Abstract
Tail-anchored (TA) membrane proteins are involved in a variety of important cellular functions, including membrane fusion, protein translocation, and apoptosis. The ATPase Get3 (Asna1, TRC40) was identified recently as the endoplasmic reticulum targeting factor of TA proteins. Get3 consists of an ATPase and α-helical subdomain enriched in methionine and glycine residues. We present structural and biochemical analyses of Get3 alone as well as in complex with a TA protein, ribosome-associated membrane protein 4 (Ramp4). The ATPase domains form an extensive dimer interface that encloses 2 nucleotides in a head-to-head orientation and a zinc ion. Amide proton exchange mass spectrometry shows that the α-helical subdomain of Get3 displays considerable flexibility in solution and maps the TA protein-binding site to the α-helical subdomain. The non-hydrolyzable ATP analogue AMPPNP-Mg2+- and ADP-Mg2+-bound crystal structures representing the pre- and posthydrolysis states are both in a closed form. In the absence of a TA protein cargo, ATP hydrolysis does not seem to be possible. Comparison with the ADP·AlF4−-bound structure representing the transition state (Mateja A, et al. (2009) Nature 461:361–366) indicates how the presence of a TA protein is communicated to the ATP-binding site. In vitro membrane insertion studies show that recombinant Get3 inserts Ramp4 in a nucleotide- and receptor-dependent manner. Although ATP hydrolysis is not required for Ramp4 insertion per se, it seems to be required for efficient insertion. We postulate that ATP hydrolysis is needed to release Get3 from its receptor. Taken together, our results provide mechanistic insights into posttranslational targeting of TA membrane proteins by Get3.
Keywords: ATP-binding protein, crystal structure, Asna1/Trc40, Get pathway, posttranslational targeting
Insertion of proteins into the membrane of the endoplasmic reticulum (ER) can proceed by different pathways (1–2). The best understood one is the signal recognition particle (SRP)-dependent pathway in which the N-terminal signal sequence as part of the ribosome nascent chain complex is targeted to the ER membrane and insertion through the Sec61 translocation channel proceeds cotranslationally (3–5). Different pathways are used by tail-anchored (TA) proteins that are anchored to the membrane by a single C-terminal transmembrane domain (TMD) (6–8). They are excluded from cotranslational targeting used by most membrane proteins by the C-terminal position of their targeting signal. Members of this protein class are components of the ER translocation site like Sec61β and the ribosome-associated membrane protein 4 (Ramp4), SNARE proteins involved in vesicular trafficking, or apoptotic proteins of the Bcl2 family (7, 9–11). One pathway of membrane insertion of TA proteins involves a highly conserved ATPase, called Asna1 or TRC40 (10). Mammalian Asna1/TRC40 (12) shows sequence similarity to bacterial ArsA, the catalytic subunit of the Escherichia coli arsenite resistance pump but has evolved a different function. Asna1/TRC40, like its yeast homolog Get3, associates in the cytosol with the TMD of TA proteins, targets their cargos to the ER, and mediates insertion into the ER membrane. Efficient membrane insertion was found to be ATP-dependent (9, 10). In vivo analyses show that deletion of Get3 in Saccharomyces cerevisiae is not lethal but is sensitive to heat and metal stress (13, 14), whereas knockout of the Get3 homolog Asna1 in mice results in early embryonic lethality (15).
In addition to Get3, 4 other proteins of the Get pathway were detected recently using genetic and physical interaction analysis. It was shown that Get1 and Get2 together form a receptor at the ER membrane (16), whereas Get4 and Get5 interact with Get3 in the cytosol to form the yeast TMD recognition complex (17). In the absence of its receptor Get1/Get2, Get3-TA protein complexes aggregate in the cytosol and delivery of TA proteins to the ER fails (16). Thus, Get3 seems to function also as a membrane protein chaperone that keeps TA proteins in an insertion-competent state (18).
Up to now, the molecular mechanism of TA protein recognition and binding by Get3 remained unclear. TA protein insertion is regulated by ATP binding and hydrolysis in Get3. The sequence similarities observed between Get3 and bacterial ArsA suggest that they share a similar fold. ArsA is a member of the SRP, MinD, BioD (SIMIBI) class of nucleotide-binding proteins, which is characterized by the formation of nucleotide-dependent dimers (19). The SIMIBI class contains ATP- and GTP-binding proteins, and all members contain insertions of different length in the nucleotide-binding domain (NBD) that are connected to the switch I and/or switch II region. The crystal structures of ArsA show conformational changes related to the nucleotide load, which propagate to the helical insertions that carry an arsenite-binding site (20, 21). In the well-characterized SRP GTPases (22–24), a unique insertion in the switch I region plays an essential regulatory role in the activation of GTP hydrolysis during cotranslational protein targeting (25–28).
To understand the molecular mechanisms of Get3-directed TA protein insertion, we determined the crystal structure of Get3 from a thermophilic fungus in the non-hydrolyzable ATP analogue AMPPNP-Mg2+- and ADP-Mg2+-bound states. Recently, two reports on the crystal structure of Get3 were published (29, 30). These studies showed the nucleotide-free and ADP structures of Get3 in an open state and the ADP·AlF4−-bound structure in a closed state. However, important intermediate states of the catalytic cycle of the Get3 ATPase were still missing. The two structures determined in our study represent the pre- and posthydrolysis states and complete the series of snapshots along the catalytic cycle of the Get3 ATPase. We used amide hydrogen exchange mass spectrometry (HX-MS) (31, 32) to analyze the conformational dynamics of Get3 in solution and to determine the TA-binding site. Furthermore, we performed in vitro membrane insertion studies and show that TA protein insertion by Get3 depends on nucleotide and a membrane receptor. ATP hydrolysis is not a prerequisite for TA protein insertion per se but is required for efficient insertion into the ER membrane. Taken together, our functional data and comparisons of the pre- and posthydrolysis structures with the transition state (29) allow us to propose a model for Get3-mediated TA protein targeting.
Results and Discussion
Overall Structure of the Get3 Dimer.
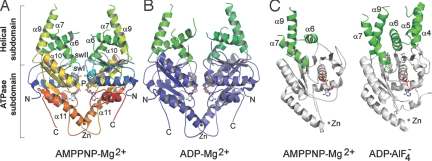
To understand the molecular mechanism of Get3-dependent TA protein insertion, we aimed to determine the crystal structure of Get3. We took advantage of the thermophilic fungus Chaetomium thermophilum (C. therm.) and cloned the get3 gene from this organism (see SI Text). C. therm. Get3 shares 53% sequence identity and 71% similarity with Homo sapiens Asna1 (49% and 66%, respectively, with S. cerevisiae Get3) (Fig. S1). Using the ATPase domain of ArsA (PDB ID code 1f48) as a search model, the AMPPNP-Mg2+- and ADP-Mg2+-bound Get3 structures were solved by molecular replacement at resolutions of 3 and 3.5 Å, respectively. Crystallographic statistics are given in Table S1. The asymmetric unit contains one Get3 homodimer with dimensions of 70 × 60 × 50 Å3 (Fig. 1 A and B), which correlates with the dimer observed as the major form of Get3 in solution. The homodimer encloses a continuous interface of 1,275 Å2 of buried surface area in the AMPPNP-Mg2+-bound structure (1,125 Å2 for the ATPase subdomain) and 1,241 Å2 in the ADP-Mg2+-bound structure as calculated with PISA (33). Each Get3 monomer consists of two domains, an ATPase and an α-helical subdomain. The C. therm. AMPPNP-Mg2+- and ADP-Mg2+-bound Get3 structures represent the pre- and posthydrolysis states and are very similar overall (rmsd of 0.5 Å over 259 residues; Fig. S2 A and D). In contrast to the ADP structure (30), the additional presence of the Mg2+ ion leads to a closed structure that involves a rotation of the two monomers toward each other similar to ArsA (20), NifH (34), or the NBDs of ABC transporter proteins (35, 36). Therefore, both structures show the Get3 dimer in a closed state, as indicated by the close proximity of the two P-loops [distances between Gly-36 are 4.1 Å in Get3 with AMPPNP-Mg2+ and 4.5 Å with ADP-Mg2+; 3.6 Å in Saccharomyces cerevisiae with ADP·AlF4− (29)]. The open state is indicated by longer distances [e.g., 13.8 Å in ADP (30) and 16.9 Å in the Schizosaccharomyces pombe apo form (29)]. The nucleotides are arranged head-to-head in the dimer interface. The ATPase subdomains of the homodimer are clamped together by a zinc ion, which is coordinated by two conserved cysteine residues (C281 and C284) from helix α11 (Fig. 1 A and B and Fig. S2B). The presence of a zinc ion in heterologously expressed C. therm. Get3 was confirmed by several independent methods (Fig. S2B). The functional importance of the zinc ion was recently demonstrated in yeast, because Get3 variants that are not able to bind zinc are monomers and cannot complement growth defects of a Δget3 yeast strain (37). The high conservation of Get3 suggests a similar functional role of the zinc ion in C. therm. Get3. The zinc ion might act as a hinge that allows opening and closing of the dimer depending on cargo binding or on the nucleotide load, as proposed previously (29). In contrast, isolated NBDs of ABC transporter proteins that are not held together by a metal ion are typically monomers in the absence of nucleotide, whereas in the full transporters, they stay associated as dimers independent of the nucleotide (35).
Fig. 1.
Overall structure of Get3. (A) AMPPNP-Mg2+-bound structure of C. therm. Get3. The dimer is in the closed state. N- and C-termini, the P-loop, swI, swII, and the Zn ion and secondary structure elements mentioned in the text are labeled. Coloring of secondary structure elements is done in a ramp from blue (N-terminus) to red (C-terminus). (B) ADP-Mg2+-bound structure of C. therm. Get3. The α-helical (green) and ATPase (blue) subdomains for the 2 chains are colored in similar shades. (C) Overall changes in the α-helical subdomains in different nucleotide states. Monomeric Get3 is shown with AMPPNP-Mg2+ of C. therm. (Left; same for ADP-Mg2+) and with ADP·AlF4− of S. cerevisiae (Right; PDB code 2woj). The α-helical and ATPase subdomains are colored in green and gray, respectively. In the AMPPNP-Mg2+-bound (and ADP-Mg2+-bound) states, the α-helical subdomains are only partially folded.
Get3 shares the fold of the SIMIBI class of nucleotide-binding proteins (19). Using the DALI server (38), the highest structural similarities are found with ArsA [Fig. S2C; rmsd of 1.88 Å over 208 residues (20)], Soj (39), MinD (40), and NifH (34), followed by other SIMIBI class members BioD (41) and the 3 SRP GTPases Ffh (22), FtsY (23), and FlhF (24). The α-helical subdomain of Get3 is composed mainly of 2 parts (helices α4–6 and α7–9), which are inserted adjacent to the switch I and switch II regions (Fig. 1). Although an α-helical subdomain is also present in NBDs of ABC transporter proteins, the Get3 homologs contain an additional insertion between helices α7 and α9 [TRC40 insert (29)]. The Get3 α-helical subdomain is enriched in methionine and glycine residues (Fig. S1) and has been proposed as a putative TA protein-binding site (29, 30). Similar to the proposed signal sequence-binding site in the M domain of SRP54 (42, 43), the sequence conservation of these regions is low. The physicochemical properties of methionine make this residue excellently suited to interact with hydrophobic regions without sequence specificity. In both structures of C. therm., Get3 helices α6, α7, and α9 form about half of the α-helical subdomain and are ordered similar to the ADP-bound structure of Get3 from S. cerevisiae (30). The hydrophobic groove, as observed in the ADP·AlF4−-bound structure (29), is therefore not fully assembled. A comparison of the different Get3 structures shows that arrangement of the helices varies significantly (Fig. 1C). The α-helical subdomain therefore seems highly dynamic and may adopt a defined structure only upon TA protein binding.
Localization of Fast-Exchanging Regions in Get3.
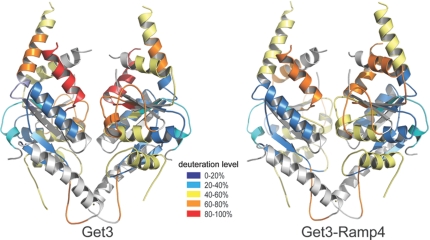
To understand the conformational flexibility of Get3, particularly of the α-helical subdomain in solution, we performed continuous-labeling 1H/2H HX-MS experiments, as described previously (44, 45). With this approach, the global stability and different conformational states of a protein can be analyzed. It also allows determining protein–protein interaction sites (31). Get3 in its apo form [corresponding to the open form observed previously (29, 30)] exchanged about 57% of its exchangeable amide hydrogens within 10 s in D2O and about 82% within 1 h. Well-folded globular proteins generally exchange about 50% of their exchangeable amide hydrogens within 1 h (46). The α-helical subdomain, which is only partially ordered in the crystal structures, comprises about 30% of Get3. Therefore, the overall exchange characteristics observed for Get3 indicate a high degree of solvent accessibility, which correlates nicely with what is expected for a protein with highly dynamic regions. To localize regions within Get3 that contain slow- and fast-exchanging amide hydrogens, the protein was digested after the continuous-labeling HX reaction under quench conditions (47). The peptides generated were subsequently analyzed by electrospray ionisation mass spectrometry (ESI-MS). This procedure allows identifying structural elements in Get3 that are tightly folded or are in a rapid unfolding-folding equilibrium. The obtained peptides cover about 80% of the Get3 sequence, with an average length of about 12 residues (Fig. 2; the analysis is shown in detail in Fig. S3 and Fig. S4A). The N-terminus of Get3 shows a fast exchange, indicating a highly dynamic structure accessible to solvent, which might explain why Get3FL protein crystals did not diffract to high resolution. The ATPase subdomain shows less deuteron incorporation, indicating a high level of structural rigidity (Fig. 2). Overall, the protection against HX agrees nicely with the crystallographic data. The well-folded secondary structure elements of the ATPase subdomain provide a stable structural framework for Get3. Highly dynamic or solvent-accessible regions indicated by fast exchange (e.g., residues 105–126, residues 139–149, residues 156–183, residues 218–235) belong mainly to the α-helical subdomain (helices α4–6 and α7–9). In solution, helices α4–9 show a fast exchange, indicating that these helices are in a rapid unfolding-folding equilibrium, and helix α7, which is adjacent to switch II, shows almost complete deuteration within 10 s (for the kinetics of deuteron incorporation, see Fig. S4B). Helices α6, α7, and α9 contribute about half of the putative TA protein-binding site and are present in all crystal structures of Get3. However, high B-factors and partial disorder have been observed (29). In the crystal structure of apoGet3, helix α7 is partially dissolved (29). In the AMPPNP-Mg2+- and ADP-Mg2+-bound structures (this study), it is present but poorly ordered, and in the ADP·AlF4−-bound structure, it is well ordered (29). In all previous crystal structures, these helices are involved in crystal contacts, and are therefore stabilized to a different extent. The HX-MS data support the notion that the differences in helix arrangement observed in the crystal structures are an intrinsic property of the highly dynamic α-helical subdomain.
Fig. 2.
Localization of Ramp4-induced changes in Get3 using HX-MS. Ramp4-dependent changes in HX kinetics of Get3 segments are projected onto the crystal structure. Get3 secondary structure is colored according to the deuteron incorporation shown in Fig. S3 for Get3 alone (Left) and the Get3–Ramp4 complex (Right): blue, 0–20%; cyan, 20–40%; yellow, 40–60%; orange, 60–80%; and red, 80–100%. It should be noted that helix α7 gets significantly stabilized.
Localization of Ramp4-Induced Changes in Get3.
The crystal structures of Get3, together with mutagenesis data, pull-down experiments, and complementation assays, suggest that the α-helical subdomain is involved in TA protein binding (refs. 29, 30; this study). To characterize the interaction of Get3 with TA proteins, we coexpressed and copurified C. therm. Get3 with C. therm. Ramp4 similar to the human Asna1–Ramp4 complex. The Get3–Ramp4 complex does not contain bound nucleotide as analyzed by HPLC analysis. We performed an HX-MS experiment of the Get3–Ramp4 complex similar as before for Get3 alone. To localize TA protein-induced changes in Get3, we compared deuteron incorporation into different segments of the Get3–Ramp4 complex with Get3 alone (Fig. 2 and Fig. S3, Middle and Right). The presence of Ramp4 had a small but distinct effect. Protection of amide hydrogens was observed specifically in the α-helical subdomain (residues 156–183 and residues 218–235), corresponding to helices α7, α8, and α9. A small but significant deprotection was observed at the end of helix α6 (residues 139–152). Ramp4 binding had no significant effect on the ATPase subdomain, indicating that cargo remains bound in the absence of nucleotide and does not induce a completely closed form as observed in the nucleotide-bound Get3 crystal structures. The specific protection of the α-helical subdomain suggests that it is indeed involved in TA protein binding.
Binding of Ramp4 Can Induce Oligomerization of Get3.
Get3 forms a stable dimer in solution independent of the nucleotide load. Because higher oligomeric forms of Get3 homologs were described previously (48, 49), we analyzed C. therm. Get3 and the Get3–Ramp4 complex by analytical ultracentrifugation (AUC; Fig. S5). For Get3 alone, 3 species were identified in the c(s)/c(M) distributions computed from the sedimentation velocity analysis: monomer (48%, 3.09S), dimer (51%, 4.45S), and tetramer (1.5%, 7.73S), with corresponding molecular masses of 37.5, 62.7, and 144.0 kDa, respectively. The ratio between monomer and dimer varies, with 50% being the lowest dimer fraction observed. In the presence of ADP or AMPPNP, the Get3 dimer shows a lower sedimentation coefficient, which suggests a more compact structure (Fig. S5B). This observation is in line with the crystal structures, which show that binding of nucleotide and Mg2+ induces a closed state of Get3. The copurified Get3–Ramp4 complex displays a behavior in AUC [and size exclusion chromatography (SEC)] very different from that of Get3 alone and is present mainly as a higher oligomer with a sedimentation coefficient of 7.39S (corresponding to a molecular mass of 136 kDa), indicative of a tetramer (Fig. S5 C and D). Negligible amounts of a higher oligomeric state were also observed that might represent a hexamer. TA protein binding seems therefore able to induce oligomerization of Get3. Because the recombinant Get3–Ramp4 complex does not contain nucleotide, TA proteins remain bound to Get3 in the absence of nucleotide. The relevance of oligomerization for the in vivo function of Get3 remains to be shown. However, in Δget1/2 yeast strains (16), Get3 is found in large protein aggregates in the cytosol, which suggests that disruption of membrane insertion induces aggregation of Get3–substrate complexes. The observed tetrameric Get3–Ramp4 complex that is formed during coexpression in E. coli might represent a stable intermediate that is formed because of the absence of the membrane components required for TA protein insertion. It underlines the chaperone function of Get3, which keeps TA proteins in solution before membrane insertion.
Recombinant Get3–Ramp4 Complex Is Functional in Membrane Insertion.
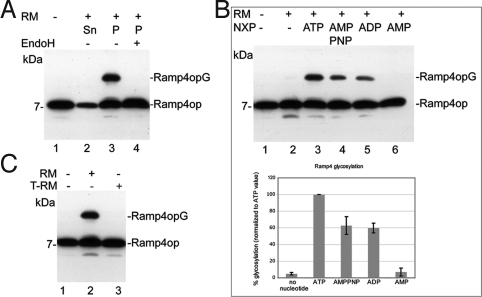
To test whether C. therm. Get3 can mediate insertion of Ramp4 into ER-derived membranes, we performed membrane insertion assays using a chimeric complex of C. therm. Get3-mammalian Ramp4 and rough microsomal membranes (RMs; Fig. 3). The chimeric complex was chosen because the membrane insertion of mammalian Ramp4 has been analyzed in detail. Ramp4 as part of the Get3–Ramp4 complex (Fig. 3A, lane 1) is shifted to a higher molecular weight form after incubation with RMs (lane 3), indicating membrane insertion and glycosylation of Ramp4. Disappearance of this band on deglycosylation (lane 4) confirms that the slower migrating band indeed represents a glycosylated form of Ramp4. Membrane insertion of Ramp4 requires the presence of nucleotides (Fig. 3B), because membrane insertion is not observed in the absence of ATP (lane 2). In addition to ATP, the nonhydrolyzable ATP analogue AMPPNP and ADP can promote significant membrane insertion of Ramp4 (lanes 3–5) but not AMP (lane 6), suggesting that the presence of nucleotide (with both the α- and β-phosphates) is required for membrane insertion of Ramp4. Membrane insertion in the presence of both ADP and AMPPNP reaches about 60% of the level obtained with ATP (Fig. 3B, Lower). Because ADP and AMPPNP promote membrane insertion but ATP is more efficient, hydrolysis is not required for insertion per se. We speculate that the release of Get3 from a membrane receptor, similar to the situation in the SRP system (25–27), might depend on hydrolysis, so that efficient insertion is only observed with ATP. To test whether Get3-dependent insertion into mammalian RMs requires a proteinaceous receptor, we treated RMs with trypsin to remove domains of membrane proteins exposed to the cytosol (Fig. 3C). Such treated RMs were no longer capable of promoting membrane insertion and glycosylation of Ramp4 (lanes 2 and 3). A proteinaceous component is therefore crucial for membrane insertion. Using the chimeric C. therm. Get3–Ramp4 complex and mammalian RMs, the inserted fraction was similar to that obtained with the homologous system. C. therm. Get3 can therefore functionally interact with the membrane components present in mammalian RMs. The high efficiency of membrane insertion using this heterologous system underlines the high degree of conservation of both the insertion machinery and the molecular mechanism of insertion.
Fig. 3.
Functional characterization of the Get3–Ramp4 complex. (A) Membrane insertion and glycosylation of Ramp4. The chimeric C. therm. Get3-mammalian–Ramp4op complex (for definition of Ramp4op, see SI Materials and Methods) was incubated in the presence of ATP without (lane 1) or with (lanes 2 to 4) RMs. The mixtures were separated into a supernatant (Sn) fraction and a pellet (P) fraction. Endoglycosidase H (EndoH) was added to one-half of the pellet fraction (lane 4). (B) Nucleotide requirements for membrane insertion of Ramp4op. (Upper) Get3–Ramp4op complexes were incubated with RMs in the absence or presence of adenosine nucleotides as indicated. (Lower) Amounts of glycosylated Ramp4op observed with different nucleotides are shown. (C) Dependence of Ramp4op membrane insertion on a membrane proteinaceous factor. Get3–Ramp4op complex was incubated with RMs (lane 2) or with trypsin-treated RMs (T-RMs; lane 3) in presence of ATP.
Communication Between the Nucleotide and the TA Protein-Binding Sites.
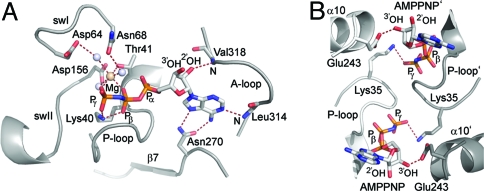
We have shown that the recombinant Get3–Ramp4 complex is capable of promoting efficient membrane insertion in the presence of nucleotide. To understand the communication between the nucleotide and the TA protein-binding sites in Get3, we analyzed the pre- and posthydrolysis states of Get3 (AMPPNP-Mg2+- and ADP-Mg2+-bound, respectively) in more detail and compared them with the recent structure of the transition state [ADP·AlF4−-bound (29)]. In the AMPPNP-Mg2+-bound (and in the ADP-Mg2+-bound) Get3 structure, the dimer is in a closed state. The α-helical subdomain is not stably assembled; therefore, it does not significantly contribute to the dimer interface. Comparison with the apo- and ADP-bound open structures (29, 30) shows that the presence of the Mg2+ ion induces the closed state. The nucleotide is primarily bound by one monomer but interactions in trans are present (Fig. 4). In the AMPPNP-Mg2+-bound structure, the A-loop and Asn-270 adjacent to strand β7 interact with the adenine ring of the nucleotide. The ribose interacts with Val-318 (main chain) of the A-loop in cis and with Glu-243 from the start of helix α10 in trans. The P-loop residues Gly-37, Gly-39, Lys-40, Thr-41, and Thr-42 interact with the α- and β-phosphates of AMPPNP (and ADP) in cis, whereas the conserved Lys-35 interacts with the γ-phosphate in trans (Fig. 4B). Corresponding interactions are also present in the ADP·AlF4−-bound structure (29). The Mg2+ ion is octahedrally coordinated in the AMPPNP-Mg2+-bound (and ADP·AlF4−-bound) structures by 3 water molecules, the β- and γ-phosphate oxygens and Thr-41 (Figs. 4A and 5B). However, the switch I and switch II regions are not well defined and are too far apart to allow ATP hydrolysis. The closed state of Get3 observed in the AMPPNP-Mg2+-bound structure is therefore not sufficient for ATP hydrolysis.
Fig. 4.
Nucleotide-binding site of AMPPNP-Mg2+-bound Get3. (A) Key interactions in the nucleotide binding site in cis. Important elements such as the A-loop, P-loop, swI, and swII are labeled. AMPPNP and side chains of interacting amino acids are shown in sticks, Mg2+ as a wheat-colored sphere, and water molecules as gray spheres. (B) Key interactions in the nucleotide binding site in trans. Lys-35 from the P-loop and Glu-243 from helix α10 interact with AMPPNP in trans. Secondary structure elements belonging to the second monomer are indicated by a single quotation mark, such as α10′ and P-loop', in Figs. 4 and 5.
Fig. 5.
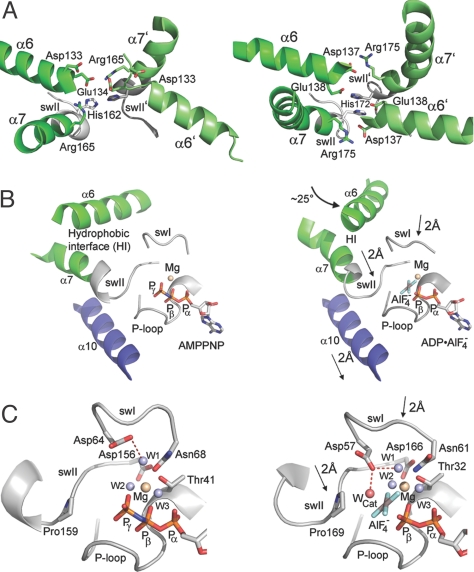
Communication between the TA protein and the nucleotide-binding sites in Get3. The Get3 prehydrolysis state (Left, AMPPNP-Mg2+) is compared with the transition state (Right, ADP·AlF4−, PDB ID code 2woj). (A) Active site closure viewed from the TA binding site. In the AMPPNP-Mg2+-bound structure, helices α6 and α7 pack loosely against each other, leaving the nucleotide solvent exposed. In the transition state, they close tightly over the nucleotide binding site. (B) Differences in the α-helical subdomain and in nucleotide-binding motifs between the prehydrolysis and the transition states are shown with the following changes observed in the transition state: helix α6 rotates around 25°, and helix α7, swI, swII, and helix α10 move toward the nucleotide. Helices α6 and α7 interact by means of a hydrophobic interface (HI). (C) Detailed view of the active site. The closing up in the transition state is clearly visible. The catalytic water (WCat) can be placed by the conserved aspartate from swI only in the transition state.
Major differences between the prehydrolysis (and posthydrolysis) state and the transition state are observed (Fig. 5). In the latter, the α-helical subdomain is more stably assembled and the dimer interface is closed up further. The “ordering” of the α-helical subdomain contributes to a significant increase of dimer interface by formation of a putative composite TA-binding site. Closing up the dimer in the transition state establishes symmetrical interactions across the dimer interface that are not present in the pre- and posthydrolysis states (Fig. 5A). Switch II and helices α6 and α7 play an important role, and the residues involved are highly conserved. In the ADP·AlF4−-bound structure, His-172 stacks to His-172′ across the dimer interface and interacts with Glu-138′ in trans. A salt bridge is formed across the dimer interface between Asp-137 and Arg-175′. In the AMPPNP-Mg2+-bound structure, these interactions are not present, because the interface is not fully closed. In C. therm. Get3, His-162 is rotated with respect to His-162′ and they are located further apart. His-162 does not interact with Glu-134, and the salt bridge between Arg-165 and Asp-133 is not formed because of the different orientation of helix α6 at the bottom of the TA protein-binding site. For example, in a superposition of the AMPPNP-Mg2+- and ADP·AlF4−-bound structures, the Cα-atoms of Glu-134 and the corresponding Glu-138 are more than 5 Å apart.
Closing up is communicated to the active sites in the ATPase subdomains, as indicated by a shorter distance between the two P-loops across the dimer interface. Whereas in the AMPPNP-Mg2+-bound (and ADP-Mg2+-bound) structure, the nucleotides are still accessible from the solvent, they are completely shielded in the transition state. In the fully closed transition state, helix α7 and the switch regions are well ordered. Helices α7 and α10 are moved in by 2 Å, and helix α6 reorients by about 25° (Fig. 5B). A hydrophobic interface between helices α6 and α7 takes part in the transition and suggests how closing up could be triggered by the hydrophobic TMD of a TA protein. Moving in the switch I and switch II regions by about 2 Å results in the formation of a water-filled cavity. An intricate hydrogen-bonding network of the enclosed water molecules is set up, with the putative catalytic water being oriented by Asp-57 from the switch I region (Asp-64 in C. therm.; Fig. 5C). Therefore, the closing up of the active sites in the transition state organizes the environment of the γ-phosphate as required for efficient ATP hydrolysis.
A detailed analysis of the different catalytic states of Get3 demonstrates how the dimer interface is used to communicate between the TA protein-binding site and the active site of the ATPase. The differences observed, particularly for helices α6 and α7, suggest how cargo binding could induce the closing up of the dimer, which is required for ATP hydrolysis. Thereby, futile cycles of ATP hydrolysis by Get3 are prevented in the absence of TA proteins. The release of inorganic phosphate after hydrolysis seems blocked by the presence of a TA protein, because closing up the interface, as seen in the transition state, completely shields the nucleotide-binding sites.
Integrated Model for TA Protein Targeting and Insertion.
Based on two crystal structures of Get3 representing the open and fully closed states, a plausible model was proposed for how TA protein insertion is regulated by Get3 (29). In the cytosol, ATP binding to Get3 drives the dimer to the closed state so that TA proteins can interact with their cognate binding site in the α-helical subdomain. The Get3-TA protein complex may stay in the closed state, which is not competent in ATP hydrolysis, until it reaches the Get1/2 receptor at the ER membrane. On interaction with the receptor and/or other partners, the fully closed state (transition state) is established and ATP gets hydrolyzed. Relaxation to the closed state after release of inorganic phosphate triggers TA protein release to the membrane. Conversion to the open state induces dissociation of Get3 from the membrane. In this model, membrane insertion would be strictly dependent on ATP hydrolysis.
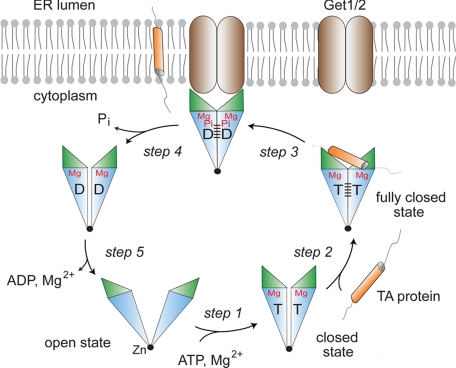
Having now in hand structural snapshots of almost the complete catalytic cycle of the Get3 ATPase as well as functional data on membrane insertion, a modified and integrated model can be proposed for how TA protein binding and insertion are linked to the ATPase cycle of Get3. We have shown that membrane insertion by Get3 does not depend on ATP hydrolysis per se, because significant insertion is also observed with ADP and AMPPNP. This is somewhat reminiscent of cotranslational targeting by the SRP system, which is regulated by GTPases that are also members of the SIMIBI class. Here, the activation of GTP hydrolysis in the targeting complex is required for the release of SRP from its receptor (25, 26). Applied to Get3 and TA protein insertion, this would translate to a scenario in which ATP hydrolysis is required for the dissociation of Get3 from the Get1/2 receptor. We therefore favor the following mechanism (Fig. 6). TA proteins bind to Get3 in the closed state (ATP-, AMPPNP-, or ADP-Mg2+-bound) (step 2). Subsequent interaction with the Get1/2 receptor triggers TA protein release to the membrane (step 3). In case of the Get3-ATP-Mg2+ complex, the transition to the fully closed state allows ATP hydrolysis but ADP and inorganic phosphate stay trapped in the closed active site. The chemical energy of ATP hydrolysis is stored in a strained conformation. After release of the TA protein, ADP and inorganic phosphate can be released and relaxation of Get3 to the open state induces dissociation from the receptor (step 4). A somewhat similar mechanism based on related structural differences as observed here for Get3 has been reported for NBDs of ABC transporter proteins (35, 50). In case of a Get3-AMPPNP-Mg2+ or Get3-ADP-Mg2+ complex, TA proteins are also inserted into the membrane. However, Get3 stays trapped at the Get1/2 receptor, because the energy of ATP hydrolysis is not available to drive dissociation. This model explains the lower efficiency of TA protein insertion observed with ADP and AMPPNP. AMP does not support membrane insertion because it does not drive the dimer to the closed state. Whether ATP hydrolysis occurs immediately on TA protein binding (step 2) or on Get1/2 receptor interaction (step 3) is not clear at present. It will therefore be exciting in the future to delineate the molecular details of how TA protein binding regulates the ATPase cycle of Get3 and how it modulates the interaction with the Get1/2 receptor.
Fig. 6.
Model for TA binding and insertion. In the cytosol, the nucleotide-free (and ADP-bound) Get3 dimer is open. ATP binding to Get3 drives closure of the dimer (step 1) and allows TA protein binding. TA binding induces the fully closed hydrolysis-competent state (step 2). Get1/2 receptor docking occurs before or after ATP hydrolysis, with ADP and inorganic phosphate staying trapped in the closed active site (step 3). After TA protein release, inorganic phosphate is released and Get3 dissociates from the Get1/2 receptor (step 4). The Get3 dimer adopts an open structure on release of Mg2+ (step 5).
Methods
Summary.
Detailed methods on cloning, purification, crystallization, structure determination, and biochemical characterization (HX-MS, AUC, and membrane insertion studies) are provided in SI Materials and Methods. Briefly, C-terminally His-tagged Get3 from C. therm. was cloned into the pet24d vector and expressed in E .coli. Recombinant Get3 was purified with Ni-affinity and SEC. In the presence of AMPPNP-Mg2+ or ADP-Mg2+, C. therm. Get3 crystallized in the primitive orthorhombic space group P212121. The AMPPNP-Mg2+- and ADP-Mg2+-bound Get3 structures were solved by molecular replacement using the ATPase domain of ArsA (PDB ID code 1f48) as a search model.
Supplementary Material
Acknowledgments.
We thank the European Synchrotron Radiation Facility for access to beamlines ID23–1 and ID23–2 and for excellent user support. We thank Matthias Mayer (Zentrum für Molekulare Biologie der Universität Heidelberg) for providing access to the HX-MS setup and initial training, Jürgen Kopp and Claudia Siegmann of the Heidelberg University Biochemistry Center–CellNetworks crystallization platform, Volker Windeisen and Patrick König for help with the AUC experiments, and Michael Krachler for induced coupled plasma MS (Environmental Geochemistry, Heidelberg University). This work was supported by a collaborative research grant from the Deutsche Forschungsgemeinschaft (SFB 638) and the Graduiertenkolleg GRK1188, the Boehringer Ingelheim Fonds, and the German-Israeli-Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3IQW and 3IQX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910223106/DCSupplemental.
References
- 1.Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 2.Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 3.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 4.Wild K, Rosendal KR, Sinning I. A structural step into the SRP cycle. Mol Microbiol. 2004;53:357–363. doi: 10.1111/j.1365-2958.2004.04139.x. [DOI] [PubMed] [Google Scholar]
- 5.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Abell BM, Rabu C, Leznicki P, Young JC, High S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J Cell Sci. 2007;120:1743–1751. doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- 9.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Janiak F, Glover JR, Leber B, Rachubinski RA, Andrews DW. Targeting of passenger protein domains to multiple intracellular membranes. Biochem J. 1994;300:191–199. doi: 10.1042/bj3000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurdi-Haidar B, et al. Isolation of the ATP-binding human homolog of the arsA component of the bacterial arsenite transporter. Genomics. 1996;36:486–491. doi: 10.1006/geno.1996.0494. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals. 2003;16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- 14.Zuniga S, Boskovic J, Jimenez A, Ballesta JP, Remacha M. Disruption of six Saccharomyces cerevisiae novel genes and phenotypic analysis of the deletants. Yeast. 1999;15:945–953. doi: 10.1002/(SICI)1097-0061(199907)15:10B<945::AID-YEA394>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay R, Ho YS, Swiatek PJ, Rosen BP, Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580:3889–3894. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabu C, High S. Membrane protein chaperones: A new twist in the tail? Curr Biol. 2007;17:R472–R474. doi: 10.1016/j.cub.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: The catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, Radaev S, Rosen BP, Gatti DL. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J Biol Chem. 2001;276:30414–30422. doi: 10.1074/jbc.M103671200. [DOI] [PubMed] [Google Scholar]
- 22.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 23.Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 24.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc Natl Acad Sci USA. 2007;104:13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bange G, Wild K, Sinning I. Protein translocation: Checkpoint role for SRP GTPase activation. Curr Biol. 2007;17:R980–R982. doi: 10.1016/j.cub.2007.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C, Connolly T, Morrison T, Gilmore R. Integration of membrane proteins into the endoplasmic reticulum requires GTP. J Cell Biol. 1988;107:69–77. doi: 10.1083/jcb.107.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol Chem. 2009;390(8):775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- 29.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009 doi: 10.1038/nature08319. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suloway CJ, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. Analyst. 2003;128:623–628. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- 32.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Georgiadis MM, et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 35.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem. 2006;281:410–417. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 38.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: Structure and DNA binding of the Soj dimer—A conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi I, Oyama T, Morikawa K. Structural and functional studies of MinD ATPase: Implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 2001;20:1819–1828. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kack H, Gibson KJ, Lindqvist Y, Schneider G. Snapshot of a phosphorylated substrate intermediate by kinetic crystallography. Proc Natl Acad Sci USA. 1998;95:5495–5500. doi: 10.1073/pnas.95.10.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 43.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rist W, Jorgensen TJ, Roepstorff P, Bukau B, Mayer MP. Mapping temperature-induced conformational changes in the Escherichia coli heat shock transcription factor sigma 32 by amide hydrogen exchange. J Biol Chem. 2003;278:51415–51421. doi: 10.1074/jbc.M307160200. [DOI] [PubMed] [Google Scholar]
- 45.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 46.Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc Natl Acad Sci USA. 2001;98:956–961. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Post CB, Smith DL. Amide hydrogen exchange determined by mass spectrometry: Application to rabbit muscle aldolase. Biochemistry. 1996;35:779–791. doi: 10.1021/bi952227q. [DOI] [PubMed] [Google Scholar]
- 48.Kurdi-Haidar B, Heath D, Aebi S, Howell SB. Biochemical characterization of the human arsenite-stimulated ATPase (hASNA-I) J Biol Chem. 1998;273:22173–22176. doi: 10.1074/jbc.273.35.22173. [DOI] [PubMed] [Google Scholar]
- 49.Wang HW, et al. Trimeric ring-like structure of ArsA ATPase. FEBS Lett. 2000;469:105–110. doi: 10.1016/s0014-5793(00)01257-6. [DOI] [PubMed] [Google Scholar]
- 50.Zaitseva J, et al. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 2006;25:3432–3443. doi: 10.1038/sj.emboj.7601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.