Abstract
Time of day-dependent variations of immune system parameters are ubiquitous phenomena in immunology. The circadian clock has been attributed with coordinating these variations on multiple levels; however, their molecular basis is little understood. Here, we systematically investigated the link between the circadian clock and rhythmic immune functions. We show that spleen, lymph nodes, and peritoneal macrophages of mice contain intrinsic circadian clockworks that operate autonomously even ex vivo. These clocks regulate circadian rhythms in inflammatory innate immune functions: Isolated spleen cells stimulated with bacterial endotoxin at different circadian times display circadian rhythms in TNF-α and IL-6 secretion. Interestingly, we found that these rhythms are not driven by systemic glucocorticoid variations nor are they due to the detected circadian fluctuation in the cellular constitution of the spleen. Rather, a local circadian clock operative in splenic macrophages likely governs these oscillations as indicated by endotoxin stimulation experiments in rhythmic primary cell cultures. On the molecular level, we show that >8% of the macrophage transcriptome oscillates in a circadian fashion, including many important regulators for pathogen recognition and cytokine secretion. As such, understanding the cross-talk between the circadian clock and the immune system provides insights into the timing mechanism of physiological and pathophysiological immune functions.
Keywords: adrenalectomy, LPS, IL-6, microarray, TNF-α
A 24-h periodicity in the environment has led to the evolution of molecular circadian clocks in organisms ranging from cyanobacteria to humans. Circadian rhythms display a near 24-h period and persist even in the absence of external timing information. In mammals, a small hypothalamic region, the suprachiasmatic nucleus (SCN), has been identified as the master pacemaker regulating circadian rhythms in physiology, metabolism, and behavior (1). Recent evidence shows that also peripheral organs such as liver, heart, kidney, skin, and even cultured cell lines contain circadian oscillators. Although the SCN probably sets the phase of these peripheral clocks (by as yet unknown means), recent reports implicate peripheral clocks in the regulation of local physiology (2–4). The fundamental mechanism of rhythm generation is cell autonomous and highly conserved in SCN and peripheral cells: Interlocked transcriptional/translational feedback loops involving clock genes, such as Per1–3, Cry1–2, Clock, Bmal1, and Rev-Erbα create oscillations on the molecular level (reviewed in ref. 2).
In the immune system, many functions and parameters have been described to be time-of-day dependent, e.g., lymphocyte proliferation (5), natural killer (NK) cell activity (6), humoral immune response (7), rhythms in absolute and relative numbers of circulating white blood cells and their subsets (8), cytokine levels (9), and serum cortisol (10) (reviewed in ref. 11). In addition, time-of-day variation in susceptibility to infection (12), course of disease [rheumatoid arthritis (13) or asthma (14)], parameters in clinical diagnostics as well as pharmacological therapy uncover the integral role the circadian system plays in immunological responses (11). Although both cell-autonomous and systemic pathways have been discussed to relay timing information (15), it is largely unknown how the circadian system and the immune system are communicating. Circadian-controlled humoral factors such as cortisol and melatonin or innervations by the autonomic nervous system may regulate gene expression and protein activity (16), but it is also possible that local clocks in immune cells directly control cellular immune functions. Therefore, the aim of this study was to gain a deeper understanding of the mechanisms regulating circadian immunological rhythms on a cellular as well as on a systemic level. To this end, we systematically investigated (i) whether cells of the immune system possess an autonomous circadian clock, (ii) whether such a clock regulates circadian immune functions, and (iii) to what extent such a clock regulates the expression and possibly also function of immune system components. Answers to those questions will likely change our view on the dynamics and pathophysiology of immune responses.
Here, we first established that spleen, lymph nodes, and isolated macrophages do contain autonomous cellular oscillators that even operate without systemic time information. These clocks regulate circadian rhythms in cytokine secretion upon stimulation with bacterial endotoxin. Using adrenalectomized mice, we show that glucocorticoid hormones are necessary neither for circadian cytokine secretion rhythms nor for the phase of the immune clock. Global transcriptome analysis indicates that circadian regulation of immune functions likely occurs at many levels within regulatory networks of the immune system: ≈8% of all genes expressed in peritoneal macrophages are rhythmically transcribed, including essential elements in the LPS-triggered toll-like-receptor 4 (TLR4)/TNF-α pathway, e.g., the LPS coreceptor Cd14, Mapk14, AP-1 subunits Jun and Fos as well as Adam17.
Results
Circadian Clock Genes Are Rhythmically Expressed in Spleen, Lymph Nodes, and Peritoneal Macrophages.
Circadian clock genes are expressed in a number of immunological compartments (17–19); however, it remains unclear whether they constitute a functional autonomous circadian clockwork able to operate without exogenous Zeitgeber information. To test the former, we first analyzed the expression levels of the key clock components Period2 (Per2) and Rev-Erbα—representatives of two major feedback loops within the circadian clockwork—in spleen and lymph nodes of mice kept in constant conditions. Mice were entrained to a 12-h light/12-h dark (LD) cycle for 2 weeks and then transferred to constant conditions [i.e., constant darkness (DD), water and food ad libitum], and subsequently killed at regular intervals for two consecutive days. Expression of clock genes in spleen, inguinal lymph nodes, and CD11b+ peritoneal macrophages was determined. In spleen and lymph nodes both Per2 and Rev-Erbα transcripts showed high-amplitude circadian oscillations with peak-to-trough ratios of ≈4 for Per2 and ≈20 for Rev-Erbα (Fig. 1). In peritoneal macrophages, amplitudes of Per2 and Rev-Erbα mRNA rhythms were much higher (peak-to-trough ratio of ≈100 and 300, respectively). The expression peaked around circadian time (CT) 6–9 for Rev-Erbα and around CT 12–15 for Per2—similar to other peripheral tissues, such as in liver, heart, or kidney (Table S1 and ref. 17).
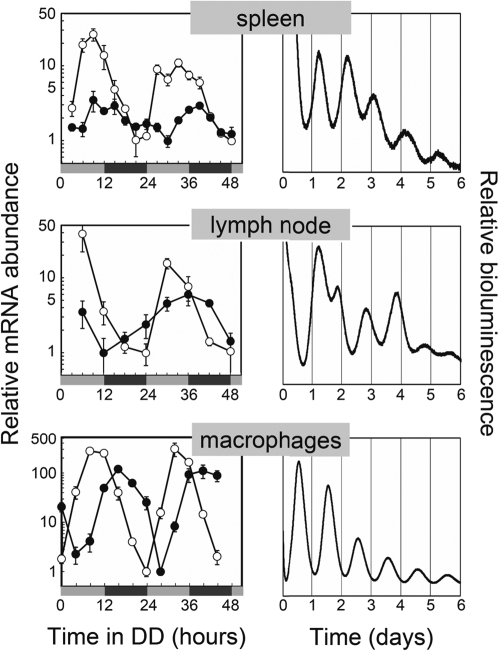
Fig. 1.
Fully competent circadian clocks in tissues and cells of the immune system. (Left) Circadian clock genes Per2 (filled circles) and Rev-Erbα (open circles) are rhythmically expressed in murine spleen cells, lymph nodes, and peritoneal macrophages. Tissues and cells were harvested at regular intervals over the course of the first 2 days after transfer of the mice from a LD cycle to DD. Gray and black bars refer to the previous light and dark periods, respectively. CT 0 corresponds to the time in DD when the light would have turned on in the prior LD cycle. Transcript levels were analyzed by using quantitative RT-PCR. Displayed are the means ± SEM. (spleen: n = 3–6; lymph nodes: n = 3–4 except for n = 2 at CT 6/day 2; macrophages: n = 3–4) normalized to nonoscillating Gapdh expression levels. (Right) Autonomous clock gene oscillation in in vitro conditions. A small piece of spleen, superficial inguinal lymph nodes as well as peritoneal macrophages from PER2::LUC knockin mice (20) were cultured in medium containing luciferin. Circadian bioluminescence was continuously recorded for ≈1 week by using photomultiplier tubes. Representative time series for at least three independent experiments are shown.
To test whether these oscillating clock components are part of an autonomous functional clockwork, we used a knockin mouse model, where—instead of the clock protein PER2—a PER2::LUCIFERASE (PER2::LUC) fusion protein is expressed (20). The circadian system in these PER2::LUC animals is unaffected, therefore circadian properties of various tissues can be analyzed by monitoring bioluminescence emission of tissue explants cultured in constant conditions. Spleen and lymph node explants of these mice show persistent circadian oscillation of bioluminescence for more than a week (Fig. 1). Similar results were obtained by using a related rat model, i.e., the Per1::luc rat (4), where luciferase expression is driven by the circadian Period1 promoter (Fig. S1). To rule out the possibility that these oscillations are solely derived from rhythms of connective tissue cells, we monitored peritoneal macrophages from PER2::LUC mice ex vivo. These cells display a high-amplitude circadian rhythmicity for more than a week (Fig. 1). Together, these results indicate that, in immune cells, a circadian clock is able to operate autonomously without the requirement of systemic drivers.
Circadian TNF-α and IL-6 Secretion Rhythms in Lipopolysaccharide (LPS)-Challenged Spleen Macrophages.
The existence of a cellular clock in tissues and cells of the immune system suggests that this peripheral clock fulfils a local regulatory function as described for other peripheral clocks (e.g., ref. 3). We asked whether this hypothetical regulation might also include immune cell-specific mechanisms like responses to surface receptors triggering cytokine secretion or cellular trafficking. Macrophages were found to represent an interesting model to study these questions: Already in 1960, Halberg and colleagues (21, 22) found that the mortality upon LPS-induced endotoxic shock in mice depends highly on the time of day when LPS is administered, suggesting a circadian clock regulation of macrophage-dependent cytokine secretion.
Therefore, we tested whether induced cytokine secretion from spleen cells (i.e., splenocytes—mononuclear white blood cells extracted from spleen tissue) is regulated by the circadian oscillator using splenic macrophages as a model system. To this end, spleens were harvested at 4-h intervals from C57BL/6 mice kept in constant conditions, and single cell suspensions of the splenocytes were stimulated with LPS. Subsequently, TNF-α and IL-6 levels were quantified in culture supernatants. Both TNF-α and IL-6 secretion of toll-like-receptor 4 (TLR4)-expressing spleen cells (i.e., mostly monocytes/macrophages) displayed a significant circadian oscillation (Fig. 2A) with an ≈2-fold peak-to-trough ratio and a peak phase in the subjective day (i.e., around CT8–12). We can exclude the possibility that these rhythms are primarily due to a time of day-dependent variation in the cellular composition of the spleen, because the amount of TNF-α and IL-6 per macrophage is also rhythmic, yet with a different phase (Fig. 2C). To calculate cytokine secretion per macrophage, we analyzed the cellular composition of the spleen in a time-dependent manner. Interestingly, we found circadian fluctuations in relative number of T cells (CD90+) and monocytes/macrophages (CD11b+/CD14+) as well as in absolute number of B cells and monocytes/macrophages. In addition, the number of total splenocytes was rhythmic in a circadian manner (Fig. 2B), indicating that also immune cell trafficking is under circadian regulation. Because TNF-α and IL-6 levels normalized to monocyte number is still rhythmic, it seems reasonable to assume that cytokine secretion is regulated by the circadian clock within these cells.
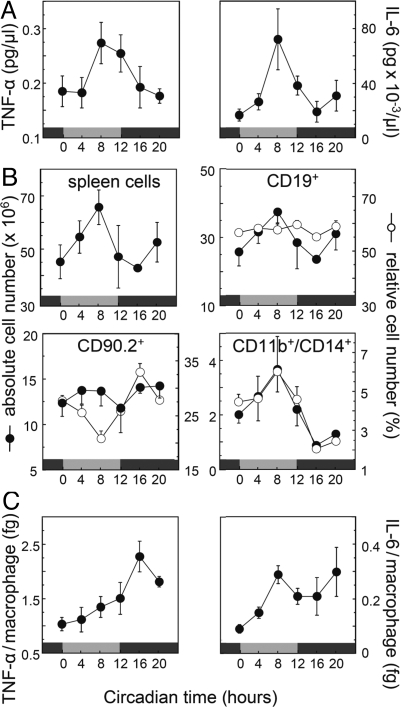
Fig. 2.
Circadian cytokine secretion upon challenge with bacterial endotoxin. (A) Spleens from C57BL/6 mice transferred in DD were harvested at regular 4-h intervals. After stimulation with LPS, TNF-α (Left) and IL-6 (Right) secretion was determined by ELISA. Gray and black bars refer to the previous light and dark periods, respectively. Presented are the means ± SEM (n = 4–5). Circadian oscillations are statistically significant as analyzed with CircWave (TNF-α and IL-6: P < 0.05). (B) Cellular composition of the spleen is time-of-day dependent. The same samples as in A were analyzed with cell-counting chamber and flow cytometry. CD19, CD90.2, and CD11b in combination with CD14 were used as characteristic surface markers of B cells, T cells, and monocytes/macrophages, respectively (for FACS gate settings and surface marker expression levels, see Fig. S4). (C) Cytokine response as in A with respect to numbers of CD11b/CD14-positive spleen cells from B lower right.
Alternatively, rhythmic humoral signals, such as circadian variation in cortisol levels, might primarily control the observed rhythmic cytokine secretion pattern. Cortisol is a particularly plausible candidate because, first, it displays high amplitude circadian rhythms in serum abundance (10), and, second, it has a prominent inhibitory function on proinflammatory cytokine transcription (23). To test a putative regulatory role of glucocorticoid rhythms on the circadian control of cytokine secretion, we performed a second LPS stimulation experiment with spleen cells from adrenalectomized (ADX) mice. These mice are glucocorticoid deficient because of excision of the adrenal glands. As in nonoperated mice, LPS stimulated spleen-derived macrophages of ADX animals also showed a clear, statistically significant circadian pattern in TNF-α and IL-6 secretion with an ≈3-fold peak-to-trough ratio (Fig. 3A). The total levels of TNF-α and IL-6 as well as the phase of oscillation were similar to nonoperated mice (compare Figs. 2C and 3A). These results demonstrate that circadian cytokine response upon LPS stimulation does not depend on circadian cortisol levels but is likely due to functional circadian clocks within immune cells.
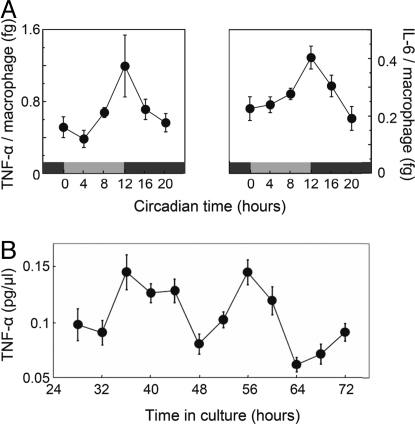
Fig. 3.
A macrophage intrinsic clockwork regulates circadian TNF-α and IL-6 secretion upon LPS stimulation. (A) Circadian modulation of LPS-induced cytokine response is independent of systemic cortisol. Spleens from adrenalectomized C57BL/6 mice were harvested and analyzed as described in Fig. 2. TNF-α and IL-6 cytokine secretion per macrophage was determined via ELISA by taking the absolute number of monocytes/macrophages of the spleen into account (see also Figs. S2 and S4A and Materials and Methods). Circadian oscillations are statistically significant as analyzed with CircWave (P < 0.001 and P < 0.05 for TNF-α and IL-6, respectively). (B) TNF-α response upon LPS stimulation is regulated by a cell-intrinsic, local clock. Spleen cells from 20 C57BL/6 mice were harvested, pooled, and plated for tissue culture. Individual wells were stimulated for 4 h with LPS at indicated times, and supernatants were collected thereafter. TNF-α levels in supernatant were determined by ELISA and tested for statistical significance with CircWave (presented are means ± SEM, n = 15, P < 0.0001).
Still, systemic time cues other than glucocorticoids might drive circadian cytokine output of monocytes/macrophages. To exclude such a possibility, we analyzed TNF-α secretion ex vivo in primary cell culture of splenic and peritoneal macrophages. Every 4 h, one sample of 2-day primary macrophages cultured in parallel were treated with LPS. Four hours later, TNF-α levels in supernatants were determined by ELISA. The results show that cytokine secretion in primary macrophages and primary spleen cell culture is significantly rhythmic in a circadian manner (Fig. 3B and Fig. S3, respectively) underlining the importance of the macrophage clockwork for rhythmic LPS-induced cytokine secretion.
Approximately 8% of the Macrophage Transcriptome Is Under Circadian Regulation.
Regulation of gene expression is a major output mechanism of the circadian clock (24). To investigate how the circadian system interacts with and modulates immune functions, we performed a global circadian gene expression profiling experiment in macrophages. We sorted peritoneal CD11b+ macrophages sampled every 4 h over 48 h. Total RNA from indicated times was pooled and used for microarray-based genome-wide transcriptional profiling. A total number of 17,308 genes were detected to be expressed in peritoneal macrophages. Fourier score analysis for determination of circadian expression patterns identified 1,403 (i.e., 8.1%) genes to be rhythmically expressed in a circadian manner (Fig. 4A and Fig. S6). Among these were, as expected, canonical clock genes such as Bmal1, Cry1, Cry2, Per1, Per2, Per3, Rev-Erbα, and Rev-Erbβ as well as the clock-controlled genes Dbp and Nfil3. These data were confirmed by quantitative PCR analyses (Fig. 4B and Table S1).
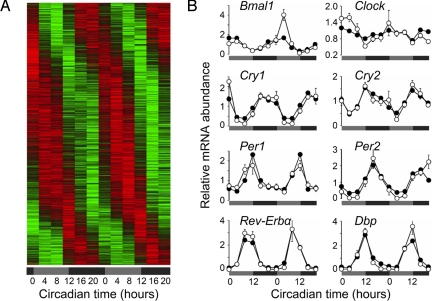
Fig. 4.
Eight percent of all transcripts in macrophages are expressed with a circadian rhythm. (A) Phase-sorted heat map of genes transcribed in a circadian manner in peritoneal macrophages. Cells harvested via peritoneal lavage from four C57BL/6 mice every 4 h were magnetically purified for CD11b surface expression (see Fig. S5). Three individual RNA samples of each time were pooled and subjected to global gene transcription measurement by using Affymetrix chips. The analysis on circadian rhythmicity was done with CircWaveBatch. Lfdrs were determined and cutoff value was arbitrarily set to 0.1 as a measure for rhythmic versus nonrhythmic transcripts (see Fig. S7). Genes expressed in a circadian manner were plotted phase-sorted in a heat-map style (colors indicate min–max normalized relative expression: green, minimum expression; red, maximum expression). (B) Canonical clock gene expression in peritoneal macrophages. Individual datasets from A were plotted (filled circles) together with data obtained by a quantitative RT-PCR assay of the same samples (open circles, means ± SEM, n = 4, except for times CT 24 and 28, n = 3). Statistical analysis for qPCR data and chip data were performed with CircWave and CircWaveBatch, respectively (microarray: P ≤ 0.0001: Bmal1, Cry1, Per2; P ≤ 0.01: Rev-Erbα, Dbp, Cry2; P ≤ 0.05: Per1 and Clock; qPCR: P ≤ 0.0001: Bmal1, Cry1, Per1/2, Clock, Rev-Erbα, Dbp; P ≤ 0.001: Cry2).
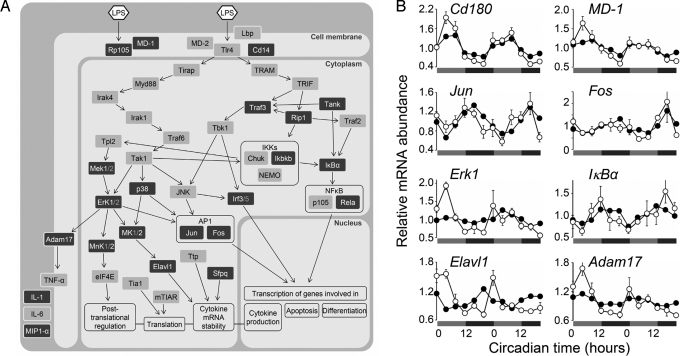
Using these data, we next investigated whether circadian TNF-α release upon LPS stimulation might be controlled by transcriptional regulation of components involved in TLR4 signaling. This pathway is complex and includes transcriptional but also posttranscriptional, translational, and posttranslational events (25). We find circadian transcriptional regulation at every level of LPS-induced immune response (Fig. 5 and Table S2): (i) components regulating LPS binding to TLR4, or the homodimerization of TLR4 (MD-1 and CD180/RP105); (ii) components of the MAPK pathway controlling multiple downstream levels including transcription factor activation (e.g., ERK1) and cytokine protein processing (e.g., MEK1); (iii) subunits and regulatory components of NFκB and AP-1—transcription factors involved in proinflammatory cytokine transcription (NFκB1, RELA, IκBα, JUN, FOS); (iv) components regulating cytokine mRNA stability and localization (ELAVL1, SFPQ) as well as protein processing (ADAM17). The pervasive nature of transcriptional circadian control within this pathway makes it very likely that the observed rhythmic immune functions are at least in part controlled by circadian clocks present in immune cells.
Fig. 5.
Circadian transcription of genes contributing to LPS response. (A) Gene regulatory network forming the LPS-triggered cytokine response. Dark gray boxes indicate circadian transcriptional regulation of the respective gene (P ≤ 0.05). Arrows indicate molecular interaction of genes involved in LPS response generation. For detailed information about circadian transcripts in this network, see Table S2. (B) Selected circadian transcriptional profiles of genes participating in LPS-triggered signaling cascade. Individual datasets from microarray analysis were plotted (filled circles) together with data obtained by a quantitative RT-PCR assay of the same samples (open circles, means ± SEM, n = 4, except of times CT 24 and 28, n = 3). Statistical analysis for qPCR data and chip data were performed with CircWave and CircWaveBatch, respectively (microarray: P ≤ 0.0001: Jun, Adam17, Cd180; P ≤ 0.01: IκBα, MD-1, Elavl1, Fos, Erk1; qPCR: P ≤ 0.0001: Adam17, Elavl1, Cd180, Erk1, MD-1; P ≤ 0.01: IκBα; P ≤ 0.05: Jun).
Discussion
Time of day-dependent changes in various parameters have long been known in mammalian physiology including the immune system. Rhythms in immune cell number, cytokine concentration, surface marker abundance, and immunological effector functions etc. support the argument for a pervasive regulation of the immune system by the circadian system. Recent years have witnessed an enormous progress in our understanding of the molecular basis of the circadian clock and its importance for health and disease (26). We know now that many peripheral tissues have autonomous circadian clocks, and we also know that these peripheral clocks are essential regulators of normal peripheral physiology (3, 27).
Here, we provide evidence that fully operational autonomous circadian clockworks exist in immunological tissues like spleen, lymph nodes, and resident peritoneal macrophages. Focusing on macrophages, we find that a crucial feature of innate immunity—the recognition of pathogens and subsequent initiation of defense strategies—is strongly regulated by a macrophage-intrinsic clock. Specifically, the strength of proinflammatory cytokine production of macrophages in response to bacterial endotoxin is determined by the circadian phase of the macrophage clock rather than by systemic circadian modulators such as rhythmic cortisol levels (10). Using systematic transcriptome analysis of peritoneal macrophages—showing ≈8% of transcripts rhythmically expressed—we uncover multiple possible control points in the LPS response pathway that link the macrophage-intrinsic circadian clock with crucial immunological effector functions.
Fully functional circadian clockworks within immunological compartments like spleen and lymph nodes (Fig. 1) put these tissues in line with other peripheral organs such as liver, kidney, heart, and lung. The phase of clock gene expression rhythms parallels that in other tissues (17), which may indicate similar input pathways. However, the cellular composition of immunological tissues does not remain constant over the period of 1 day as it seems to be the case for nonimmunological tissues (Fig. 2), making immunological tissues even more dynamic. This time-dependent heterogeneity together with the different cell types in immunological tissues (such as granulocytes, T cells, B cells, myeloid lineage derived cell lines, and others) underlines the fact that clock gene dynamics are representing an average of many possibly distinct clocks in different cell populations. A number of publications report clock gene expression in some of the cell types and subpopulations thereof (6, 28, 29). Yet, comparing these data is difficult because of large differences in the experimental conditions (tissue/compartment, species, light schedule, etc.). Thus, much more work has to be done to accumulate a comprehensive picture of clock gene expression within different populations and compartments of the immune system.
What are the functions of such local, immune cell intrinsic clocks? Several studies report circadian variation in cytokine levels, lytic activity, and phagocytosis in NK cells and macrophages (6, 28). From these in vivo and ex vivo experiments, it is not clear, however, whether local or systemic timing signals are responsible for the observed fluctuations. One candidate for a systemic timing signal—cortisol—is secreted in a circadian manner by the adrenal gland (10) and has been proposed to (i) drive circadian rhythms of cytokines and other immune functions (30) and (ii) to participate in the synchronization of peripheral oscillators (31). Here, we show that glucocorticoids are necessary neither for rhythmic LPS-induced cytokine secretion nor for daily resetting by the master clock. Circadian oscillations in spleen cell numbers, their cellular composition, and cytokine secretion are very similar in adrenalectomized mice compared with nonoperated mice with respect to phase and amplitude (compare Figs. 2, 3, and S2). We cannot exclude, however, that systemic time cues such as glucocorticoids, melatonin, or adrenergic/noradrenergic hormones play a role in synchronization or shaping of an immune response in the intact organism. Still, the fact that circadian cytokine secretion rhythms persist in constant in vitro culture conditions (Fig. 3 and Fig. S1) strongly indicates that a local macrophage-intrinsic clock rather than systemic cues is predominantly regulating these immune cell rhythms.
How does the molecular circadian clock regulate rhythmic macrophage functions? Transcriptional regulation is one of the major output routes of the circadian system (24). In many tissues, a substantial fraction (5–10%) of the transcriptome is controlled by the circadian clock, with most transcripts oscillating in a tissue-specific manner. Here, we demonstrate in peritoneal macrophages that ≈8% of the expressed genes show circadian modulation. When analyzing specifically the LPS immune response pathway, we discovered circadian expression at multiple levels ranging from signal reception via signal transduction to response generation (Fig. 5). Interestingly, for proteins acting in a complex like AP-1, or the TLR4 inhibitory molecules CD180 and MD-1, the phase of their gene transcription is similar, indicating that circadian transcription is precisely controlled and therefore has, most likely, a functional impact within this signaling pathway. The sheer amount of circadian transcription uncovered here points to a regulatory role of the circadian clock in many other functional aspects (including phagocytosis, antigen presentation, and immune regulation). Among the genes expressed with high amplitude, we found members of the stress response (Hspa1, Hspd1, Hspa5, Hsp110, Hsp90aa1), immune regulation (Cd59a, Cd69, Cd86, Cd200r1 and 4), components involved in phagocytosis [Vamp8, scavanger receptors (Slc7a8, Slc27a1, Slc25a1, Slc2a9, Slc41a3, Slc9a9, Slc9a8, Slc29a1, Slc22a15, Slc9a3r2, Slc39a1), Tlr1, lectins (Clec5a, Clec2i, Lman2, Lgals9, Siglec1, Lman1, Clec4d) and integrins (Itga5, Itfg2)], and genes involved in wound healing and extracellular matrix homeostasis like Mmp9 and P4ha1 and 2.
At present, it is unclear how and to what extent these transcriptional rhythms are conveyed into rhythmic immune cell outputs. Further experiments (e.g., studying the regulation downstream of transcription) are required to unravel the mechanistic details. In the case of the phagocytosis function of macrophages, a circadian control is very likely, because a diurnal regulation has been reported recently (but not yet a circadian regulation, i.e., in constant environmental conditions) (28). Another immune cell population of the innate immune system—the NK cells—show circadian rhythmicity on functional and molecular levels (refs. 6 and 32 and reviewed in ref. 33), again underlining the pervasiveness of circadian control in the immune system.
Why should the response to bacterial endotoxin in macrophages be regulated in a time of day-dependent manner? The dramatic diurnal variations in survival rate observed in an endotoxic shock mouse model first described by Halberg and colleagues (21, 22, 34) suggest a biological significance of the timing of immune functions. Although we cannot formally exclude other influences, we speculate that the high amplitude in mortality is due to circadian variation in proinflammatory cytokine secretion. In fact, the TNF-α and IL-6 secretion patterns observed in this study from ex vivo stimulated spleen cells peak in subjective day, when the mortality rate of endotoxic shock is highest (21). At the same time overall spleen cell content and relative as well as absolute numbers of monocytes/macrophages have their maxima (Fig. 2 A and B). Evolutionary adaptation to time of day-dependent pathogen pressure or activity correlated infection risk is an overt interpretation of these phenomena. Because macrophages are critical elements of the first line of defense against bacterial infections, anticipation of daily variation of infection risk is probably of great advantage. On the other hand, excessive responsiveness of these cells may be detrimental for the organism, and thus a tight regulation of its timing is likely to be beneficial.
A local circadian clock in cells of the immune system might enable the organism to integrate various environmental time cues (such as light conditions, food availability, physical activity) to better adapt to the requirements of individual habitats as it has been suggested for other peripheral clocks (35). Local clocks modulated by systemic timing cues offer the possibility to influence phases, phase relations, and amplitudes as well as expression levels of functional groups of genes. Although this study focuses on the level and mechanism of circadian regulation of immune functions, evidence exists that the relationship of both systems is bidirectional, i.e., that also immune system parameters can modulate the circadian clock (reviewed in refs. 36 and 37). For example, LPS can phase shift the clock in mice (38), and proinflammatory cytokines (such as TNF-α) both can alter circadian neuronal activity in the SCN (39) as well as down-regulate clock gene expression in cultured fibroblasts (40). Moreover, the impact of infections on circadian rhythms and clock-controlled behavior (such as sleep/wake patterns) is a matter of intense research (reviewed in refs. 36, 37, and 41).
Learning more about circadian immune regulation should not only have strong impact on our understanding of the pathophysiology of inflammatory responses but also on antiinflammatory drug strategies. In addition, the immune system might turn out to be a flexible and versatile model to tackle many open questions of circadian physiology on multiple scales (e.g., from behavior and disease to cellular and molecular levels) due to the availability of adoptive transfer techniques and large numbers of conditional Cre-expressing mouse lines. Thus, we anticipate that not only chronobiology will profit from immunological tools and techniques but also that immunologists will increasingly appreciate yet another level of dynamics in immunological responses and functions.
Materials and Methods
Bioluminescence Recordings.
A small piece of spleen and inguinal lymph nodes of PER2::LUC mice (20) and a Per1:luc rat (4) were cultured individually on Millicell membranes (Millipore). Peritoneal macrophages were cultured in Petri dishes. Bioluminescence was recorded using medium containing 0.1 mM beetle luciferin (Promega) and single photomultiplier tubes (Hamamatsu Photonics) (42).
Cytokine Secretion Assay.
Ex vivo assay.
Splenocytes from wild-type and adrenalectomized mice were harvested every 4 h after transfer in DD and treated with 5 μg/mL LPS (Alexis, from Escherichia coli serotype R515) for 4 h.
In vitro assay.
Splenocytes and peritoneal macrophages were collected and cultured in parallel by using microtiter plates. Every 4 h cells were treated with LPS (3 μg/mL) for 4 h starting at 24 h after seeding. Cell-free supernatants were harvested, frozen at −20 °C, and analyzed at the end of the time series. TNF-α and IL-6 contents in the supernatant were analyzed by ELISA.
Analysis of the Circadian Transcriptome in Macrophages.
Peritoneal cells were harvested from four mice every 4 h on two consecutive days after transfer in DD. Macrophages were purified by magnetic cell sorting (see SI Text for details), and total RNA was isolated by using RNeasy Mini kit (Qiagen). Gene expression profiles were determined by using GeneChip Mouse Gene 1.0 ST arrays (Affymetrix) and corresponding software packages. Analysis on circadian gene transcription was performed with CircWaveBatch v3.3 software (R. A. Hut) (43) and controlled for multiple testing errors by estimation of the local false-discovery rate (44).
Additional information about materials and methods used is available in SI Text.
Supplementary Material
Acknowledgments.
We thank J. Klank for help in clock gene transcription analysis of lymph nodes and A. Friedrich, M. Seifert, B. Sawitzki, K. Mäthner, H. Herzel, R. Kuban, R. Hut, and D. Hanson for their expert technical advices and all members of A.K.′s laboratory for help and discussions. We are grateful to J. S. Takahashi (University of Texas, Dallas) and H. Tei (Mitsubishi Kagaku Institute of Life Sciences, Tokyo) for providing PER2::LUC reporter mice and Per1::luc reporter rats, respectively. This work was supported by the Deutsche Forschungsgemeinschaft (A.K.) and National Institutes of Health Grant MH63104 (to E.H.). Research in A.K.'s laboratory was supported by the 6th European Union framework program EUCLOCK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906361106/DCSupplemental.
References
- 1.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U. Circadian time keeping: The daily ups and downs of genes, cells, and organisms. Prog Brain Res. 2006;153:271–282. doi: 10.1016/S0079-6123(06)53016-X. [DOI] [PubMed] [Google Scholar]
- 3.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 5.Esquifino AI, et al. Twenty-four-hour rhythms in immune responses in rat submaxillary lymph nodes and spleen: effect of cyclosporine. Brain Behav Immun. 1996;10:92–102. doi: 10.1006/brbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 6.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes G, Halberg F, Yunis EJ, Good RA. Circadian rhythmic plaque-forming cell response of spleens from mice immunized with Srbc. J Immunol. 1976;117:962–966. [PubMed] [Google Scholar]
- 8.Kawate T, Abo T, Hinuma S, Kumagai K. Studies of the bioperiodicity of the immune response. II. Co-variations of murine T and B cells and a role of corticosteroid. J Immunol. 1981;126:1364–1367. [PubMed] [Google Scholar]
- 9.Young MR, et al. Circadian rhythmometry of serum interleukin-2, interleukin-10, tumor necrosis factor-alpha, and granulocyte-macrophage colony-stimulating factor in men. Chronobiol Int. 1995;12:19–27. doi: 10.3109/07420529509064496. [DOI] [PubMed] [Google Scholar]
- 10.Krieger DT. Rhythms of ACTH and corticosteroid secretion in health and disease, and their experimental modification. J Steroid Biochem. 1975;6:785–791. doi: 10.1016/0022-4731(75)90068-0. [DOI] [PubMed] [Google Scholar]
- 11.Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16:581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- 12.Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: Influence of light and adrenocortical secretions. Science. 1973;182:285–287. doi: 10.1126/science.182.4109.285. [DOI] [PubMed] [Google Scholar]
- 13.Cutolo M, et al. Circadian rhythms in RA. Ann Rheum Dis. 2003;62:593–596. doi: 10.1136/ard.62.7.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland ER. Nocturnal asthma. J Allergy Clin Immunol. 2005;116:1179–1186. doi: 10.1016/j.jaci.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, et al. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boivin DB, et al. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 19.Chen YG, et al. Expression of mPer1 and mPer2, two mammalian clock genes, in murine bone marrow. Biochem Biophys Res Commun. 2000;276:724–728. doi: 10.1006/bbrc.2000.3536. [DOI] [PubMed] [Google Scholar]
- 20.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes PJ, Adcock I. Anti-inflammatory actions of steroids: Molecular mechanisms. Trends Pharmacol Sci. 1993;14:436–441. doi: 10.1016/0165-6147(93)90184-l. [DOI] [PubMed] [Google Scholar]
- 24.Schibler U. The daily timing of gene expression and physiology in mammals. Dialogues Clin Neurosci. 2007;9:257–272. doi: 10.31887/DCNS.2007.9.3/uschibler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storch KF, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30:621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 29.Dimitrov S, et al. Cortisol and epinephrine control opposing circadian rhythms in T-cell subsets. Blood. 2009;113:5134–5143. doi: 10.1182/blood-2008-11-190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrovsky N, Harrison LC. The chronobiology of human cytokine production. Int Rev Immunol. 1998;16:635–649. doi: 10.3109/08830189809043012. [DOI] [PubMed] [Google Scholar]
- 31.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 32.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469–476. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Arjona A, Sarkar DK. Are circadian rhythms the code of hypothalamic-immune communication? Insights from natural killer cells. Neurochem Res. 2008;33:708–718. doi: 10.1007/s11064-007-9501-z. [DOI] [PubMed] [Google Scholar]
- 34.Hrushesky WJ, Langevin T, Kim YJ, Wood PA. Circadian dynamics of tumor necrosis factor alpha (cachectin) lethality. J Exp Med. 1994;180:1059–1065. doi: 10.1084/jem.180.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornmann B, et al. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:319–330. doi: 10.1101/sqb.2007.72.041. [DOI] [PubMed] [Google Scholar]
- 36.Coogan AN, Wyse CA. Neuroimmunology of the circadian clock. Brain Res. 2008;1232:104–112. doi: 10.1016/j.brainres.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 37.Majde JA, Krueger JM. Links between the innate immune system and sleep. J Allergy Clin Immunol. 2005;116:1188–1198. doi: 10.1016/j.jaci.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Marpegan L, Bekinschtein TA, Costas MA, Golombek DA. Circadian responses to endotoxin treatment in mice. J Neuroimmunol. 2005;160:102–109. doi: 10.1016/j.jneuroim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Kwak Y, et al. Interferon-gamma alters electrical activity and clock gene expression in suprachiasmatic nucleus neurons. J Biol Rhythms. 2008;23:150–159. doi: 10.1177/0748730407313355. [DOI] [PubMed] [Google Scholar]
- 40.Cavadini G, et al. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci USA. 2007;104:12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10:199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe M, et al. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 44.Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.