Abstract
The σ-like factor YvrI and coregulator YvrHa activate transcription from a small set of conserved promoters in Bacillus subtilis. We report here that these two proteins independently contribute σ-region 2 and σ-region 4 functions to a holoenzyme-promoter DNA complex. YvrI binds RNA polymerase (RNAP) through a region 4 interaction with the β-subunit flap domain and mediates specific promoter recognition but cannot initiate DNA melting at the -10 promoter element. Conversely, YvrHa possesses sequence similarity to a conserved core-binding motif in σ-region 2 and binds to the N-terminal coiled-coil element in the RNAP β′-subunit previously implicated in interaction with region 2 of σ-factors. YvrHa plays an essential role in stabilizing the open complex and interacts specifically with the N-terminus of YvrI. Based on these results, we propose that YvrHa is situated in the transcription complex proximal to the -10 element of the promoter, whereas YvrI is responsible for -35 region recognition. This system presents an unusual example of a two-subunit bacterial σ-factor.
Keywords: RNA polymerase, initiation, promoter, open complex
Transcription is catalyzed by an evolutionarily conserved enzyme complex comprising a core catalytic complex together with accessory specificity factors. In bacteria, the RNA polymerase (RNAP) core enzyme contains, minimally, the ββ′α2-assembly, but promoter recognition and transcription initiation additionally require a σ-factor. The σ-factor binds RNAP and targets the resulting holoenzyme to cognate promoter sites (1).
All bacteria encode a primary σ-factor (called RpoD or σ70 in Escherichia coli and σA in Bacillus subtilis) that is responsible for the transcription of most genes expressed during vegetative growth. Primary (or group 1) σ-factors consist of four functional regions called regions 1, 2, 3, and 4 (2–5). Group 2 σ-factors are structurally similar to group 1 σ-factors but are not essential to the cell. Group 3 σ-factors retain only regions 2–4, whereas group 4 [or extracytoplasmic function (ECF)] σ-factors carry only the two most functionally important and strongly conserved regions, 2 and 4. Regions 2 and 4 are responsible for recognition of the -10 and -35 sequence elements found in most bacterial promoters (6).
Region 2 is the most highly conserved region in bacterial σ-factors (4, 7) and can be further subdivided into regions 2.1, 2.2, 2.3, and 2.4. Amino acids in regions 2.1 and 2.2 form a pair of α-helices that mediate core binding by virtue of interaction with a coiled-coil element in the amino terminus of the β′-subunit of core RNAP (8). This interaction is critical for holoenzyme formation, because a number of mutations in these interacting elements negatively influence σ-binding to core (9–15). This interaction is also important for σ-region 2.3–2.4 recognition of the single- and double-stranded -10 element within promoter DNA (16, 17). Finally, amino acids in region 2.3 play a critical role in recognizing and binding single-stranded nontemplate DNA encompassing the -10 region (18–20). In doing so, region 2.3 stabilizes the transcription bubble during open-complex formation (19).
Region 4 is also a highly conserved and functionally important region in σ-factors. Region 4.2 includes a helix-turn-helix (HTH) domain that recognizes and binds the -35 element. Region 4 also interacts with hydrophobic amino acids in the β-flap domain of the core RNAP β-subunit (21), and this interaction properly orients the HTH domain for interaction with -35 region DNA (22). Additionally, region 4 is a frequent target for regulators that modulate RNAP activity through interaction with σ (23).
Recently, an alternative σ-like regulator named YvrI was identified in B. subtilis (24). YvrI carries a carboxy-terminal σ70-family region 4 but is quite divergent in its amino terminus. Indeed, domain prediction programs fail to identify a σ70-family region 2 in YvrI despite this region being the most highly conserved segment in σ-factors (4, 7). In contrast, the other 17 σ70-family σ-factors in B. subtilis all carry a predicted region 2. YvrI nevertheless copurifies with RNAP and activates transcription from at least three conserved promoters in B. subtilis (24). YvrI activation in the cell is linked both to challenge with certain cell wall-acting antibiotics (25) and to growth under acidic conditions (24, 26). Activation of YvrI during growth under low pH conditions results in the accumulation of a secreted oxalate decarboxylase (OxdC) as the most abundant protein in the cell wall (27). YvrI-activated transcription additionally requires the 9.5-kDa YvrHa protein (24), although the nature of this requirement has not been previously defined.
Here, we demonstrate that YvrHa carries a σ70-family region 2, the same region poorly represented in YvrI, and that this region interacts with the β′-coiled-coil element. YvrI carries region 4, which interacts with the β-flap-tip helix. In single-subunit σ-factors, the homologous regions are known to contact these same regions of β and β′. Thus, YvrI and YvrHa reconstitute σ-factor form and function, enabling efficient promoter melting and transcription initiation.
Results
Previously, we demonstrated that both YvrI and YvrHa are required for activation of transcription from a group of conserved promoters in B. subtilis and other closely related species (24, 26). Sequence analysis suggests a model in which YvrHa contains σ70-like regions 2.1, 2.2, and 2.3, whereas YvrI carries a typical region 4 and a highly divergent region 2 (possibly including a functional region 2.4). Here, we present an analysis of protein–protein and protein–DNA interactions that, together with in vitro and in vivo transcription results, support this model.
YvrI Region 4 Interacts with β-Flap.
YvrI binds RNAP (24) and is predicted to carry a σ-region 4. Previous results have demonstrated that region 4 of σ70 specifically contacts the β-flap domain in the core enzyme. To test whether similar contacts are made by YvrI, we used a bacterial two-hybrid (BACTH) system. In this system, test polypeptides are expressed as fusions with either a DNA-binding domain (λcI) or the amino-terminal domain (NTD) of the α-subunit, and positive interactions are manifest as an increase in β-galactosidase activity from an E. coli host strain (28).
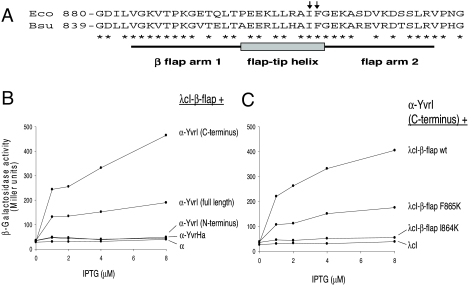
To monitor interactions with the B. subtilis β-flap domain, amino acids 817–896 (homologous to E. coli β-flap amino acids 858–937) were fused to λcI and tested for interacting partners fused to the α-subunit NTD (Fig. 1A). YvrI, but not YvrHa, interacts with β-flap (Fig. 1B). Genetic bisection of YvrI into amino- and carboxy-terminal segments [supporting information (SI) Fig. S1] revealed that this interaction is specifically mediated through the C-terminal portion of YvrI corresponding to region 4 (Fig. 1B). Because the β-subunit sequence is largely conserved between E. coli and B. subtilis (Fig. 1A), we introduced mutations in the B. subtilis β-flap that are known to affect interaction negatively between the E. coli β-flap (I905K and F906K; Fig. 1A) and σ70-region 4 (21). As predicted, the corresponding B. subtilis flap mutations (I864K and F865K) dramatically reduced the observed interaction between region 4 of YvrI and the β-flap (Fig. 1C). These results support a model in which the YvrI-core interaction is analogous to that observed between conventional σ-factors and the β-subunit of core RNAP.
Fig. 1.
YvrI region 4 interacts with the β-flap-tip helix. (A) Relevant amino acid sequence of E. coli and B. subtilis β-flap-tip regions (21). Two tip helix mutations (I905K and F906K) that impair in E. coli binding by σ70-region 4 (21) are indicated (arrows). For interaction assays (BACTH), the region from amino acid 817 to 896 was fused to λcI. (B) BACTH analysis to test YvrI and YvrHa binding to B. subtilis β-flap. (C) BACTH analysis of the effect of I864K and F865K flap-tip mutations on the B. subtilis β-flap–YvrI region 4 interaction.
YvrI region 4.2 carries a predicted HTH motif that aligns with those in σ-factors (e.g., B. subtilis σX and σW; Fig. S2A). We mutated 2 residues (Q166 and S169) in the YvrI HTH recognition helix to alanine (based on their strong conservation among YvrI homologs; Fig. S3), and, despite similar expression levels (Fig. S2C), both mutations strongly impaired in vivo transcription (Fig. S2B). These data, coupled with the observation that YvrI-dependent promoters possess a conserved -35 promoter motif (TTGACW) (24, 26), suggest that YvrI region 4.2 mediates recognition of -35 promoter DNA and that this is critical for transcription.
YvrI Binds Core and Acts as a Promoter-Binding Specificity Factor.
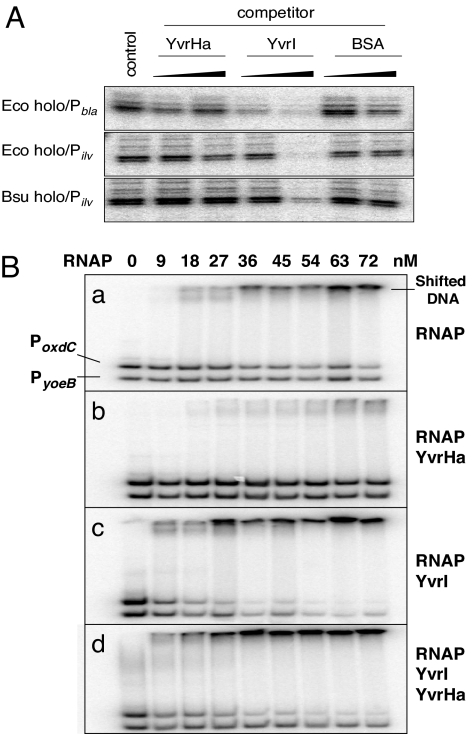
We reasoned that if YvrI interacted with core RNAP (E) at the same site as conventional σ-factors, this binding should competitively inhibit recognition of σA-dependent promoters. Indeed, YvrI inhibited promoter interaction, as judged by monitoring DNA melting using KMnO4 footprinting, by either Eσ70- or EσA-holoenzymes at three different promoters (Fig. 2A). In contrast, YvrHa did not competitively appear to inhibit promoter recognition and melting by holoenzyme. Thus, although YvrI-dependent activation of transcription relies on YvrHa, YvrI binding to RNAP does not.
Fig. 2.
σ-Factor activities of YvrI. (A) YvrI competitively inhibits DNA melting by E. coli σ70- and B. subtilis SigA. Reactions include E. coli σ70-holoenzyme melting of the E. coli bla and B. subtilis ilv promoters and B. subtilis SigA holoenzyme melting of the ilv promoter. Competitor proteins [YvrHa, YvrI, and a control protein (BSA)] were either not added (control lane) or added at a concentration 5-fold and 10-fold over the concentration of holoenzyme (62 nM) before oxidation reaction using KMnO4. (B) YvrI is a promoter specificity factor determined using EMSA and end-labeled YvrI-dependent oxdC promoter DNA (PoxdC) and a control promoter DNA (PyoeB). Each binding reaction contained the indicated concentration of RNAP (a) and 500 nM YvrHa (b), 500 nM YvrI (c), and 500 nM YvrHa and 500 nM YvrI (d). Note that the initial binding reaction in b, c, and d containing 0 nM RNAP includes the indicated amount of YvrI and/or YvrHa to control for nonspecific binding by these proteins.
To test whether YvrI acts as a promoter-binding specificity factor, we mixed increasing concentrations of RNAP containing either YvrI or YvrHa (or both) with end-labeled PoxdC DNA (or control DNA) and monitored binding using an EMSA assay (Fig. 2B). As expected, RNAP core binds both target DNAs nonspecifically (Fig. 2B, a). Interestingly, YvrHa largely abolished this nonspecific binding (Fig. 2B, b), an activity reminiscent of σ70 (29, 30). In contrast, YvrI alone resulted in a bias toward shifting PoxdC DNA vs. the control DNA (Fig. 2B, c), suggesting that the E-YvrI complex has significant promoter recognition capability. Finally, inclusion of both YvrI and YvrHa in the binding reactions further increased the specificity of binding (Fig. 2B, d), probably because YvrHa is reducing residual nonspecific binding of the control DNA by RNAP itself. These data suggest that YvrI (in the absence of YvrHa) can act as a specificity factor and direct binding to PoxdC DNA, even though, as shown previously (24), it is insufficient to activate transcription.
YvrHa Is Also a σ-Factor-Related Protein.
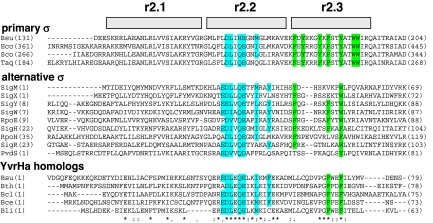
YvrHa is a 9.5-kDa protein encoded by the gene downstream of, and cotranscribed with, yvrI. A multiple sequence alignment of YvrHa and its orthologs with various σ-factors (Fig. 3) demonstrates a remarkable similarity in the region corresponding to the most highly conserved portions of region 2 (including the originally defined “RpoD box”) (31). The amino acid sequence EDLxQE within region 2.2 is the principal point of conservation and is thought to be part of a conserved σ-factor core interface (4, 7). This EDLxQE motif in YvrHa is also the most strongly conserved region among its homologs (Fig. 3, Bottom), implying functional importance. Previously, mutations in σ-factor that negatively influence holoenzyme formation (presumably by disrupting interaction with the β′-subunit coiled coil) were mapped to this subsequence (13–15). To determine if this region is also critical for YvrHa function, we engineered mutations in this motif. As predicted, mutations in this conserved region of YvrHa strongly impaired transcriptional activation from a target promoter in vivo (Fig. S4).
Fig. 3.
σ-Factor region 2-like sequence in YvrHa includes functionally important residues. Composite alignments of the relevant sequences [regions 2.1, 2.2, and 2.3 as previously defined (4)] from a selection of primary and alternative σ-factors and the complete sequences of YvrHa and four YvrHa homologs. Highly conserved amino acids in regions 2.2 are highlighted in blue, whereas positionally conserved aromatic amino acids among the subgroups are highlighted in green. ClustalW was used to generate each individual alignment, and conservation symbols (asterisks) pertain to YvrHa homolog alignment only. For the alternative σ-factors: sequences SigM, SigX, SigW, SigY, and SigH are from B. subtilis; RpoE and RpoH are from E. coli; SigR is from Streptomyces coelicolor; and PvdS is from Pseudomonas aeruginosa. For the vegetative and YvrHa proteins, abbreviations are as follows: Bsu, B. subtilis; Eco, E. coli; Sco, S. coelicolor; Taq, Thermus aquaticus; Bth, Bacillus thuringiensis; Bcl, Bacillus clausii; Bce, Bacillus cereus; Bli, Bacillus licheniformis.
YvrHa also displays similarity to conserved region 2.3, which characteristically carries positionally conserved aromatic amino acids known, in the vegetative factors of B. subtilis and E. coli, to play a role in stabilizing open-complex formation (18, 19). This region in YvrHa might also play a role in DNA melting. Note that the sequence of YvrHa ends after region 2.3, and therefore does not allow for a region 2.4-like segment.
YvrHa Copurifies with RNAP.
Previously, it was shown that overexpressed YvrI copurifies with RNAP (24). To examine whether YvrHa stably associates with RNAP in vivo, we partially purified hexahistidine-tagged RNAP (12) from B. subtilis after coinduction of epitope-tagged YvrI and YvrHa. Indeed, both YvrI and YvrHa were copurified with RNAP using nickel affinity chromatography (Fig. S5B). YvrI and YvrHa was not detected in control purifications from cells not expressing YvrI and YvrHa (Fig. S5A) or expressing WT RNAP (Fig. S5D). We also tested a variant of YvrHa carrying a mutation (D43A) in its conserved region 2.2 EDLxQE motif that resulted in decreased transcription activation in vivo (Fig. S4). Although the mutant protein was readily detected in crude lysate (Fig. S5C), it copurified with RNAP to a much lesser extent than WT YvrHa. This supports the hypothesis that this region 2.2 motif in YvrHa is involved in core binding, consistent with results for conventional σ-factors (13–15).
YvrHa Interacts with the β′-Subunit Coiled-Coil Element.
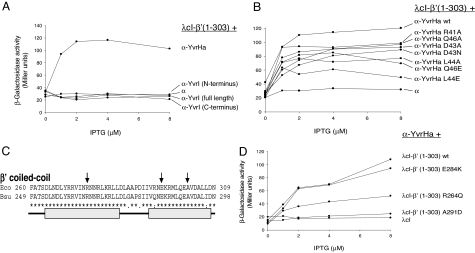
Because YvrHa carries what appears to be a σ-region 2.2 (including the EDLxQE motif), we tested whether it could interact with the initial 303 aa of the β′-subunit from B. subtilis that, by analogy to E. coli β′ (amino acids 1–314) (12), should contain the coiled-coil element critical for region 2 core interaction. Indeed, YvrHa interacted with this region (Fig. 4A). Several point mutations in this conserved region of YvrHa, many of which strongly decrease transcription in vivo (Fig. S4), also decreased the magnitude of this apparent interaction (Fig. 4B). In repeated experiments, the most dramatic impairment resulted from mutation of amino acid L44 in the YvrHa EDLxQE motif. Mutations E42A and E47A had no significant impact on this interaction, whereas E45A, D43R, and a double-mutant D43A/Q46A slightly increased the apparent interaction. In contrast, we were unable to detect an interaction between β′ (amino acids 1–303) and YvrI, despite the possible presence of a divergent region 2 in the amino terminus of YvrI (Fig. 4A).
Fig. 4.
YvrHa interacts with the coiled-coil domain in β′-subunit. (A) BACTH analysis of YvrHa and YvrI interactions with β′ (amino acids 1–303). (B) Influence of mutations in the region 2.2-like motif (Fig. 3) of YvrHa on binding with β′ (1–303). (C) Sequence of the E. coli and B. subtilis coiled-coil element in β′ as previously defined (8). Mutations in E. coli β′ (R275Q, E294K, and A302D) that impair β′-binding to σ70-region 2 are indicated with arrows. Approximate dimensions of helices that contribute to the coiled-coil domain are indicated by gray bars. (D) Influence of mutations in B. subtilis β′ (analogous to mutations shown in C) on β′-binding to YvrHa.
Several mutations in the E. coli β′-subunit coiled-coil element impair its interaction with σ-region 2 (12), including (E. coli numbering) R275Q, E295K, and A302D (Fig. 4C). The introduction of two analogous mutations in the B. subtilis β′- (amino acids 1–303) fusion protein (R264Q and A291D) reproducibly and significantly impaired the interaction with YvrHa in the BACTH system (Fig. 4D). In contrast, an E284K mutation had no significant effect. These results support a model in which the region 2.2 motif of YvrHa makes a specific interaction with the coiled-coil element of the B. subtilis β′-subunit.
YvrHa Is Required to Stabilize Open-Complex Formation.
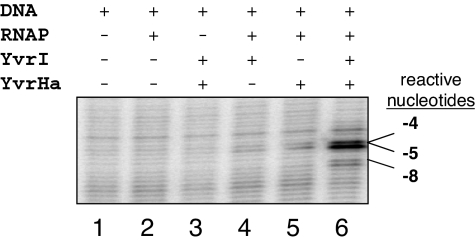
Collectively, our data indicate that the E-YvrI complex can mediate promoter binding (Fig. 2B), but previous data indicate that it cannot activate transcription (24). Therefore, we monitored the effects of YvrI and YvrHa alone and together on DNA melting of PoxdC using KMnO4 footprinting and negatively supercoiled promoter DNA (Fig. 5). Despite acting as a promoter specificity factor, YvrI itself is unable to stabilize open-complex formation (Fig. 5, lane 4). In contrast, the E-YvrI-YvrHa holoenzyme melts (minimally) a ≈12-nt region of the oxdC promoter (Fig. 5, lane 6, and Fig. S6). Thus, YvrHa is essential for DNA melting, and this can account for its essential function in transcription initiation.
Fig. 5.
YvrI and YvrHa are required for promoter melting. Permanganate sensitivity of nucleotides in the template strand of PoxdC as influenced by the addition of indicated proteins is shown. Nucleotide coordinates are shown in Fig. S6. Reactions contained B. subtilis RNAP (60 nM) with other proteins added to a final concentration of 300 nM.
YvrHa Interacts with the Amino Terminus of YvrI.
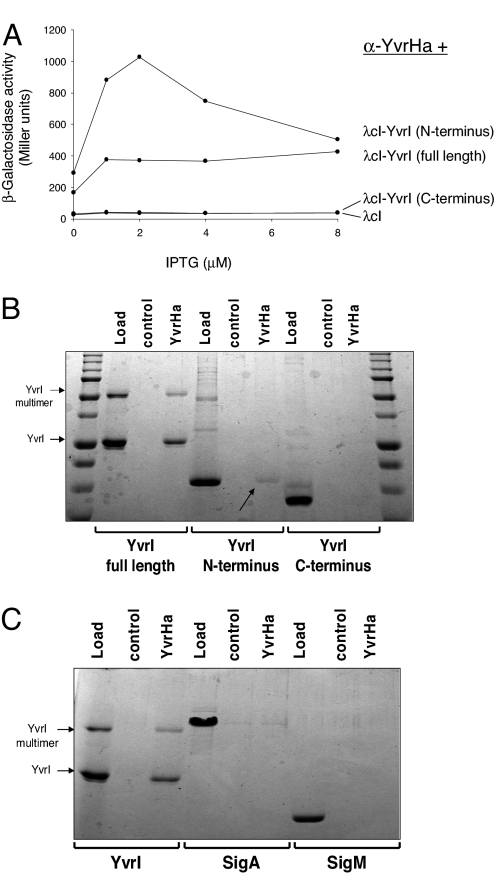
Because both YvrHa and YvrI are required for transcription (24) and both make contacts with core subunits characteristic of σ-factors, we hypothesized that they might also interact with one another. In fact, a BACTH analysis demonstrated that they do and that YvrHa interacts with the amino but not carboxy (region 4) segment of YvrI (Fig. 6A). Further, using purified YvrHa (and a control protein, lysozyme) conjugated to NHS-activated sepharose beads (Fig. 6B), we found that both full-length YvrI and the amino segment of YvrI, but not the carboxy segment, were pulled down by immobilized YvrHa, thus corroborating the BACTH data.
Fig. 6.
YvrHa interacts with the amino terminus of YvrI. (A) BACTH analysis of interactions between YvrHa and either full-length YvrI or amino- and carboxyl-terminal segments of YvrI (Fig. S1). (B) In vitro pull-down assay using NHS-conjugated YvrHa or a control protein (lysozyme). Binding reactions included either full-length YvrI or the amino- or carboxy-terminal segment of YvrI. Arrows indicate bound amino-terminal fragment of YvrI. (C) In vitro pull-down assay testing interaction between immobilized YvrHa and either YvrI, SigA, or SigM. All test proteins were added to binding reactions to a final concentration of ≈1 μM.
To check whether binding to σ-factors might be a general property of YvrHa, we repeated the in vitro interaction experiment (Fig. 6C) with purified B. subtilis σA and an ECF σ-factor, σM. Unlike with YvrI, there was no indication that YvrHa specifically interacted with either protein. Therefore, YvrHa interacts specifically with the amino region of YvrI but not with other σ-factors tested.
Discussion
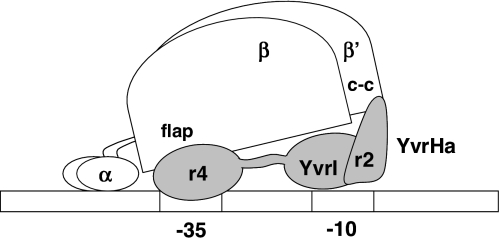
Collectively, these genetic, biochemical, and BACTH experiments support a model in which YvrI and YvrHa functionally reconstitute σ-factor activity on interaction with core RNAP. YvrHa binds the coiled-coil element in the β′-subunit and the amino-terminal segment of YvrI. The carboxy-terminal region 4 domain of YvrI interacts with the β-flap domain and contains an HTH motif postulated to mediate -35 region recognition. These results lead to a provisional model for the subunit architecture of the E-YvrI-YvrHa holoenzyme (Fig. 7).
Fig. 7.
Provisional model for a YvrI-YvrHa holoenzyme–promoter DNA closed transcription complex. Nonessential B. subtilis polymerase subunits (δ- and ω-subunits) are not included. The model depicts known interactions of YvrHa region 2-like segment (r2) with β′-coiled-coil domain (c-c), YvrHa with the amino terminus of YvrI, and YvrI region 4 (r4) with the β-subunit flap domain (flap). Drawing not to scale.
Because YvrI is about the size (23 kDa) of many alternative σ-factors and includes a divergent region 2, this model postulates that YvrI is broadly extended along a single face of core RNAP and that the amino terminus of YvrI is situated toward the leading edge of the holoenzyme. The ability to direct specific promoter DNA binding suggests that YvrI recognizes promoter DNA. This presumably includes the conserved -35 element (recognized by region 4) but may additionally include contacts to the -10 element mediated, in conventional σ-factors, by region 2.4.
Our model further proposes that YvrHa (which has sequence determinants highly similar to those of σ-factor region 2) is situated proximal to the -10 element of promoter DNA by virtue of its contacts with the N-terminal regions of YvrI and the β′-subunit (Fig. 7). In this position, it plays an essential role in open-complex formation, perhaps analogous to that played by σ-region 2 in conventional holoenzyme, which is situated proximal to, and acts on, DNA sequence in the -10 element during the transcription initiation process. Future analyses are to be directed at mapping the precise contacts between YvrHa and YvrI and promoter region DNA.
YvrI and YvrHa coregulate transcription from at least three conserved promoters in B. subtilis driving the expression of five genes, including the yvrI-yvrHa operon. Taken together, the sequence similarity of YvrHa to σ, its protein interaction patterns, and its required involvement in activating open-complex formation indicate that this “accessory” protein differs from previously characterized conventional transcription activators and also from more recently characterized regulators that function by interacting with RNAP but not DNA (32). The YvrI-YvrHa system is reminiscent of certain phage σ-factor systems that involve two proteins. For example, B. subtilis phage SPO1 proteins gp33 and gp34 are both required for activation of late genes, and both act as promoter-binding specificity factors (33), although the precise role of these proteins in transcription activation was never elucidated (34). The more thoroughly studied T4 phage regulators gp55 and gp33 participate in reconstituting σ-factor activity at late-phage promoters lacking -35 subsequences (35). In this system, gp33 (which carries no obvious sequence similarity to σ-factor) stimulates transcription by the divergent truncated σ-factor gp55 through a critical interaction with the flap domain of the β-subunit (36). Thus, gp33 partially substitutes for σ-region 4 function.
In contrast to these phage models, the YvrI-YvrHa system consists of two σ-factor-related proteins of bacterial origin that activate transcription from promoters with well-defined and conserved -10 and -35 elements (24, 26). The smaller accessory protein in this system (YvrHa) interacts with an N-terminal coiled-coil element in the β′-subunit, contacts the N-terminus of YvrI, and plays an essential and potentially direct role in DNA melting. Significantly, the yvrI-yvrHa gene order is not consistent with a simple frameshift-mediated splitting of an ancestral σ into two separately encoded polypeptides: in this system, the region 4-containing polypeptide (YvrI) is encoded upstream of the region 2-containing protein (YvrHa). We believe that YvrI-YvrHa represents the first known bipartite bacterial σ-factor. An investigation of the mechanism by which these proteins activate transcription should extend our understanding of noncanonical σ-factor systems and unusual transcription initiation pathways in bacteria.
Materials and Methods
Bacterial Strains and Plasmids.
The bacterial strains and plasmids used in this study are listed in Table S1.
In Vivo Expression and Protein Pull-Down Assays.
Relevant strains were grown overnight, and this growth was used to inoculate fresh 100-mL volumes of LB broth to OD600 of 0.05. After incubation at 37 °C with shaking to OD600 of 0.4, YvrI and YvrHa expression was induced with 2% (vol/vol) xylose for 1 h. Crude lysate was supplemented with 0.5 mL of Ni NTA beads (Qiagen) and incubated at 4 °C with slow rotation for 1 h. Bound His-tagged RNAP and cobinding proteins were eluted with a step gradient of increasing imidazole. Crude lysate and eluate samples were separated on 12% (vol/vol) SDS/PAGE gels, and proteins were visualized by Coomassie staining or were transferred to PVDF membrane and visualized by immunoblotting.
Electrophoretic Mobility Assays.
Each 10-μL binding reaction included <100 pmol end-labeled target DNA carrying either PoxdC (150 nt) or a control promoter (σA-dependent PyoeB) on a 110-nt fragment, constant amounts of YvrI and YvrHa, and increasing concentrations of RNAP (see legend for Fig. 2). Samples were loaded onto a 6% (vol/vol) nondenaturing polyacrylamide gel running at 100 V. After electrophoresis for 45 min, the gel was dried and exposed to a phosphor screen.
In Vitro Pull-Down Assays.
For in vitro interaction assays, 50 μg of YvrHa or a control protein was conjugated to NHS-activated beads (GE Lifesciences). After extensive washing in 100 mM Tris-HCl (pH 8.0), conjugated beads were stored in this same buffer as a 50% (vol/vol) bead slurry at 4 °C. A typical experiment involved resuspending 50 μL of the bead slurry in binding buffer and supplementing the suspension with 1 μM test protein. After extensive washing, the beads were resuspended in 40 μL of SDS/PAGE sample buffer, beads were sedimented, and 30 μL of sample buffer containing dissolved proteins was analyzed by SDS/PAGE using a 14% (vol/vol) polyacrylamide gel. For each experiment, conjugated lysozyme beads were used to control for incidental protein carryover and nonspecific binding of test proteins to conjugated protein, the sepharose bead matrix, or the reaction tube wall.
In Vivo Two-Hybrid Assay (BACTH).
The E. coli reporter strain FW Kan OL2–62 lac (37) was cotransformed with relevant recombinant bait and prey plasmids. Single colonies from each strain were used to inoculate a series of 2-mL volumes of LB broth containing ampicillin (100 μg/mL); kanamycin (50 μg/mL); chloramphenicol (25 μg/mL); and either 0, 1, 2, 4, or 8 μM isopropyl-β-d-thiogalactopyranoside (IPTG). After overnight growth at 37 °C, these cultures were used to inoculate fresh 5-mL volumes of the same broth series to OD600 of 0.05. When cultures reached OD600 of 0.3 to 0.5, 1-mL volumes were extracted, cells were collected by centrifugation, and pellets were frozen overnight. β-galactosidase assays were conducted as previously described (38). Each strain was tested in duplicate, and each experiment was conducted at least two times.
DNA Footprinting.
Potassium permanganate footprinting (39) was conducted using supercoiled plasmid (pSM106) DNA in a 20-μL reaction. Each reaction contained 285 nM B. subtilis RNAP and equimolar concentrations of YvrI and YvrHa. Reactions were started by the addition of 1 μL of 40 mM KMnO4 and were stopped after 3 min by the addition of 1 μL of β-mercaptoethanol. Oxidized nucleotides were detected using primer extension as previously described (24).
Supplementary Material
Acknowledgments.
The authors thank Ann Hochschild for providing reagents and advice on the BACTH system. This work was supported by the National Institutes of Health Grant GM047446.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910006106/DCSupplemental.
References
- 1.Burgess RR, Anthony L. How sigma docks to RNA polymerase and what sigma does. Curr Opin Microbiol. 2001;4:126–131. doi: 10.1016/s1369-5274(00)00177-6. [DOI] [PubMed] [Google Scholar]
- 2.Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 3.Stragier P, Parsot C, Bouvier J. Two functional domains conserved in major and alternate bacterial sigma factors. FEBS Lett. 1985;187:11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- 4.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribskov M, Burgess RR. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 7.Campbell EA, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 8.Arthur TM, Burgess RR. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta' subunit. J Biol Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- 9.Shuler MF, Tatti KM, Wade KH, Moran CP., Jr. A single amino acid substitution in sigma E affects its ability to bind core RNA polymerase. J Bacteriol. 1995;177:3687–3694. doi: 10.1128/jb.177.13.3687-3694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo DM, Nolte A, Calendar R, Zhou YN, Jin DJ. Multiple regions on the Escherichia coli heat shock transcription factor sigma32 determine core RNA polymerase binding specificity. J Bacteriol. 1998;180:1095–1102. doi: 10.1128/jb.180.5.1095-1102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesley SA, Burgess RR. Characterization of the Escherichia coli transcription factor sigma 70: Localization of a region involved in the interaction with core RNA polymerase. Biochemistry. 1989;28:7728–7734. doi: 10.1021/bi00445a031. [DOI] [PubMed] [Google Scholar]
- 12.Arthur TM, Anthony LC, Burgess RR. Mutational analysis of beta '260–309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. J Biol Chem. 2000;275:23113–23119. doi: 10.1074/jbc.M002040200. [DOI] [PubMed] [Google Scholar]
- 13.Sharp MM, et al. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MJ, Lamont IL. Mutational analysis of an extracytoplasmic-function sigma factor to investigate its interactions with RNA polymerase and DNA. J Bacteriol. 2006;188:1935–1942. doi: 10.1128/JB.188.5.1935-1942.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong K, Kassavetis GA, Leonetti JP, Geiduschek EP. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J Biol Chem. 2003;278:7073–7080. doi: 10.1074/jbc.M211447200. [DOI] [PubMed] [Google Scholar]
- 16.Gruber TM, et al. Binding of the initiation factor sigma(70) to core RNA polymerase is a multistep process. Mol Cell. 2001;8:21–31. doi: 10.1016/s1097-2765(01)00292-1. [DOI] [PubMed] [Google Scholar]
- 17.Young BA, Gruber TM, Gross CA. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- 18.Juang YL, Helmann JD. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 19.deHaseth PL, Helmann JD. Open complex formation by Escherichia coli RNA polymerase: The mechanism of polymerase-induced strand separation of double helical DNA. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones CH, Moran CP., Jr. Mutant sigma factor blocks transition between promoter binding and initiation of transcription. Proc Natl Acad Sci USA. 1992;89:1958–1962. doi: 10.1073/pnas.89.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates sigma(70) region 4 function. J Mol Biol. 2004;343:569–587. doi: 10.1016/j.jmb.2004.08.063. [DOI] [PubMed] [Google Scholar]
- 22.Kuznedelov K, et al. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 23.Dove SL, Darst SA, Hochschild A. Region 4 of sigma as a target for transcription regulation. Mol Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 24.MacLellan SR, Wecke T, Helmann JD. A previously unidentified sigma factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis. Mol Microbiol. 2008;69:954–967. doi: 10.1111/j.1365-2958.2008.06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLellan SR, Helmann JD, Antelmann H. The YvrI alternative sigma factor is essential for acid stress induction of oxalate decarboxylase in Bacillus subtilis. J Bacteriol. 2009;191:931–939. doi: 10.1128/JB.01435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antelmann H, Towe S, Albrecht D, Hecker M. The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J Proteome Res. 2007;6:897–903. doi: 10.1021/pr060440a. [DOI] [PubMed] [Google Scholar]
- 28.Dove SL, Hochschild A. Bacterial two-hybrid analysis of interactions between region 4 of the sigma(70) subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J Bacteriol. 2001;183:6413–6421. doi: 10.1128/JB.183.21.6413-6421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bautz EK, Bautz FA, Beck E. Specificity of sigma-dependent binding of RNA polymerase to DNA. Mol Gen Genet. 1972;118:199–207. doi: 10.1007/BF00333456. [DOI] [PubMed] [Google Scholar]
- 30.Mueller K. The function of the -factor of Escherichia coli RNA polymerase in template site selection. Mol Gen Genet. 1971;111:273–296. doi: 10.1007/BF00433112. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, Shiina T, Takahashi H. Multiple principal sigma factor homologs in eubacteria: Identification of the “rpoD box.”. Science. 1988;242:1040–1042. doi: 10.1126/science.3194753. [DOI] [PubMed] [Google Scholar]
- 32.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tijan R, Pero J. Bacteriophage SP01 regulatory proteins directing late gene transcription in vitro. Nature. 1976;262:753–757. doi: 10.1038/262753a0. [DOI] [PubMed] [Google Scholar]
- 34.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 35.Kassavetis GA, Elliott T, Rabussay DP, Geiduschek EP. Initiation of transcription at phage T4 late promoters with purified RNA polymerase. Cell. 1983;33:887–897. doi: 10.1016/0092-8674(83)90031-4. [DOI] [PubMed] [Google Scholar]
- 36.Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 38.Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. Methods Mol Biol. 2004;261:231–246. doi: 10.1385/1-59259-762-9:231. [DOI] [PubMed] [Google Scholar]
- 39.Sasse-Dwight S, Gralla JD. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.