Abstract
Bacterial DNA replication requires DnaA, an AAA+ ATPase that initiates replication at a specific chromosome region, oriC, and is regulated by species-specific regulators that directly bind DnaA. HobA is a DnaA binding protein, recently identified as an essential regulator of DNA replication in Helicobacter pylori. We report the crystal structure of HobA in complex with domains I and II of DnaA (DnaAI–II) from H. pylori, the first structure of DnaA bound to one of its regulators. Biochemical characterization of the complex formed shows that a tetramer of HobA binds four DnaAI–II molecules, and that DnaAI–II is unable to oligomerize by itself. Mutagenesis and protein–protein interaction studies demonstrate that some of the residues located at the HobA-DnaAI–II interface in the structure are necessary for complex formation. Introduction of selected mutations into H. pylori shows that the disruption of the interaction between HobA and DnaA is lethal for the bacteria. Remarkably, the DnaA binding site of HobA is conserved in DiaA from Escherichia coli, suggesting that the structure of the HobA/DnaA complex represents a model for DnaA regulation in other Gram-negative bacteria. Our data, together with those from other studies, indicate that HobA could play a crucial scaffolding role during the initiation of replication in H. pylori by organizing the first step of DnaA oligomerization and attachment to oriC.
Keywords: DiaA, isothermal titration calorimetry, replication regulators, X-ray crystallography
In all living organisms, DNA replication involves the formation of a complex between a chromosomal origin of replication and initiator proteins. The formation of this complex leads to the loading of the replication machinery and the formation of an active replication fork (1). In bacteria, the initiator protein is DnaA, a member of the AAA+ family of ATPases. DnaA binds to specific sequences on the origin of replication oriC, thus forming the orisome [protein–oriC complexes, reviewed in (2, 3)]. Escherichia coli DnaA comprises four domains (DnaAI–DnaAIV) (4). DnaAI (≈80 residues) is important for DnaA multimerization (5, 6) and association with the helicase DnaB (7, 8). DnaAII (≈40 residues) function is unknown, whereas DnaAIII (≈220 residues) forms the ATPase domain and DnaAIV (≈120 residues) binds DNA. NMR structures of E. coli and Mycoplasma genitalium DnaA fragments showed that DnaAI adopts a K homology (KH) fold, whereas DnaAII is assumed to be a flexible linker, as it was disordered in the structures (9, 10). The crystal structure of an ADP-bound DnaAIII-IV from Aquifex aeolicus identified the boundaries between the ATPase domain (DnaAIII) and the helix-turn-helix–containing DNA-binding domain (DnaAIV) (11).
Additional studies have shed light on the molecular mechanisms by which DnaA plays its role during chromosome replication. The DNA-bound structure of E. coli DnaAIV identified the mode of binding onto the DnaA box sequence (12), whereas the structure of A. aeolicus DnaAIII–IV bound to a nonhydrolyzable ATP analog was shown to form a right-handed super-helical structure (13). Taken together, these structures suggest a model in which a helix of ATP-bound DnaA could be the oligomeric form onto which DNA wraps, thereby facilitating the opening of AT-rich regions of the oriC at the so-called DNA unwinding elements (DUE) (2, 13). This open helical structure might also explain how the regulatory inactivation of DnaA (RIDA) occurs (2, 13). RIDA requires a DnaA related ATPase, Hda, and the β-sliding clamp (2, 14). These two proteins facilitate the hydrolysis of ATP by DnaA which would lead to a disassembly of the helical arrangement and hence to the inactivation of DnaA. In E. coli, the formation of the orisome is assisted by several accessory factors such as IHF, SeqA, HU, and DiaA [reviewed in (3)], with the latter two binding directly to DnaA (15, 16). The DnaA paralogue DnaC is necessary to load the DnaB helicase. DnaC can form helical structures and can specifically bind to ATP-DnaA (17). This suggests that in E. coli, DnaC is a molecular adaptor that uses DnaA as a docking platform to load DnaB on the replication fork (17).
Although DnaA- and DnaB-mediated orisome construction is conserved in all bacteria, species-specific proteins can also play key roles in replication (18). In Bacillus subtilis, YabA is a negative regulator of initiation of chromosomal replication that downregulates initiation as part of a multimeric complex with DnaA and DnaN (19, 20). B. subtilis DnaA activity is also controlled by the Soj (ParA) protein, which couples DNA replication with cell division (21, 22). In the human pathogen Helicobacter pylori, no sequence homologues have been identified for E. coli proteins DnaC, Hda, Dam, SeqA, or for B. subtilis YabA. However, a comprehensive yeast two-hybrid study (2HB) identified HP1230 [where HPXXXX denotes the ORF number as defined in (23)], as a putative DnaA-binding protein, a potential interaction that was confirmed in vitro (24, 25). The protein HP1230 is absolutely required for the survival of H. pylori, forms complexes with DnaA in vivo, and modifies the structure of the orisome (26). HP1230 was named HobA, for Helicobacter orisome binding protein A (26). HobA and DnaA are expressed in similar amounts in H. pylori cells (26). Flow cytometry experiments revealed that, similar to DnaA, the absence of HobA inhibits replication at the initiation stage (26). The crystal structure of HobA showed that the protein is a structural homologue of E. coli DiaA despite a complete lack of sequence similarity (27). In E. coli, DiaA stimulates ATP-DnaA assembly on oriC and regulates the timing of replication (16, 28). HobA and DiaA adopt the Sugar ISomerase (SIS) fold, form compact tetramers, and bind to DnaAI–II, suggesting a common DnaA regulation mechanism (16, 24, 27, 28).
To gain further insight into this mechanism, we have solved the crystal structure of HobA in complex with DnaAI–II from H. pylori and validated the observed crystallographic stoichiometry by isothermal titration calorimetry (ITC) and analytical gel filtration. Our studies of the oligomerization state of H. pylori DnaAI–II show that, in contrast to E. coli DnaAI–II, these domains do not self-interact. The study of selected mutants in vitro and in vivo provides insight into DnaA regulation by HobA and demonstrates the essential role of the HobA-DnaA interaction in H. pylori, suggesting that HobA-mediated DnaA scaffolding observed in the crystal structure is an important step during DNA replication in this bacterium.
Results
Architecture of the HobA/DnaAI–II Complex.
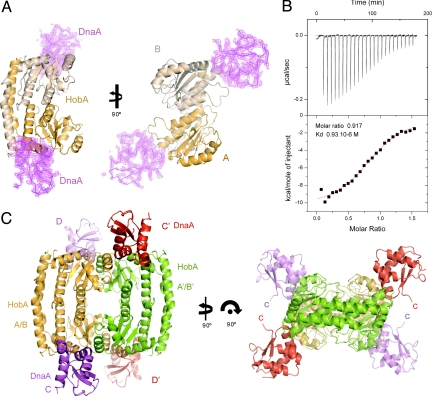
The asymmetric unit of the crystals of the HobA/DnaAI–II complex contains two molecules of HobA (chains A and B in Fig. 1A), which form a dimer, and two molecules of DnaAI–II (chains C and D in Fig. 1A) located at opposite poles of the HobA dimer. The HobA dimer is almost identical to that seen in the crystal structure of HobA in the absence of DnaA (RMSD 0.78Å, for 359 Cα) (27). In the crystal structure of the complex, one DnaAI–II molecule binds to each of the HobA monomers, with chain C bound to HobA chain A and chain D bound to chain B. The DnaA chains C and D are very similar, as are the binding sites of each DnaAI–II molecule onto HobA. To determine the molar ratio of the complex in solution, we performed ITC experiments on HobA using DnaAI–II as a ligand (Fig. 1B). The molar ratio of the complex is 1:1 (experimentally determined stoichiometry is 0.9), thereby validating the stoichiometry observed in the crystal structure. The ITC experiment also yielded a Kd of 0.93 × 10−6 M. Because HobA alone forms tetramers (27), we investigated whether the HobA/DnaAI–II complex was formed on a tetramer of HobA. The molecular weights of HobA, HobA tetramer, and DnaAI–II are 22, 88, and 13 kDa, respectively. Therefore a complex consisting of two DnaAI–II with two HobA molecules (HobA2(DnaAI–II)2), as seen in the asymmetric unit, should be of 70 kDa. Gel filtration experiments were performed and showed that the HobA/DnaAI–II complex eluted earlier than HobA and therefore formed a complex of higher molecular weight than the HobA tetramer or a HobA2/(DnaAI–II)2 complex (Fig. S1A). Thus, analytical gel filtration combined with the ITC experiments indicated that, in solution, the complex consisted of four molecules of DnaAI–II bound to one tetramer of HobA. Examination of the crystal structure of HobA/DnaAI–II complex fully confirms these data. Symmetry-related dimers form tetramer to which four molecules of DnaAI–II are bound (Fig. S1B). The symmetry related subunits, chains A′ and B′ of HobA also bind to DnaAI–II molecules C and D, respectively. DnaAI–II molecules are, in fact, positioned at the dimer–dimer interface of the tetramer, with each of the DnaAI–II bound to two adjacent, symmetry-related monomers of HobA (chains A and B′). This suggests that the HobA dimer–dimer interface is needed for DnaA interaction (Fig. 1C).
Fig. 1.
Structure of the HobA/DnaA complex. (A) Diagrams representing two different views of the asymmetric unit of the crystal structure of the HobA/DnaAI–II complex. The HobA dimer is represented in ribbon, with subunits A and B colored in light orange and wheat, respectively. Electron density map, contoured at 1σ level is shown for the two DnaAI–II molecules (magenta chicken wire) together with their Cα traces. (B) Data obtained from the isothermal calorimetry titration of purified HobA using injection of DnaAI–II (top panel) and fitting curve with the estimated molar ratio of 0.9 and the Kd of 0.93 micro molar (bottom panel; also SI Text). (C) Ribbon diagrams of the structure of HobA4(DnaAI–II)4 complex. HobA dimers are colored in orange (chain A and B) and green (chain A′ and B′), DnaAI–II monomers are colored in magenta (chain C and D) and red (chain C′ and D′). Figures were produced using Pymol (www.pymol.org).
Structure of HobA Bound DnaA I–II.
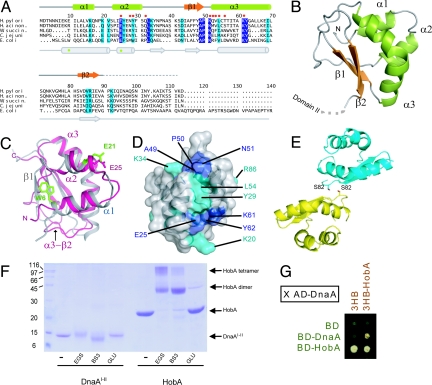
NMR structures of DnaAI–II from E. coli and M. genitalium showed that DnaAI consists of around 80 residues, whereas DnaAII is disordered (9, 10). In our crystal structure, no electron density was interpretable for most of the DnaAII (residue 91 onward) for both the DnaAI–II molecules in the asymmetric unit. The absence of a structured DnaAII is in agreement with the NMR structures of DnaA from E. coli and M. genitalium. The ordered regions of H. pylori DnaAI–II that we observe adopt a KH fold similar to that described for E. coli DnaAI–II, albeit with significant variations in the length and relative positions of secondary structural elements (Fig. 2 A and B). In particular, the conformation of the β1 strand in E. coli DnaAI–II is markedly modified in H. pylori DnaAI–II by the presence of a proline (P37) causing the β-sheet to be disrupted (Fig. 2 A and C). As a consequence, DnaAI–II from H. pylori shows only two antiparallel β-strands, β1 and β2, corresponding to β2 and β3 of E. coli DnaA, respectively. The β-sheet packs against α3, an extended helix with a 45° kink at residue Y62 (Fig. 2B). This feature has been observed before in the NMR structures of DnaAI–II. Our crystal structure suggests that a kinked α3 is important for DnaA to form the complex with HobA, as a nonkinked α3 would cause steric clashes at the HobA dimer–dimer interface (Fig. 3 B and D). The KH fold is completed by two small α helices, α1 and α2, positioned perpendicularly to α3 and the β-sheet (Fig. 2B). A sequence comparison of the DnaAI–II from E. coli and H. pylori reveals that most of the conserved residues reside in the α2–α3 region (Fig. 2 A and D), as are residues involved in the formation of the complex of DnaAI–II with HobA. Also worth mentioning is the conservation of a residue E25 (E21 in E. coli) identified as critical for the interaction between DnaA and DnaB in E. coli (10).
Fig. 2.
Structure and oligomerization state of DnaAI–II from Helicobacter pylori. (A) Sequence alignment of DnaAI–II from H. pylori with DnaAs from H. acinonichis, Wolinella succinogenens, Campylobacter jejuni, and E. coli. Identical and conserved residues are indicated in dark and light blue, respectively. Residues involved in complex formation with HobA are indicated by red dots. Residues important for self-oligomerization (W6) and interaction with DnaB (E21) in E. coli DnaA are indicated by green dots. Secondary elements of DnaAI–II (pdb code 1EOG) proteins are indicated above (H. pylori) and below (E. coli) the alignment. (B). Ribbon representation of H. pylori DnaAI–II crystal structure. (C) Superimposition of DnaAI from E. coli (gray ribbon) and from H. pylori (pink ribbon). Side chains of W6 and E61 from E. coli DnaA are colored green and displayed in ball-and-stick representation as well as E25 from H. pylori DnaA. (D) Surface representation of H. pylori DnaAI–II illustrating identical (dark blue) and conserved (light blue) residues identified in (A). (E) Diagram depicting a possible dimer C/C′ of DnaAI–II as seen in the crystal packing of the HobA/DnaA complex, mediated by interactions between each Ser-82. (F) SDS/PAGE gel of cross-linking experiments with DnaAI–II and HobA using EGS, BS3, or glutaraldehyde (GLU) cross-linking agents. (G) Yeast triple hybrid experiment carried out between BD-DnaA and AD-DnaA in the absence (3HB empty vector) or in the presence of HobA (3HB-HobA). Haploid strains carrying both the BD and 3HB vector derivatives are here organized as a small, 2:2 matrix and mated against an haploid strain expressing the AD-DnaA fusion (SI Text). A binary interaction with AD-DnaA was observed with BD-HobA but not with BD-DnaA (column 3HB), in agreement with the 2HB. An interaction phenotype could be detected between BD-DnaA and AD-DnaA when HobA is expressed constitutively (column 3HB-HobA).
Fig. 3.
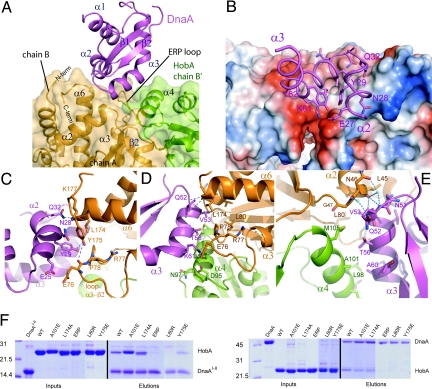
Interactions mediating HobA/DnaAI–II complex formation. (A) Cartoon representation of DnaAI–II/HobA interface with the same color coding as in Fig. 1C showing helices α2 and α3 in the pocket formed by two adjacent HobA molecules. (B) Opposite view of the binding of α2 and α3 helices of DnaA onto HobA with a surface representation of HobA colored according to its electrostatic potential. DnaAI–II carbons are colored in magenta, nitrogen in blue, and oxygen in red. Backbone carbons of DnaAI–II α2 and α3 are displayed in ribbon; side chains of the residues from DnaA forming the interface with HobA are shown in sticks. (C) Close-up view of the interface between DnaAI–II α2 helix and the binding site of HobA. In the next three panels, the cartoon backbones are colored as in (A): DnaAI–II in magenta, Chain A of HobA in orange and chain B′ in green. Side chains participating in the interface are displayed as sticks and labeled in the color corresponding to their chains. Hydrogen bonds between atoms are depicted by blue dashes. (D) Close-up view of the interactions between DnaAI–II α3 helix and HobA ERP loop, α6, and α2-β1 from Chain A and α4 from Chain B′. (E) Opposite view of α3 interactions with HobA; waters mediating interactions between different chains are indicated by light blue spheres. Residues from HobA α4 are mainly involved in hydrophobic interactions with α3. (F). Pull-down assays of HobA and HobA mutants using His6-DnaAI–II (left) or His6-DnaA (right).
E. coli DnaA has been shown to form oligomers in vitro (5, 6) with residues located in helix α1 being essential for self-interaction, in particular W6 (6), part of an hydrophobic surface that could provide an interaction site seen in other KH fold proteins (10). However, in H. pylori DnaA, helix α1 is shorter, W6 is not conserved, and the corresponding interacting area is positively charged. In the crystal structure we report, the only interaction observed between DnaAI–II is a single hydrogen bond between symmetry-related S82 residues (Fig. 2E). This suggests that H. pylori DnaAI–II does not self-associate. We therefore investigated the oligomeric state of DnaAI–II from H. pylori and found that it did not form oligomers in either gel filtration (Fig. S1) or in cross-linking experiments using three different cross-linking agents (Fig. 2F). On the other hand, cross-linking experiments identified HobA oligomers (Fig. S2G) and the HobA/DnaAI–II complex (Fig. S3). In a yeast 2HB experiment (Fig. S2), DnaA fused to the activating domain of Gal4 (AD-DnaA) did not interact with DnaA fused to the DNA binding domain of Gal4 (BD-DnaA), whereas HobA and DnaA interacted strongly, whether fused with the Gal4 AD or BD domains (25) (Fig. S2 and Fig. 4). We tested the ability of HobA to promote DnaA–DnaA interaction using a yeast three-hybrid (3HB) assay in which both AD- and BD-DnaA fusions were co-expressed in the diploid cells in the presence or in the absence of HobA (Fig. 2G). We observed that the co-expression of the HobA protein triggered an interaction phenotype, whereas no interaction was detected in the absence of HobA, in agreement with the 2HB data. We conclude that, in contrast to the E. coli homologue, H. pylori DnaAI–II does not form dimers, a feature observed also for M. genitalium DnaAI–II, possibly because of the structural differences observed in α1. Together, these results suggest that in H. pylori, HobA is an important determinant for DnaA self-oligomerization.
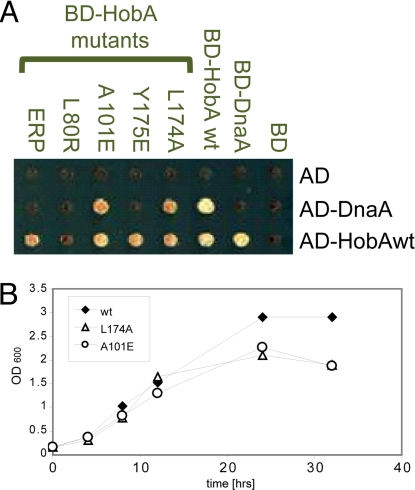
Fig. 4.
In vivo study of HobA mutants. (A) The HobA wild-type protein and mutant derivatives were tested for their ability to self-interact and to interact with DnaA using the yeast two-hybrid phenotypic interacting assay. Each HobA derivative was expressed in yeast haploid cells as protein fusion with the BD domain of Gal4 (columns, in green) and mated with complementary mating-type cells carrying AD-DnaA or AD-HobA protein fusions (lines, in black). BD and AD indicate cells harboring empty vectors as control. The interacting phenotypes were monitored by the ability of the diploid cells to grow on –LUA– selective media. BD-HobA mutants containing the Y175E and ERP substitutions are defective for interaction with AD-DnaA while staying proficient for self-interaction. L80R mutant does not interact with either AD-DnaA or AD-HobA. Substitutions L174A and A101E have no effect on HobA interaction phenotypes. (B) Growth curves of H. pylori 26695 wild-type, HobA-A101E, and HobA-L174A mutants. Results are representative of three independent experiments.
Binding Mode of DnaAI–II onto HobA.
In the HobA4(DnaAI–II)4 complex, each of the DnaAI–II molecules interacts with two adjacent subunits of HobA, and the interface buries a total of ≈660 Å2 of HobA solvent-accessible surface area (Fig. 3 A and B). The interaction between each DnaAI–II and HobA requires the binding of helices α2 and α3 of DnaAI to two separate binding cavities on each side of the long α3-β2 loop of HobA (Fig. 3A). Interestingly, this loop contains three residues E76, R77, and P78 (ERP loop) conserved between HobA and E. coli DiaA (27). In the first binding cavity, α2 from DnaAI–II sits in a small crevice in HobA delimited by the ERP loop and the C terminus of α6. Residues from α2 of DnaAI–II bind predominantly by polar contacts, with the major interactions being two hydrogen bonds between DnaAI–II Y29 and the backbone carbonyl of E76 and hydroxyl group of Y175 from HobA (Fig. 3C). The π-electron ring of Y175 also interacts with the hydrophobic part of N28 of DnaAI–II, the latter also interacting with the K177 side chain from HobA. Q32 of DnaAI–II makes a hydrogen bond with the backbone carbonyl of L174 from HobA α6 (Fig. 3C).
In the second cavity, the α-helical residues preceding the 45° kink in α3 from DnaAI–II sit in a hydrophobic cleft delimited by loops α2–β1 and the ERP loop from one HobA subunit and helix α4 from an adjacent subunit (Fig. 3C). In addition to hydrophobic interactions, binding of α3 is secured by hydrogen bonds between K61 (DnaAI–II) and E76 from the HobA ERP loop (Fig. 3D) and backbone atoms of V53 and Q52 (DnaAI–II) and L45 from HobA. DnaAI–II V53 side chain inserts into in a small hydrophobic cavity formed by L170, L80, L174, and L45 of HobA (Fig. 3 D and E). Several water-mediated hydrogen bonds involving Q52, N51 from DnaAI–II, and G47, N46, and L45 from HobA also strengthen the interaction (Fig. 3 D and E). The position of α3 is secured by hydrophobic interactions of T56 and A60 of DnaAI–II with M105, A101, and L98 from helix α4 of HobA.
Mutational Analysis of the HobA/DnaA Interaction.
In addition to the structure determination, we used PCR random mutagenesis to generate single amino acid changes in HobA. Loss of HobA–DnaA interaction while retaining HobA–HobA self-interaction in yeast 2HB was monitored (see SI Text). This random mutational mapping of HobA identified three mutations that specifically abolished HobA-DnaA interaction (SI Text and Fig. S2). These mutations, L45P, L174P, K177* (* stop codon) are in agreement with the crystal structure, since these residues are all directly involved in the HobA-DnaAI–II interface. However, further biochemical study of these mutations proved to be impossible because of poor solubility of the mutants (L45P and K177*) or failure to remove the His6-tag from the purified protein (L174P), a step necessary for binding studies using our pull-down assay (27). As a result of our structural analysis, we designed further mutants of HobA: the single mutants L80R, A101E, L174A, Y175E, and a triple mutant of the ERP loop (E76A/R77A/P78A, named ERP mutant hereafter). The purified untagged mutant proteins were then used in a pull-down assay in which they were mixed with His6-DnaAI–II or full length His6-DnaA. Complex formation was achieved by pull-down on Talon resin (Fig. 3F). Wild-type HobA (wt-HobA), A101E, and L174A mutants were able to interact with both His6-DnaAI–II and His6-DnaA. In contrast, HobA ERP and L80R mutants were completely defective. Y175E mutant interacted weakly with His6-DnaAI–II and not with His6-DnaA (Fig. 3F). Cross-linking experiments with His6-DnaAI–II confirmed that interaction with HobA was abolished for ERP, L80R, and Y175E. The results obtained with L174A and A101E suggest that these mutants form a complex with His6-DnaAI–II, although with less efficiency than wt-HobA (Fig. S3). Interestingly, all of the HobA mutants retained their capacity to form tetramers except L80R and ERP, suggesting that HobA tetramer formation might be important for interaction with DnaA.
In Vivo Study of HobA Mutants.
As mentioned previously, HobA, fused to Gal4 BD, was able to interact with DnaA fused to Gal4 AD in yeast haploid strains (Fig. 4). We therefore tested the BD-fused HobA mutants for their ability to interact with AD-DnaA. As seen in Fig. 4A, ERP, L80R, and Y175E mutants lost their ability to interact with DnaA but L174A and A101E mutants did not. These results are in agreement with those observed in our pull-down assays and cross-linking experiments (discussed above). Interestingly, L80R mutant interaction with wt-HobA was also altered, confirming the results obtained in the cross-linking experiments (Fig. S3). The ERP HobA mutant could interact with wt-HobA: thus, the loss of tetramer observed in the cross-linking experiments described above is observed only when each subunit contains the ERP mutation.
We then used a gene replacement approach to exchange wt-HobA by HobA mutants in H. pylori cells strain 26695 (SI Text and Fig. S4). HobA mutants A101E and L174A were introduced successfully in H. pylori. The growth curves of bacteria carrying HobA mutants A101E and L174A were similar to those carrying wt-HobA (Fig. 4B), showing that these mutations that have no effect on DnaA binding also have no effect on bacterial growth. For the ERP, L80R, and Y175E mutants, the aph-3 cassette was introduced but the sequences of hobA were that of the wild type. This suggests that alternative recombination events (shortening the homological arms and/or performing two independent crossings-over), took place in the bacteria, to avoid the lethal changes introduced by the designed mutations (SI Text and Fig. S4). This strongly suggests that introducing mutations in HobA that affect DnaA binding are lethal for H. pylori. Although we cannot rule out that the loss of HobA tetramerization observed in cross-linking for the ERP and L80R mutants could also be involved in the effect observed, this is not the case for Y175E (Fig. S3). We therefore conclude that the interaction of HobA and DnaA is required for H. pylori viability.
Structural Conservation of DnaA Binding Mode of HobA in E. coli DiaA.
We previously showed that HobA and DiaA are structural homologues (27) (Fig. S5A). DiaA is a DnaA binding protein that promotes formation of the ATP-bound DnaA/oriC complex in vitro and controls the timing of initiation (16, 28). HobA and DiaA share the same SIS fold and have a similar tetrameric architecture (Fig. S5B). Conserved, surface-accessible residues include Leu-45 (Leu-39 in DiaA), Asn-46 (Asn-40 in DiaA), the ERP loop (E70, R71, P72 in DiaA), and L174 (L190 in DiaA). In DiaA, these residues form a cleft similar to that seen in HobA (Fig. S5 C and D). Previous studies had shown that the DiaA residues L190, F191 and P72 are necessary for DnaA binding, as is the intact DiaA tetramer interface for the assembly of DnaA on the oriC (16, 28). These results correlate remarkably with the results we report here for the HobA4(DnaAI–II)4 complex, in particular for the observation that ERP (E70, R71, P72 in DiaA) and Y175E (F191 in DiaA) HobA mutants can no longer interact with DnaA (Fig. 3 and Fig. S5). Many residues of H. pylori DnaA involved in complex formation with HobA are also conserved in E. coli DnaA (Fig. 2 A and D). Thus the binding mechanism of DnaA to HobA is likely to be conserved in E. coli DnaA and DiaA.
Discussion
The DnaA AAA+ ATPase is the initiator of replication in bacteria and functions via multiple protein–protein or protein–DNA interactions (2, 3, 17). Its modular architecture allows for interactions with different binding partners during the various steps of the formation of the active replication forks and coupling to other cellular events (3, 15, 17, 20, 22, 29, 30). In E. coli, DnaAIII interactions with itself, Hda, or DnaC are important for the spatial positioning of the ATP-DnaA molecules and their recycling (13, 17, 31, 32). Protein–protein interactions involving DnaAI–II play critical roles in DnaA self-oligomerization, helicase loading (7, 10, 17), and regulation of the replication timing (16, 28). However, structural studies have so far failed to capture the mode of action of the DnaAI–II and its regulators (33). The work presented here shows the structure of a DnaA domain in complex with one of its regulators and provides new insight into the roles of DnaAI–II during DNA replication in H. pylori.
HobA is a structural homologue of DiaA, a protein-stimulating ATP-DnaA/oriC complex formation and regulating the timing of replication in E. coli (10, 27). We show here that the residues of HobA involved in DnaAI–II binding are conserved in DiaA, as are residues of H. pylori and E. coli DnaA that participate in this mechanism. During the preparation of this manuscript, a study by Keyamura et al. (34) identified residues of DnaA interacting with DiaA (34). In particular, W46 in E. coli DnaA was found to be critical for DiaA binding via the residues L190 and F191. The equivalent residues in H. pylori DnaA are V53 (Fig. 2A) that inserts into a cavity formed by the ERP loop and L80, L174 (L190 in DiaA), and Y175 (F191). The crystal structure of HobA4(DnaAI–II)4 that we report here is not only consistent with the studies on DiaA/DnaA interaction but also adds new and important details of the DnaA binding interface.
DiaA promotes replication in E. coli but is not necessary for viability, whereas HobA interaction with DnaA is necessary for H. pylori survival. DiaA appears to contribute to the organization of the E. coli orisome by binding to a subgroup of DnaA molecules, especially those binding to regions near the DNA unwinding elements (DUE) (34). Interestingly, DnaB loading is inhibited by DiaA binding to this subgroup of DnaA molecules (34). Keyamura et al. thus propose that DiaA promotes the formation of an open complex while preventing the loading of DnaB molecules. Another (unknown) factor would disassemble DiaA molecules from the orisome and allow DnaB loading on oriC by DnaA, thereby controlling the timing of replication (34). The replication machinery in H. pylori remains poorly understood compared with the extensively studied E. coli system. Whereas H. pylori DnaA has a similar domain architecture to E. coli DnaA (35), the oriC region is different, as it comprises two pairs of closely spaced, high affinity DnaA boxes, one separate low affinity DnaA box, and no clear DUE region (35). H. pylori DnaB also displays unique oligomerization properties that allows it to complement DnaB-DnaC–depleted E. coli cells (36, 37). HobA and DnaA are present in similar amounts and form complexes in H. pylori; the absence of HobA inhibits replication at the initiation stage (26). This suggests that the HobA/DnaA complex plays a critical role during replication initiation. We show here that DnaA binds to HobA with micromolar affinity and in a 4:4 stoichiometry. The crystal structure of HobA4(DnaAI–II)4, together with the biochemical data presented, shows that HobA acts as a hub onto which four DnaA molecules assemble. The HobA4(DnaAI–II)4 complex is highly organized with symmetrically positioned DnaAs. This suggests that the HobA4DnaA4 may provide a preformed, oligomeric state of DnaA that could facilitate the formation of ATP-DnaA/oriC complexes by positioning ATP-DnaA on the two pairs of DnaA boxes that are specific to the H. pylori oriC. This scaffolding role of HobA is in agreement with its remodeling effect on the orisome, observed by electron microscopy (26). DnaAI self-oligomerization is a prerequisite for replication initiation in E. coli and loading of the DnaB helicase (6, 38). Our study shows that, in contrast to E. coli DnaA, H. pylori DnaAI–II does not form oligomers, suggesting that the presence of HobA is necessary for DnaAI oligomerization, which is likely to be the main function of HobA in the bacteria. This hypothesis is supported by the HobA-dependent DnaA self-oligomerization observed in yeast 3HB and may explain why HobA is absolutely required for H. pylori survival (26). Future studies to identify the molecular interplay between DnaA, DnaB, HobA, and oriC will help understand how DNA replication is initiated in H. pylori. It should also be noted that DnaA has recently been shown to be a suitable target for drug design (39). As the HobA–DnaAI–II interaction appears to be essential for H. pylori viability, the structure of the HobA4(DnaAI–II)4 complex that we present also provides a new and attractive target for the design of drugs against H. pylori infection.
Methods
Cloning, Protein Expression, and Purification.
For the purification of the HobA/DnaAI–II complex, hobA gene was cloned into the pRSF-Duet1 vector (Novagen) and co-transformed with pET151-Nterm, a vector expressing the N-terminal fragment of DnaA (DnaAI–II, residues 1–112) fused to a TEV-cleavable histidine tag (27). Briefly, the complex was purified on a Ni-NTA column and the His-tag was removed by TEV cleavage overnight at 4 °C. Pure HobA/DnaAI–II complex was collected in the flow-through fraction obtained by reloading the Ni-NTA column with the cleaved protein solution. Details of the expression and purification of the proteins and protein complexes can be found in the SI Text.
Crystallization, Structure Determination, and Refinement.
Crystals of the HobA/DnaAI–II were grown by vapor diffusion in hanging drops by mixing 1 μl protein complex (at 5 mg/ml) to 1 μl of a reservoir solution containing 19–21% (wt/vol) polyethylene glycol (PEG) 3350, 200 mM potassium acetate, and 50 mM Tris-HCl, pH 8.0. Crystals were transferred into a solution reservoir solution supplemented with 20% glycerol (wt/vol). Dataset was collected at the ESRF beamline ID14EH4, intensities were integrated with MOSFLM (30) and scaled with SCALA (31). Crystals of HobA/DnaAI–II diffracted to a resolution of 2.67 Å, and belonged to the spacegroup C2, with cell dimensions of a = 127.6 Å b = 55.9 Å and c = 96.3 Å, α = γ = 90.00 and β = 97.9 (Table S1). Molecular replacement was done with HobA monomer and a DnaAI–II homology model using PHASER (32). Refinement using PHENIX (33) converged to a model with R and Rfree values of 22.04% and 26.4% at 2.6 Å resolution with good geometry (Fig. S6 and Table S1). The coordinates of the HobA/DnaAI–II structure have been deposited in the PDB (entry codes 2wp0). Details of the method can be found in SI Text.
Details of materials and methods used are given in SI Text. Methods include ITC, site-directed mutagenesis, pull-down, chemical cross-linking, analytical gel filtration assays, yeast two- and three-hybrid system, and genetic complementation in H. pylori.
Supplementary Material
Acknowledgments.
We thank members of the MX group at ESRF for help during data collection and the Partnership for Structural biology (PSB) for support. We also thank G. Waksman, J. Timmins, and G. Leonard for critical comments on the manuscript. This work was supported by the ESRF “In House Research” program. Support was provided by the Foundation for Polish Science (to A.Z.P.) and by a grant from Iceland, Lichtenstein, and Norway through the EEA Financial Mechanism (to F.N.P.) and the Ministry of Science and Higher Education (project N N301 029334).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates, phases, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2wp0).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908966106/DCSupplemental.
References
- 1.Kaguni JM, Kornberg A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell. 1984;38:183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- 2.Mott ML, Berger JM. DNA replication initiation: Mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 3.Kaguni JM. DnaA: Controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–371. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 4.Messer W, et al. Functional domains of DnaA proteins. Biochimie. 1999;81:819–825. doi: 10.1016/s0300-9084(99)00215-1. [DOI] [PubMed] [Google Scholar]
- 5.Weigel C, et al. The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol Microbiol. 1999;34:53–66. doi: 10.1046/j.1365-2958.1999.01568.x. [DOI] [PubMed] [Google Scholar]
- 6.Simmons LA, Felczak M, Kaguni JM. DnaA Protein of Escherichia coli: Oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol Microbiol. 2003;49:849–858. doi: 10.1046/j.1365-2958.2003.03603.x. [DOI] [PubMed] [Google Scholar]
- 7.Seitz H, Weigel C, Messer W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol Microbiol. 2000;37:1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x. [DOI] [PubMed] [Google Scholar]
- 8.Sutton MD, Carr KM, Vicente M, Kaguni JM. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 9.Lowery TJ, et al. NMR structure of the N-terminal domain of the replication initiator protein DnaA. J Struct Funct Genomics. 2007;8:11–17. doi: 10.1007/s10969-007-9022-7. [DOI] [PubMed] [Google Scholar]
- 10.Abe Y, et al. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282:17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 11.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujikawa N, et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 14.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 16.Keyamura K, et al. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 19.Soufo CD, et al. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev Cell. 2008;15:935–941. doi: 10.1016/j.devcel.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Noirot-Gros MF, et al. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103:2368–2373. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 24.Terradot L, et al. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: Strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol Cell Proteomics. 2004;3:809–819. doi: 10.1074/mcp.M400048-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Rain JC, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 26.Zawilak-Pawlik A, et al. HobA—a novel protein involved in initiation of chromosomal replication in Helicobacter pylori. Mol Microbiol. 2007;65:979–994. doi: 10.1111/j.1365-2958.2007.05853.x. [DOI] [PubMed] [Google Scholar]
- 27.Natrajan G, Hall DR, Thompson AC, Gutsche I, Terradot L. Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli. Mol Microbiol. 2007;65:995–1005. doi: 10.1111/j.1365-2958.2007.05843.x. [DOI] [PubMed] [Google Scholar]
- 28.Ishida T, et al. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J Biol Chem. 2004;279:45546–45555. doi: 10.1074/jbc.M402762200. [DOI] [PubMed] [Google Scholar]
- 29.Flatten I, Morigen, Skarstad K. DnaA protein interacts with RNA polymerase and partially protects it from the effect of rifampicin. Mol Microbiol. 2009;71:1018–1030. doi: 10.1111/j.1365-2958.2008.06585.x. [DOI] [PubMed] [Google Scholar]
- 30.Chodavarapu S, Gomez R, Vicente M, Kaguni JM. Escherichia coli Dps interacts with DnaA protein to impede initiation: A model of adaptive mutation. Mol Microbiol. 2008;67:1331–1346. doi: 10.1111/j.1365-2958.2008.06127.x. [DOI] [PubMed] [Google Scholar]
- 31.Ozaki S, et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 32.Su'etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- 33.Katayama T. Roles for the AAA+ motifs of DnaA in the initiation of DNA replication. Biochem Soc Trans. 2008;36:78–82. doi: 10.1042/BST0360078. [DOI] [PubMed] [Google Scholar]
- 34.Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J Biol Chem. 2009;24:24. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zawilak A, et al. Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 2001;29:2251–2259. doi: 10.1093/nar/29.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soni RK, Mehra P, Mukhopadhyay G, Dhar SK. Helicobacter pylori DnaB helicase can bypass Escherichia coli DnaC function in vivo. Biochem J. 2005;389:541–548. doi: 10.1042/BJ20050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitharwal RG, et al. The domain structure of Helicobacter pylori DnaB helicase: The N-terminal domain can be dispensable for helicase activity whereas the extreme C-terminal region is essential for its function. Nucleic Acids Res. 2007;35:2861–2874. doi: 10.1093/nar/gkm167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- 39.Fossum S, et al. A robust screen for novel antibiotics: Specific knockout of the initiator of bacterial DNA replication. FEMS Microbiol Lett. 2008;281:210–214. doi: 10.1111/j.1574-6968.2008.01103.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.