Abstract
Bacterial alkaline phosphatases (APases) are important enzymes in organophosphate utilization in the ocean. The subcellular localization of APases has significant ecological implications for marine biota but is largely unknown. The extensive metagenomic sequence databases from the Global Ocean Sampling Expedition provide an opportunity to address this question. A bioinformatics pipeline was developed to identify marine bacterial APases from the metagenomic databases, and a consensus classification algorithm was designed to predict their subcellular localizations. We identified 3,733 bacterial APase sequences (including PhoA, PhoD, and PhoX) and found that cytoplasmic (41%) and extracellular (30%) APases exceed their periplasmic (17%), outer membrane (12%), and inner membrane (0.9%) counterparts. The unexpectedly high abundance of cytoplasmic APases suggests that the transport and intracellular hydrolysis of small organophosphate molecules is an important mechanism for bacterial acquisition of phosphorus (P) in the surface ocean. On average, each marine bacterium possessed at least one suite of uptake of glycerol phosphate (ugp) genes (e.g., ugpA, ugpB, ugpC, ugpE) for dissolved organic phosphorus (DOP) transport, but only half of them had ugpQ, which hydrolyzes transported DOP, indicating that cytoplasmic APases play a role in hydrolyzing transported DOP. The most abundant heterotrophic marine bacteria, α- and γ-Proteobacteria, might hydrolyze DOP outside the cytoplasmic membrane, but the former could also transport and hydrolyze DOP in the cytoplasm. The abundant extracellular APases could provide bioavailable P for organisms that cannot directly access organophosphates, and thereby increase marine biological productivity and diversity.
Keywords: PhoD, PhoX, PhoA, uptake of glycerol phosphate, protein localization
Phosphorus (P) is an essential element for life, and P cycling is intimately linked to carbon and nitrogen dynamics in the ocean (1). Although inorganic phosphate (Pi) is the preferred P source for microbial growth (2), it frequently becomes depleted in surface waters of many oceanic regions (2, 3). Dissolved organic phosphorus (DOP) dominates the total dissolved P pool in the surface ocean (4). Thus, the ability to use the DOP pool would be ecologically advantageous for marine microorganisms.
Alkaline phosphatases (APases) occur in a broad diversity of microorganisms and are important in the utilization of phosphoesters, one of the most abundant groups of DOP compounds in the ocean (5). To date, at least 3 prokaryotic APase gene families have been recognized (i.e., PhoA, PhoD, PhoX). They differ in substrate specificity and requirements of specific metal ions for their activities [supporting information (SI) Table S1]. Many marine microorganisms use APase enzymes to release Pi from phosphoesters, and thereby fulfill their P requirement for growth and reproduction (6).
APases have been reported primarily to be periplasmic in Gram-negative bacteria (7–10), but they also occur on the cell surface and extracellularly (11, 12). Because of their critical ecological role in organic P processing, efforts have been made to distinguish periplasmic and cell surface APase activities in marine bacteria (7). Quantification of the subcellular localizations of APases would provide valuable insights about the ecology of marine bacteria and enhance our understanding of the marine P cycle.
The Global Ocean Sampling (GOS) metagenomic database (13) provides an opportunity to investigate the distribution patterns of subcellular localizations of APases in the marine bacterial community. In this study, we developed a bioinformatics pipeline to identify APase peptide sequences and designed an algorithm to predict their subcellular localizations. The ecological and biogeochemical significance of our findings is discussed.
Results and Discussion
APase Distribution in World Ocean.
A total of 935 PhoA, 887 PhoX, and 1,911 PhoD homologs were identified in this survey of GOS sampling sites. Normalizing APases to recA, a single-copy gene (14), indicated 0.50 and 0.32 APase per genome in the open ocean and coastal waters. This is consistent with previous observations in isolates in which 63% of open ocean bacterial isolates expressed APase compared with only 11–53% of coastal isolates (15, 16).
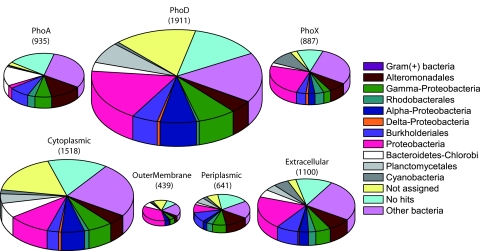
MEtaGenome Analyzer (MEGAN) (17) analysis binned the majority of APases as uncharacterized taxonomic groups (Fig. 1). This is in contrast to species distribution data, in which only 16.3% of the 16S rRNA was attributed to unclassified Proteobacteria and other bacteria (13). The substantial amount of unclassified APases indicated that APases in marine bacteria were highly diverged from their counterparts in characterized bacteria. Although α-Proteobacteria are more abundant than γ-Proteobacteria in the GOS samples (13), the latter made a greater contribution to the APase gene pool (Fig. 1). Planctomycetes accounted for 0.1% of the bacteria in the GOS samples, but they appeared to be an important source of APases (Fig. 1). In contrast, few characterized APases were affiliated with Gram-positive bacteria (Fig. 1), which represent 12% of bacteria in the GOS samples (13). Alteromonadales, Burkholderiales, and Rhodobacterales were consistently represented in all three APase families, whereas Planctomycetes, Bacteroidetes, and Cyanobacteria were overrepresented in PhoD, PhoA, and PhoX, respectively (Fig. 1). All six phylotypes were well represented in cytoplasmic, periplasmic, and extracellular APases (Fig. 1).
Fig. 1.
Taxonomic distribution of APase genes recovered from the GOS metagenomic database. APases were sorted by family type and by subcellular localizations. The chart size indicates the relative abundance of APases, and the number of genes is shown. The γ-Proteobacteria category does not include Alteromonadales, which is shown separately. The α-Proteobacteria category does not include Rhodobacterales, which is shown separately. Proteobacteria only include those APase genes that cannot be assigned to any known phylotypes within Proteobacteria.
Examining the distribution of the individual APase families in the 46 open ocean and coastal water samples (Dataset S1), the mean numbers of PhoA and PhoX per genome were not significantly different (t test, P = 0.09), whereas the mean number of PhoD per genome was significantly greater than the mean number of either PhoA or PhoX per genome (t test, P < 10−6 in either case). The mean number of each APase type per genome was greater in the open ocean than in coastal waters, but these differences were not significant (t test, P > 0.05). Strong correlations were found between the number of PhoA, PhoD, and PhoX, respectively, and the total number of sequences sampled (r = 0.94, 0.91, and 0.92, respectively) (Dataset S1), indicating that each APase type is uniformly distributed across a variety of open ocean and coastal waters.
A recent study showed that PhoX is more abundant in the ocean than previously considered (18). We observed that PhoD is even more abundant than PhoX across a wide variety of marine habitats. Moreover, 4 pairs of PhoX and PhoD peptides were mapped to four paired reads and one pair of PhoX and PhoD was mapped to a single read (Dataset S2), indicating that PhoX and PhoD can co-occur in marine bacteria. Both PhoX and PhoD are activated by Ca2+, an abundant ion in the ocean, whereas PhoA requires Zn2+ (Table S1), which often occurs at subnanomolar concentrations (18, 19). The replacement of Zn2+ with Ca2+ could be an important factor in the selection of PhoX and PhoD over PhoA in the ocean. We applied a loose but reliable searching criterion and identified more PhoA and PhoX homologs than previously shown (18). The abundant nature of PhoD genes suggests that they may play an important role in organophosphate hydrolysis in the surface oceans.
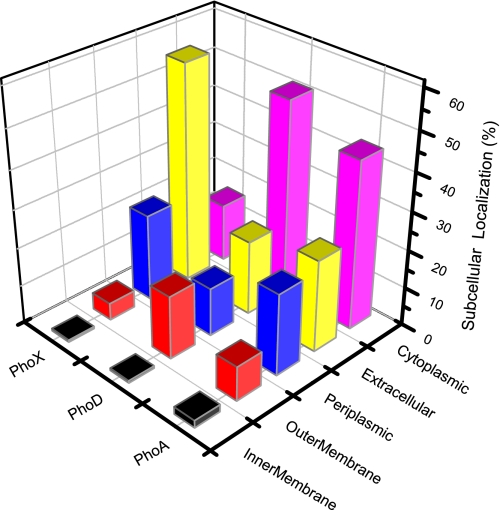
We then examined the subcellular localization of the hydrolysis of organophosphates. The MetaP algorithm was applied to the GOS datasets to sort APase sequences by their subcellular localizations. In total, there were 1,518 cytoplasmic, 641 periplasmic, 1,100 extracellular, 439 outer membrane, and 35 inner membrane APases. The mean number of cytoplasmic APases per cell across the open ocean and coastal waters was significantly greater than that of any other localization type per cell (t test, P < 0.001 in each case). The mean number of extracellular APases per cell was slightly greater than the mean number of the ectoenzymatic APases per cell, but this difference was not significant (t test, P = 0.50). Our finding of more periplasmic than outer membrane APases per cell is consistent with experimental evidence showing that more APase activity was in the periplasm than in the cell surface (7), but this difference was not significant (t test, P = 0.28). No significant differences were found between the mean number of each localization type per genome in the open ocean and in coastal waters (t test, P > 0.05). There was a significant correlation between the number of cytoplasmic APases and the total number of sequences sampled from the open ocean and coastal waters (r = 0.81). The same pattern was found for periplasmic and extracellular APases (r = 0.93 and 0.94, respectively). Membrane proteins were not included in the correlation analysis because of their low abundance. This indicates that the subcellular distribution of APases is generally homogeneous between the open ocean and coastal waters.
Examining individual APase families revealed different patterns in their subcellular localization. In both PhoA and PhoD, cytoplasmic proteins dominate over other proteins (one-sample proportion test, P < 0.05), whereas extracellular proteins comprised the majority of PhoX (one-sample proportion test, P < 0.05) (Fig. 2). The numbers of periplasmic proteins in both PhoA and PhoX were greater than in outer membrane proteins, although the inverse was observed for PhoD (Fig. 2). These patterns indicate PhoA, PhoD, and PhoX could be employed in different ecological strategies for using organophosphate in P-depleted surface waters.
Fig. 2.
Subcellular localization distributions of APases recovered from the GOS metagenomic database.
Cytoplasmic APases and Uptake of Gllycerol Phosphate System for DOP Uptake.
A substantial fraction (41%) of APases from the GOS database was located in the cytoplasm. Although no experimental evidence has shown the existence of cytoplasmic APases in isolated bacteria, it is possible that a greater diversity of APase subcellular localizations and metabolic pathways occurs in the ocean, because the GOS study found that over half of the 16S rRNA ribotypes at the species level (<97% identical) were not reported previously (13). A substantial number of highly diverged APases in the GOS database indicate that APases in marine bacteria could have evolved other functions (e.g., subcellular localizations).
Many bacteria possess the ability to take up monoesters and diesters of glycerol phosphate directly via the uptake of glycerol phosphate (ugp) system in P-limiting environments (20). Glycerol phosphate is the diacylation product of phospholipids (20), which are ubiquitous in bacteria and eukarya, accounting for ≈15–20% of total cellular P (1). In Escherichia coli, the ugp system consists of ugpB, ugpA, ugpE, ugpC, and ugpQ (20). UgpB is a specific binding protein, and ugpA and ugpE are membrane-imbedded proteins. UgpC is an ATP-binding protein that has a strong homology with the functionally exchangeable protein (MalK) for maltose transport (21). UgpQ is a glycerol phosphoryl phosphodiesterase that only hydrolyzes the assimilated diesters at the inner surface of the cytoplasmic membrane, and thus is not required for glycerol phosphate transport (20).
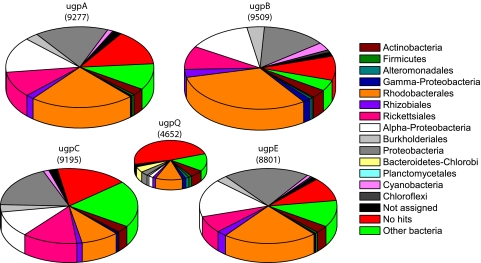
A substantial number of ugp transporter genes were identified in the GOS database (Table S2). Normalizing to the number of genomes represented by the recA gene, we found that each marine bacterial genome contained, on average, at least one suite of the ugp transport genes (Dataset S1), indicating that the ugp system may be prevalent in the ocean. In addition, the number of ugp systems per genome was significantly greater than the number of APases (sum of PhoA, PhoX, and PhoD) per genome (t test, P < 10−16) across the GOS open ocean and coastal water samples, suggesting that the ugp system is more widespread than APases and that direct transport of small molecular DOP could be substantial in the ocean. Moreover, the mean number of ugp transporter genes (sum of ugpA, ugpB, ugpC, and ugpE) per genome in the open ocean was significantly greater than that in coastal waters (t test, P < 0.05), suggesting that transport of glycerol phosphate could be more substantial in the open ocean than in coastal waters. MEGAN analysis revealed that α-Proteobacteria was the dominant phylotype in the ugp gene pool, producing approximately half of the ugp genes, whereas γ-Proteobacteria made limited contributions (Fig. 3). This pattern was in sharp contrast to APase taxonomic distribution (Fig. 1), suggesting that these two most abundant marine bacterial phylotypes (13) mediate DOP utilization by distinct biological mechanisms. Likewise, Bacteroidetes and Planctomycetes were important sources for APases, but they produced few ugp transporter genes.
Fig. 3.
Taxonomic distribution of ugp genes recovered from the GOS metagenomic database. The chart size indicates the relative abundance of ugp genes, and the number of genes is shown. The γ-Proteobacteria and Proteobacteria categories are the same as described for Fig. 1. The α-Proteobacteria category does not include Rhodobacterales, Rhizobiales, and Rickettsiales, which are shown separately.
The function of the ugp system implies that the subcellular localizations of its component proteins are associated with the cytoplasmic membrane. We confirmed this hypothesis by examining ugp proteins from the GOS metagenome. Most metagenomic ugpA (92%) and ugpE (94%) were predicted to be inner membrane proteins. Most ugpC (94%) and ugpQ (97%) were cytoplasmic proteins, and periplasmic proteins comprised the majority (72%) of ugpB (Dataset S3), suggesting that the ugp system is linked to a transport and subsequent cytoplasmic hydrolysis strategy. An intriguing observation was that the number of ugpQ per genome was only about half that of the ugp transporter gene per genome, indicating that many marine bacteria possess ugp transporter genes but lack ugp hydrolyzer genes. Some marine bacteria might substitute cytoplasmic APases for ugpQ. Taxonomic binning of ugpQ and cytoplasmic APases showed that these 2 cytoplasmic phosphoester hydrolyzers dominate in different taxonomic groups (Figs. 1 and 3), which lends strong support for the occurrence of cytoplasmic APases in marine bacteria and strengthens the hypothesis that cytoplasmic APases play a similar role as ugpQ in hydrolyzing transported DOP in marine bacteria. Moreover, the mean sum of cytoplasmic APase and ugpQ per genome is significantly smaller than the mean number of the ugp systems per genome (t test, P < 10−12), indicating that the identified number of cytoplasmic APases could be a conservative estimate. Cytoplasmic APases are likely associated with the inner surface of the cytoplasmic membrane and contribute to intracellular hydrolysis of transported phosphoesters, a scenario observed for ugpQ (20). This avoids hydrolysis of the cytoplasmic phosphoesters that are essential for metabolism.
Ecological Implications for the Ugp System and Cytoplasmic APases.
Utilization of single or multiple phosphoester substrates by a marine microorganism depends on 2 independent mechanisms: the occurrence of periplasmic or cell surface-bound phosphoesterases or the presence of phosphoester transporters embedded in the cytoplasmic membrane (1). However, the relative importance of these pathways remains unknown. Few studies have quantified the direct uptake of phosphoesters by marine bacteria. Our finding of the presence of a large number of phosphoester transporter genes in the GOS metagenome indicates that direct transport of dissolved phosphoesters by bacteria could be an important mechanism of P acquisition in the ocean. A large number of cytoplasmic APases and ugpQ suggests that intracellular hydrolysis of the exogenous DOP might be significant. However, considering the complex cellular metabolism and regulation by internal P compounds in the cytoplasm, these cytoplasmic APases could play a role in internal organophosphate hydrolysis. To support this shift in our changing perception of how marine bacteria acquire P requires additional field and laboratory studies regarding the intracellular fate of small phosphoester molecules in marine bacteria.
APase activity has been widely used as an indicator of P limitation in bacterioplankton (16, 22). The analog substrate 4-methylumbelliferyl phosphate (MUF-P) is commonly used to determine potential APase activity (22). Because MUF-P is not transported across the cytoplasmic membrane, such measurements cannot quantify the activity of cytoplasmic APases. The limitation of current APase activity measurements and the possibility of direct phosphoester uptake could potentially explain the previous observations that phosphate concentrations and APase activity are not inversely related in some aquatic ecosystems (23).
Phosphoesters are probably selectively transported by their uptake systems, because membrane transporters have specificity in substrate transport. Laboratory studies have shown that many phosphoesters cannot be taken up without hydrolysis in the periplasm or cell surface (24). The selective uptake of phosphoesters and potential subsequent intracellular dephosphorylation indicate that cytoplasmic APases could differ in substrate specificity from secreted APases. The wide distribution of bacterial cytoplasmic APases and limitations with current APase activity measurements suggest that marine bacteria may have even a greater role in the phosphoester utilization and the P cycle.
Ectoenzymatic and Extracellular APases.
Ectoenzymes function in the periplasmic space and on the cell surface, whereas extracellular enzymes are released from cells (22). Marine bacterial APases are primarily considered as ectoenzymes rather than extracellular enzymes (7, 22). However, a substantial fraction (30%) of marine bacterial APases appears to be extracellular, indicating that the function of extracellular APases in substrate processing is important. This corresponds well with the results of some laboratory and field studies showing that dissolved APase accounts for ≈30% of total APase activity (16, 25, 26). However, the fraction of extracellular APase activity is highly variable (16, 26), and extracellular APase activity has been considered to occur primarily as a result of grazing, viral lysis, or filtration artifacts. Hence, it is reasonable to propose that the observed cell-free APase activity in the ocean is a combination of signal-targeted APases and APases released as a result of the above factors.
Although we cannot account for the expression and kinetics of these gene products, the subcellular locations of the enzymes have important ecological ramifications. The hydrolytic activities of extracellular APases and subsequent release of Pi can benefit source and neighboring cells, whereas cell-associated APases mainly provide Pi for the source cells that synthesized them. Differences in subcellular localization can lead to a variety of ecological relations. However, it is important to note that such factors as diffusion and a cell's microenvironment may influence the coupling of DOP hydrolysis and Pi uptake regardless of whether the enzyme is cell-associated or cell-free. The occurrence of extracellular APase might be important for Pi utilization by marine microorganisms lacking the enzymatic capability for using DOP in the surface ocean.
Microorganisms are found in much greater abundance on organic aggregates in comparison to the surrounding seawater (27). Attached bacteria actively transform particles, gels, and colloids (28), and even though it has been proposed that “extracellular enzymes” are critical in the degradation of these matrices (28), the subcellular locations of these enzymes remain unclear. Most organic matrices are porous and can effectively retain secreted enzymes, which, in turn, readily decompose the organic matrices. Therefore, it would be ecologically advantageous for attached bacteria to secrete extracellular enzymes. In the GOS Expedition, attached bacteria were largely excluded from collection in most sampling sites. Further studies should examine the subcellular locations of APases in attached marine bacteria.
The computational prediction of APase subcellular localization indicates a potential mechanistic shift in our understanding of bacterial utilization of phosphoesters; however, it is the expression and regulation of the different APases and the DOP transport system that are important. These studies will enhance our understanding of oceanic P cycling and its interconnection with carbon cycling.
Materials and Methods
Recovery of APase and Ugp Peptide Sequences from GOS Metagenomic Databases.
Seawater samples collected from the GOS sampling sites were filtered (>0.1 μm and <0.8 μm) to concentrate marine microorganisms (13). To recover as many homologous sequences as possible, we used 2 seed queries obtained from 2 distantly related bacteria for each APase family (PhoA, PhoD, and PhoX) and ugp genes (ugpA, ugpB, ugpC, ugpE, and ugpQ). The accessions of query sequences are summarized in Table S3. We applied a position-specific iterated BLAST (29) with an expectation value of 0.1 to recover homologous sequences from the GOS database (30). In this step, some similar but nonhomologous sequences were also retrieved.
The retrieved sequences were verified using protein domain databases. We applied a reversed position-specific BLAST (29) to search all the retrieved sequences against a conserved domain database (CDD) (31). For each sequence, only the top hit known as PhoA, PhoD, and PhoX was accepted as an APase. Another putative APase family, PhoV (32), was not used in the analysis because their conserved domain has not been represented in a CDD. The same rule was applied to ugp peptides. The accessions of all APases and ugp peptides in the CDD are listed in Table S4. To identify the duplicate sequences attributable to paired reads, each APase and ugp peptide's J. Craig Venter Institute (JCVI) PEP number was mapped to JCVI Read ID and Mate ID (Datasets S2 and S3). Duplicate sequences were then assembled. All relevant information was parsed by Perl scripts. Ten GOS samples were not included in the analysis (Dataset S1), because organisms other than prokaryotes were collected. Twenty-five open ocean samples and 21 coastal water samples were used for statistical analyses. The open ocean samples from the Tropical South Pacific were not included because of their small sample sizes (Dataset S1). All statistical analyses were performed using the R statistical software package (33). Apparent taxonomic distributions of APases were estimated by MEGAN with the recommended parameter setting (min-score: 100, top-percent: 10%, min-support: 2) (17).
Subcellular Localization Prediction of APases.
Gram-negative bacteria dominate (≈90%) the marine prokaryotic community in the GOS samples (13). Archaea are nearly absent in the GOS samples, which are surface samples (13). Proteins in Gram-negative bacteria have five possible subcellular localizations: cytoplasm, inner membrane, periplasm, outer membrane, and extracellular space. Although a variety of methods are available, for the purpose of predicting fragmentary peptide sequences (e.g., GOS peptides) and discovering unrecognized localizations of APases, only algorithms using amino acid compositional bias are useful, such as CELLO, SUBLOC, and LOCTree.
In some cases, different algorithms make different predictions (Dataset S2). To reconcile this discrepancy, we developed a metaalgorithm, MetaP. It works as follows. Given a protein sequence, its localization predictions from all independent algorithms (CELLO, SUBLOC, and LOCTree) are collected and transformed into a standard format using Perl scripts. In a previous metaalgorithm (34), different locations were regarded as independent classes and performance-based weighted voting was used to summarize predictions. MetaP considers the common properties shared by sorting signals targeting neighboring subcellular locations. We used the following weighted voting to incorporate these neighborhood relations among the subcellular locations as well as suboptimal predictions by base algorithms.
The predicted location of MetaP for a sequence s is the one that has the maximum sum of weighted voting for that subcellular location. The prediction can be denoted formally as Ps = arg maxi Σj=1N P(i, j), where N is the total number of base predictors and i is the index of a predicted subcellular compartment: cytoplasmic (i = 1), cytoplasmic membrane (i = 2), periplasmic (i = 3), outer membrane (i = 4), and extracellular (i = 5). P(i, j) denotes the voting weight of the prediction(s) of the jth element predictor for compartment i. It is defined as P(i, j) = Σk=0Mj 2−|ck−i|·wk, where Mj is the number of predictions of the jth predictor. It means that the voting weight of a prediction by the jth predictor for compartment i depends on the offset of the index ck of its predicted class with regard to the index i as well as its normalized score wk.
The performance of MetaP to predict fragmentary protein sequences was evaluated using sets of testing sequences whose localizations were verified by experiments. We showed that MetaP is an accurate method in predicting cytoplasmic, periplasmic, and extracellular proteins (Table S5).
Among the three base algorithms (CELLO, SUBLOC, and LOCTree), only CELLO can predict inner membrane and outer membrane proteins, whereas membrane proteins must be removed from the datasets before analysis by SUBLOC and LOCTree (35). We showed that CELLO makes accurate predictions for membrane proteins (Table S6). We initially applied CELLO to identify membrane APase peptides, and the remaining nonmembrane sequences were predicted by all base algorithms and MetaP. For ugp proteins, CELLO was used to predict ugpA and ugpE, which are inner membrane proteins, whereas all three base algorithms and MetaP predicted ugpB, ugpC, and ugpQ, which are nonmembrane proteins. Duplicate sequences attributable to paired reads were assembled before prediction. All predictions were collected (Dataset S2). More details are provided in SI Methods.
Supplementary Material
Acknowledgments.
We are grateful to Dr. Mary Ann Moran and Shalabh Sharma (Moran laboratory, University of Georgia), Dr. Winfried Boos (University of Konstanz), Karl Kaiser (Benner laboratory, University of South Carolina), Dr. Hongmei Zhang (University of South Carolina), Dr. Jijun Tang and Yiwei Zhang (Tang laboratory, University of South Carolina), Dr. Tao Tao (National Center for Biotechnology Information), Dr. Bangqin Huang (Xiamen University), Dr. Tian Xiao (Chinese Academy of Sciences), Michael Chiu (Community Cyberinfrastructure for Advanced Marie Microbial Ecology Research and Analysis), Dr. Lukas Käll (Stockholm University), Dr. Aron Marchler-Bauer (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health), Dr. Henrik Nielsen (Technical University of Denmark), Dr. Chin-Sheng Yu (Feng Chia University), Dr. Jenn-Kang Hwang (National Chiao Tung University) and 2 anonymous reviewers' constructive comments. We especially thank Dr. Shruti Rastogi and Guy Yachdav (Rost laboratory, Columbia University) for their packaging and distributing of LOCTree software for us. J.H. was supported by the National Science Foundation (Grant BIO-DBI-0845381). R.A.L was supported by the National Science Foundation (Grant MCB-0729594). All computations were performed on a 128-core shared memory computer of the High Performance Computing Group at the University of South Carolina.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 21013.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907586106/DCSupplemental.
References
- 1.Karl DM, Bjorkman KM. Dynamics of DOP. In: Hansell DA, Carlson CA, editors. Biogeochemistry of Marine Dissolved Organic Matter. Academic; 2002. pp. 249–366. [Google Scholar]
- 2.Karl DM. Phosphorus, the staff of life. Nature. 2000;406:31–32. doi: 10.1038/35017683. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Sunda W, Boyle EA, Karl DM. Phosphate depletion in the western North Atlantic Ocean. Science. 2000;289:759–762. doi: 10.1126/science.289.5480.759. [DOI] [PubMed] [Google Scholar]
- 4.Karl DM, et al. Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA. Deep-Sea Res. 2001;48:1529–1566. [Google Scholar]
- 5.Clark LL, Ingall ED, Benner R. Marine phosphorus is selectively remineralized. Nature. 1998;393:426. [Google Scholar]
- 6.Martinez J, Smith DC, Steward GF, Azam F. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquatic Microbial Ecology. 1996;10:223–230. [Google Scholar]
- 7.Martinez J, Azam F. Periplasmic aminopeptidase and alkaline phosphatase activities in a marine bacterium: Implications for substrate processing in the sea. Mar Ecol Prog Ser. 1993;92:89–97. [Google Scholar]
- 8.Thompson LMM, MacLeod RA. Biochemical localization of alkaline phosphatase in the cell wall of a marine pseudomonad. J Bacteriol. 1974;117:819–825. doi: 10.1128/jb.117.2.819-825.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan HM, Pratt D. Biochemical and physiological properties of alkaline phosphatases in five isolates of marine bacteria. J Bacteriol. 1977;129:1607–1612. doi: 10.1128/jb.129.3.1607-1612.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doonan BB, Jensen TE. Ultrastructural localization of alkaline phosphatase in the blue-green bacterium Plectonema boryanum. J Bacteriol. 1977;132:967–973. doi: 10.1128/jb.132.3.967-973.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von-Tigerstrom RG. Production of two phosphatases by Lysobacter enzymogenes and purification and characterization of the extracellular enzyme. Appl Environ Microbiol. 1984;47:693–698. doi: 10.1128/aem.47.4.693-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole K, Hancock REW. Secretion of alkaline phosphatase and phospholipase C in Pseudomonas aeruginosa is specific and does not involve an increase in outer membrane permeability. FEMS Microbiol Lett. 1983;16:25–29. [Google Scholar]
- 13.Rusch DB, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karl DM, et al. Aerobic production of methane in the sea. Nat Geosci. 2008;1:473–478. [Google Scholar]
- 15.Kobori H, Taga N, Simidu U. Properties and generic composition of phosphatase-producing bacteria in coastal and oceanic seawater. Bulletin of the Japanese Society of Scientific Fisheries. 1979;45:1429–1433. [Google Scholar]
- 16.Hoppe H-G. Phosphatase activity in the sea. Hydrobiologia. 2003;493:187–200. [Google Scholar]
- 17.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebastian M, Ammerman JW. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME Journal. 2009;3:563–572. doi: 10.1038/ismej.2009.10. [DOI] [PubMed] [Google Scholar]
- 19.Lohan MC, Statham PJ, Crawford DW. Total dissolved zinc in the upper water column of the subarctic North East Pacific. Deep-Sea Res. 2002;49:5793–5808. [Google Scholar]
- 20.Brzoska P, Rimmele M, Brzostek K, Boos W. The Ugp paradox: The phenomenon that glycerol 3-phosphate, exclusively transported by the Escherichia coli Ugp system, can serve as a sole source of phosphate but not as a sole source of carbon is due to trans inhibition of Ugp-mediated transport by phosphate. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in Microorganisms Cellular and Molecular Biology. Washington, DC: ASM; 1994. pp. 30–36. [Google Scholar]
- 21.Hekstra D, Tommassen J. Functional exchangeability of the ABC proteins of the periplasmic binding protein-dependent transport systems Ugp and Mal of Escherichia coli. J Bacteriol. 1993;175:6546–6552. doi: 10.1128/jb.175.20.6546-6552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaas BM, Ammerman JW. Automated high resolution ectoenzyme measurements: Instrument development and deployment in three trophic regimes. Limnol Oceanogr. 2007;5:463–473. [Google Scholar]
- 23.Neddermann K, Nausch M. Effects of organic and inorganic nitrogen compounds on the activity of bacterial alkaline phosphatase. Aquatic Ecology. 2005;38:475–484. [Google Scholar]
- 24.Bengis-Garber C, Kushner DJ. Role of membrane-bound 5′-nucleotidase in nucleotide uptake by the moderate Halophile Vibrio costicola. J Bacteriol. 1982;149:808–815. doi: 10.1128/jb.149.3.808-815.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoch MP, Bronk DA. Bacterioplankton nutrient metabolism in the Eastern Tropical North Pacific. J Exp Mar Bio Ecol. 2007;349:390–404. [Google Scholar]
- 26.Labry C, Delmas D, Herbland A. Phytoplankton and bacterial alkaline phosphatase activities in relation to phosphate and DOP availability within the Gironde plume waters (Bay of Biscay) J Exp Mar Bio Ecol. 2005;318:213–225. [Google Scholar]
- 27.Alldredge AL, Cole JJ, Caron DA. Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol Oceanogr. 1986;31:68–78. [Google Scholar]
- 28.Smith DC, Simon M, Alldredge AL, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: A Community Resource for Metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, et al. CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner KU, Masepohl B, Pistorius EK. The cyanobacterium Synecchococcus sp. strain PCC 7942 contains a second alkaline phosphatase encoded by phoV. Microbiology. 1995;141:3049–3058. doi: 10.1099/13500872-141-12-3049. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 34.Liu J, Kang S, Tang C, Ellis LBM, Li T. Meta-prediction of protein subcellular localization with reduced voting. Nucleic Acids Res. 2007;35:e96. doi: 10.1093/nar/gkm562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardy JL, Brinkman FSL. Methods for predicting bacterial protein subcellular localization. Nat Rev Microbiol. 2006;4:741–751. doi: 10.1038/nrmicro1494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.