The crystal structure of the esterase EstA from a cold-adapted bacterium was determined in a form that was covalently inhibited by monoethylphosphonate.
Keywords: esterases, psychrotrophic bacteria, monoethylphosphate inhibitors
Abstract
The crystal structure of the esterase EstA from the cold-adapted bacterium Pseudoalteromonas sp. 643A was determined in a covalently inhibited form at a resolution of 1.35 Å. The enzyme has a typical SGNH hydrolase structure consisting of a single domain containing a five-stranded β-sheet, with three helices at the convex side and two helices at the concave side of the sheet, and is ornamented with a couple of very short helices at the domain edges. The active site is located in a groove and contains the classic catalytic triad of Ser, His and Asp. In the structure of the crystal soaked in diethyl p-nitrophenyl phosphate (DNP), the catalytic serine is covalently connected to a phosphonate moiety that clearly has only one ethyl group. This is the only example in the Protein Data Bank of a DNP-inhibited enzyme with covalently bound monoethylphosphate.
1. Introduction
Lipolytic enzymes hydrolyze esters of organic acids of various chain lengths and encompass lipases, esterases and phospholipases. These enzymes have been divided into eight families according to conserved sequence motifs (Arpigny & Jaeger, 1999 ▶). All lipolytic enzymes belong to the α/β-hydrolase superfamily and contain a catalytic triad, which is usually formed by Ser, His and Asp residues. However, these enzymes vary significantly in their structures and properties. Esterases differ from lipases in substrate specificity and interfacial activation. Lipases prefer water-insoluble triglycerides of long-chain fatty acids, whereas esterases prefer smaller esters that are soluble (at least partially) in water. Various sources yield different esterases and they have acquired different nomenclatures, e.g. carboxylesterases, cholinesterases, acetylxylan esterases, phosphotriestarases, aryl esterases etc. Many esterases can hydrolyze various substrates of different types. Owing to their characteristics, such as broad substrate specificity, stereoselectivity, activity in organic solvents and lack of use of cofactors, they are widely used as biocatalysts in various biotechnological processes in the chemical, pharmaceutical and food industries (Bornscheuer, 2002 ▶; Panda & Gowrishankar, 2005 ▶).
Attention has recently been drawn to lipolytic enzymes from bacteria that live in permanently cold environments (Joseph et al., 2008 ▶; Soror et al., 2007 ▶; Kulakova et al., 2004 ▶; Suzuki et al., 2002 ▶, 2003 ▶). These cold-adapted enzymes can be applied as additives to detergents used at low temperatures and as biocatalysts for reactions involving labile substrates at low temperatures (Margesin & Schinner, 1994 ▶).
We have recently described and preliminarily characterized a novel cold-adapted esterase EstA from a psychrotrophic bacterium Pseudoalteromonas sp. strain 643A isolated from the guts of the Antarctic krill Euphasia superba Dana (Cieśliński et al., 2007 ▶; Długołęcka et al., 2008 ▶). Sequence analysis suggested that this enzyme belongs to the GDSL family and the SGNH subfamily owing to the presence of strictly conserved Ser, Gly, Asn and His residues in four conserved sequence blocks (Akoh et al., 2004 ▶). Here, the crystal structure of EstA covalently inhibited by monoethylphosphonate is presented at 1.35 Å resolution.
2. Experimental procedures
EstA was found to posses a 23-amino-acid signal peptide at its N-terminus, so the 69 nucleotides encoding this signal peptide were cut from the EstA gene. The primers used for amplification of the shortened EstA gene were Est_forward primer, 5′-TACTTCCAATCCAATGCCATGGACAACACGATTTTAATAC, and Est_reverse primer, 5′-TTATCCACTTCCAATGTTATTAGACGTTATTTAACC AC. The parts of the primer sequences that are complementary to the nucleotide sequences of the EstA gene are shown in bold.
The gene was amplified using Pfu DNA polymerase and the amplification product was analyzed by electrophoresis on 1% agarose gel with ethidium bromide. The PCR product was cloned into pRCS69 vector and the cloned gene was fused to maltose-binding protein (MBP) with a His6 tag at its N-terminus and a TEV protease-cleavage site at its C-terminus.
EstA was overexpressed in Escherichia coli BL21 (DE3) strain. Bacterial cells were induced with 0.4 mM IPTG at an OD600 of 0.8–1.0, grown overnight at 290 K and subsequently harvested by centrifugation and lysed by sonication in 20 mM HEPES buffer pH 7.5 containing 250 mM NaCl, 20 mM imidazole, 5% glycerol and 5 mM β-mercaptoethanol (buffer A). The lysate was clarified by centrifugation at 18 000 rev min−1 and the supernatant was loaded onto a 50 ml nickel column equilibrated with buffer A. Nonspecifically bound proteins were eluted with this buffer. EstA was eluted with buffer A containing 300 mM imidazole. The esterase fractions were pooled together, diluted to a final concentration of 150 mM imidazole and then mixed with TEV protease in a 1:25 ratio with the addition of 5 mM EDTA. The mixture was left overnight at 277 K in order to cleave the His6-tag-MBP. After enzymatic digestion, proteins were precipitated with ammonium sulfate (90% saturation) and spun down to remove EDTA. The proteins were then dissolved in 20 mM HEPES buffer pH 7.5 containing 250 mM NaCl, 5% glycerol and 5 mM β-mercaptoethanol (buffer B) and applied onto a second nickel column to separate EstA from His6-tag-MBP. The flowthrough EstA fractions were collected and again precipitated by ammonium sulfate (90% saturation). EstA was dissolved in 5 ml 20 mM HEPES buffer pH 7.5 with 250 mM NaCl and 5% glycerol and applied onto a Superdex200 size-exclusion column. The EstA peak fractions were concentrated using a 10 kDa cutoff Centricon (Vivaspin) to a concentration of 15 mg ml−1 as measured by Bradford assay (Bradford, 1976 ▶).
The purified EstA was preliminarily crystallized using robotic screens. After optimization, the best crystals of native EstA were obtained by mixing the protein solution with a well solution consisting of 1.6 M sodium/potassium phosphate, 0.1 M HEPES pH 7.5. Samples of inhibited EstA were obtained by soaking native EstA crystals in 1.5 mM diethyl p-nitrophenyl phosphate (DNP) solution for 2 d. Prior to X-ray diffraction measurements, the crystals were soaked in reservoir solution supplemented with 25% glycerol as a cryoprotectant.
Diffraction data were collected on the SER-CAT beamline 22BM at the Advanced Photon Source (Argonne National Laboratory) using a MAR 225 CCD detector and were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). The data statistics are shown in Table 1 ▶.
Table 1. Statistics of diffraction data and structure refinement.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P6122 |
| Unit-cell parameters | |
| a = b (Å) | 83.82 |
| c (Å) | 130.95 |
| Wavelength (Å) | 1.000 |
| Resolution limit (Å) | 30–1.35 (1.40–1.35) |
| Reflections measured | 843429 (68522) |
| Reflections | 60202 (5887) |
| Multiplicity | 14.0 (11.8) |
| Completeness (%) | 100.0 (100.0) |
| Rmerge (%) | 6.9 (68.0) |
| Average I/σ(I) | 38.8 (3.8) |
| Refinement | |
| Resolution (Å) | 30–1.35 |
| R factor (%) | 17.19 |
| Work reflections | 57091 |
| Rfree (%) | 18.89 |
| Free reflections | 3056 |
| R.m.s.d. bond lengths (Å) | 0.020 |
| R.m.s.d. bond angles (°) | 1.85 |
| Residues in favored conformation (%) | 90.2 |
| Residues in allowed conformation (%) | 9.8 |
| PDB code | 3hp4 |
The structure was solved by molecular replacement with MOLREP (Vagin & Teplyakov, 1997 ▶) using the molecule of E. coli thioesterase I (TAP; Lo et al., 2005 ▶; PDB code 1ivn) as a search model and the inhibitor was built into the difference map. Refinement and water selection were performed with REFMAC5 (Murshudov et al., 1997 ▶) and ARP/wARP (Perrakis et al., 1999 ▶), respectively. Real-space model corrections were performed with Coot (Emsley & Cowtan, 2004 ▶). The structure quality was validated with PROCHECK (Laskowski et al., 1993 ▶). The coordinates and structure factors were deposited in the Protein Data Bank and assigned the accession code 3hp4.
3. Results and discussion
The structure of EstA forms one compact domain, as is typical for SGNH hydrolases, and consists of a five-stranded parallel β-sheet with three helices at the convex side and two helices at the concave side of the sheet, with an additional couple of very short helices at the domain edges. All of the main chain is visible in the electron-density map except for the N-terminal Met and C-terminal Val. According to PROCHECK no residues are found to be in the disallowed regions of the Ramachandran plot (Ramachandran et al., 1963 ▶). 19 side chains are modeled with multiple conformations. The solvent water molecules were classified on the basis of electron density and B factors into 188 fully occupied and 69 half-occupied sites. In addition, the model contains the monoethylphosphonate moiety, which is covalently bound to the OG atom of Ser11. The structure was refined at 1.35 Å resolution to an R factor of 17.19% and an R free of 18.89% in isotropic mode with five TLS fragments (2–12, 13–38, 39–114, 115–137 and 138–184).
The structure of EstA is very similar to that of E. coli thioesterase I (TAP), which was used as a search model in molecular replacement (Lo et al., 2003 ▶, 2005 ▶). Fig. 1 ▶ presents the aligned sequences of EstA and TAP, and Fig. 2 ▶ shows these two molecules superimposed on each other. EstA has a three-residue insertion (33, 34 and 35); otherwise, the only regions that differ between these two models are the short loop fragments and both terminal chain ends. The r.m.s. difference between EstA and TAP calculated for all 178 Cα atoms corresponding in sequence is 1.50 Å and that calculated for 162 structurally corresponding Cα atoms is 0.93 Å.
Figure 1.
Sequence alignment of EstA and TAP (PDB code 1ivn). Identities are shown in red, similarities in blue and the catalytic triad Ser, His and Asp residues are shown on a gray background. The two terminal residues, shown in lower case, are not visible in the electron-density map.
Figure 2.
The structures of EstA (in green) and TAP (in brown) superimposed onto each other. The monoethylphosphonate inhibitor located at the catalytic serine in the catalytic site cleft is shown as a blue stick model.
The active site is located in a groove and contains the classic catalytic triad of Ser, His and Asp, which is identical to that in TAP. However, in contrast to the structure of TAP inhibited with DNP (PDB code 1j00; Lo et al., 2005 ▶) and all other DNP-inhibited protein models in the PDB, in the EstA structure there is clearly only one ethyl group connected to the phosphate bound to the catalytic Ser11 residue (Fig. 3 ▶). The only analogous serine residue covalently bound to the monoethylphosphate moiety is found in the structure of aged tabun-inhibited murine acetylcholinesterase (PDB code 2c0p; Ekström et al., 2006 ▶), the first conjugate of acetylcholinesterase with tabun. A non-aged covalent complex with both ethyl and dimethylamine groups connected to the catalytic serine is seen with diffraction data collected 1 h after inhibition. In the structure of the complex obtained using data collected after four weeks only the ethyl group is present and the dimethylamine moiety has been hydrolysed out. In order to check whether analogous aging takes place in EstA, several data sets were measured from crystals of EstA soaked in DNP solution for times ranging between a few minutes and several hours. However, in the crystals soaked for times between a few minutes and 3 h no significant electron density corresponding to covalent inhibitor was identified near the Ser10 OG atom. In crystals soaked for several hours the partially occupied inhibitor moiety could be observed with only one identifiable ethyl group. It seems that either the aging of the complex is relatively fast or that one of the ethyl groups is removed from the phosphate during reaction with EstA.
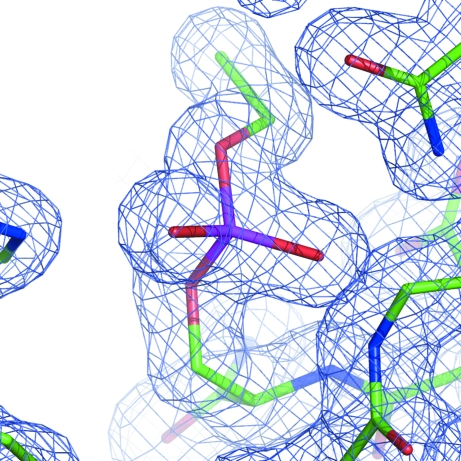
Figure 3.
The monoethylphosphonate inhibitor connected to the catalytic Ser11 is shown overlapped onto the 2F obs − F calc electron-density map contoured at the 1σ level.
Supplementary Material
PDB reference: EstA, 3hp4, r3hp4sf
Acknowledgments
This work was supported in part by Federal funds from the National Cancer Institute, National Institutes of Health contract No. NO1-CO-12400 and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government. Diffraction data were collected on the SBC 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H. & Shaw, J.-F. (2004). Prog. Lipid Res.43, 534–552. [DOI] [PubMed]
- Arpigny, J. L. & Jaeger, K.-E. (1999). Biochem. J.343, 177–183. [PMC free article] [PubMed]
- Bornscheuer, U. T. (2002). FEMS Microbiol. Rev.26, 73–81. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Cieśliński, H., Białkowska, A. M., Długołęcka, A., Daroch, M., Tkaczuk, K. L., Kalinowska, H., Kur, J. & Turkiewicz, M. (2007). Arch. Microbiol.188, 27–36. [DOI] [PubMed]
- Długołęcka, A., Cieśliński, H., Turkiewicz, M., Białkowska, A. M. & Kur, J. (2008). Protein Expr. Purif.62, 179–184. [DOI] [PubMed]
- Ekström, F., Akfur, C., Tunemalm, A.-K. & Lundberg, S. (2006). Biochemistry, 45, 74–81. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Joseph, B., Ramteke, P. W. & Thomas, G. (2008). Biotechnol. Adv.26, 457–470. [DOI] [PubMed]
- Kulakova, L., Galkin, A., Nakayama, T., Nishino, T. & Esaki, N. (2004). Biochim. Biophys. Acta, 1696, 59–65. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lo, Y.-C., Lin, S.-C., Shaw, J.-F. & Liaw, Y.-C. (2003). J. Mol. Biol.330, 539–551. [DOI] [PubMed]
- Lo, Y.-C., Lin, S.-C., Shaw, J.-F. & Liaw, Y.-C. (2005). Biochemistry, 44, 1971–1979. [DOI] [PubMed]
- Margesin, R. & Schinner, F. (1994). J. Biotechnol.33, 1–14.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Panda, T. & Gowrishankar, B. S. (2005). Appl. Microbiol. Biotechnol.67, 160–169. [DOI] [PubMed]
- Perrakis, A., Morris, R. J. H. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Ramachandran, G. N., Ramakrishnan, C. & Sasisekharan, V. (1963). J. Mol. Biol.7, 95–99. [DOI] [PubMed]
- Soror, S. H., Verma, V., Rao, R., Rasool, S., Koul, S., Qazi, G. N. & Cullum, J. (2007). J. Ind. Microbiol. Biotechnol.34, 525–531. [DOI] [PubMed]
- Suzuki, T., Nakayama, T., Choo, D. W., Hirano, Y., Kurihara, T., Nishino, T. & Eaki, N. (2003). Protein Expr. Purif.30, 171–178. [DOI] [PubMed]
- Suzuki, T., Nakayama, T., Kurihara, T., Nishino, T. & Eaki, N. (2002). J. Mol. Catal. B Enzym.16, 255–263.
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: EstA, 3hp4, r3hp4sf
PDB reference: EstA, 3hp4, r3hp4sf