [Google Scholar]
- 43.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidi S, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenow KR, Clarke AR, Jones RH. Chk1 deficiency in the mouse small intestine results in p53-independent crypt death and subsequent intestinal compensation. Oncogene. 2009;28:1443–1453. doi: 10.1038/onc.2008.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashwell S, Zabludoff S. DNA damage detection and repair pathways–recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–4037. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–2794. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 49.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.