Single orthorhombic crystals of M. genitalium protein MG289 have been grown and shown to diffract X-rays to 2.8 Å resolution with good statistics. The structure obtained from these data will help to provide insight into the function of the protein as well as improving the understanding of its role in this human pathogen.
Keywords: Mycoplasma genitalium, MG289, p37, extracytoplasmic thiamine-binding lipoprotein, Cypl
Abstract
Mycoplasma genitalium is a human pathogen that is associated with nongonococcal urethritis in men and cervicitis in women. The cloning, expression, purification and crystallization of the protein MG289 from M. genitalium strain G37 are reported here. Crystals of MG289 diffracted X-rays to 2.8 Å resolution. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 49.7, b = 90.9, c = 176.1 Å. The diffraction data after processing had an overall R merge of 8.7%. The crystal structure of Cypl, the ortholog of MG289 from M. hyorhinis, has recently been determined, providing a reasonable phasing model; molecular replacement is currently under way.
1. Introduction
Mycoplasmas (class Mollicutes) are tiny pleomorphic wall-free bacteria which survive in tandem with eukaryotes, either attached to the cell membrane or intracellularly. Known to infect plants, animals and humans, these organisms are highly pervasive and are often associated with pathological conditions. Mycoplasma genitalium is only 0.2–0.3 µm in diameter, with a genome size of 580 kb (Maniloff et al., 1992 ▶; Fraser et al., 1995 ▶). This bacterium preferentially associates with ciliated epithelial cells in the genitourinary and respiratory tracts (Baseman et al., 1988 ▶; Tully et al., 1981 ▶). It is associated with nongonococcal urethritis (NGU) in men and endocervicitis in women (Manhart et al., 2007 ▶; Gaydos et al., 2009 ▶; Anagrius et al., 2005 ▶). Studies have also indicated that M. genitalium may be a sexually transmitted infection, being 11 times more prevalent amongst people living with a sexual partner (Manhart et al., 2007 ▶). Particularly in the case of NGU, there is significant difficulty in treating this disease as tetracyclines are ineffective and there is an increasing instance of azithromycin resistance (Taylor-Robinson, 2008 ▶). It is increasingly apparent that a deeper understanding of this bacterium is needed.
M. genitalium protein MG289 is a putative substrate-binding protein located on an operon containing a theoretical ATP-binding cassette (ABC) transport system (Fraser et al., 1995 ▶). Although MG289 is one of the few proteins in M. genitalium with built-in redundancy, it is still considered to be a minimal gene and essential for life (Glass et al., 2006 ▶). This functional assignment was based on the sequence similarity of MG289 to its ortholog Cypl, also known as p37, found in M. hyorhinis (Gilson et al., 1988 ▶; Fraser et al., 1995 ▶; Dudler et al., 1988 ▶). The crystal structure of Cypl was recently determined and revealed a thiamine pyrophosphate located in a binding cleft (Sippel et al., 2008 ▶, 2009 ▶). Although there is only 32% identity between MG289 and Cypl, it is likely that they share similar secondary and tertiary structures (Adams & Oxender, 1989 ▶).
Understanding the structure of MG289 will offer new insight into the metabolism of M. genitalium and possibly offer a target to combat infection. Here, we report the cloning of MG289 from M. genitalium strain G37, as well as the expression, purification, crystallization and preliminary X-ray data analysis.
2. Methods and results
2.1. Cloning, expression and purification
The plasmid containing the coding sequence for MG289 was generated using PCR. Firstly, an N-terminally truncated form of the coding sequence (residues 26–386) was amplified using M. genitalium (strain G37) genomic DNA (the strain was a gift from Dr Joel Baseman, University of Texas and the genomic DNA was provided by Dr Leticia Reyes, Department of Infectious Disease and Pathology, College of Veterinary Medicine, University of Florida) and primers 5′-CATATGTGTGCAACCAAAAGCGATAACACCCTGATCTTTAATATTTCACTGGATCATAACGC-3′ (NdeI restriction site in bold) and 5′-CCGCTCGAGTTTTCCATAGGTTTTTTCAATTATTTCAACAACT-3′ (XhoI restriction site in bold). The N-terminal truncation was made to improve the solubility of the protein by removing a putative signal sequence and lipoprotein moiety. The PCR product was purified by agarose gel electrophoresis, digested with NdeI and XhoI and then ligated into a pET31f1m1 expression vector. Additionally, the TGA codon, which codes for Trp in mycoplasma but is a stop codon in Escherichia coli, was mutated to TGG. The plasmid was sequenced to verify that it was identical to published sequences (Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, Florida, USA).
2.2. Expression and purification
The protein was expressed and purified as described previously (Ketcham et al., 2005 ▶). Briefly, the plasmid was transformed into BL21(DE3)pLysS cells. 1 l LB supplemented with 100 mg l−1 ampicillin was inoculated and cultured until it reached an OD600 of 0.7–1.0. Isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM and the cells were allowed to grow for an additional 3 h before harvesting. The cells were lysed using a French press in a 1/100 volume of 20 mM phosphate buffer pH 7.95.
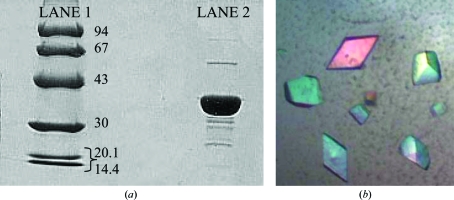
The lysate was centrifuged at 40 000g for 20 min at 277 K, after which the supernatant was applied onto a 50 ml Bio-Rad Q anion-exchange column with a 5 ml Bio-Rad Econo-Pac S cation-exchange column attached to the bottom to filter out highly basic proteins. Both columns were pre-equilibrated with 20 mM phosphate buffer pH 7.95. Approximately 125 mg total protein was applied to the column followed by a wash with the equilibration buffer. The flowthrough was collected, its pH was adjusted to 6.1 with acetic acid and it was loaded onto another 5 ml Bio-Rad Econo-Pac S cation-exchange column equilibrated with 20 mM sodium acetate pH 6.1. The column was washed with this buffer and the protein was eluted isocratically using 15% 1 M NaCl in 20 mM sodium acetate pH 6.1 and 85% 20 mM sodium acetate pH 6.1. The eluted sample was buffer-exchanged and concentrated into 50 mM Tris–HCl using a Centriprep 10 spin column (Millipore, Bedford, Massachusetts, USA). Purity was confirmed by 10% SDS–PAGE stained with Coomassie Brilliant Blue (Fig. 1 ▶ a).
Figure 1.
Purification and crystallization of MG289. (a) SDS–PAGE (10% SDS–PAGE stained with Coomassie Brilliant Blue) of the purified MG289 used for crystallization. Lane 1, low-molecular-weight standards (labeled in kDa); lane 2, MG289 sample (∼8 mg ml−1 in 50 mM Tris–HCl pH 7.5). (b) Crystals of MG289 grown using the hanging-drop vapor-diffusion method.
2.3. Crystallization and X-ray analysis
Preliminary crystallization screening was performed via the hanging-drop vapor-diffusion method using Crystal Screen I from Hampton Research (McPherson, 1982 ▶). A crystallization hit was obtained in a drop containing 2 µl purified protein (∼8 mg ml−1 in 50 mM Tris–HCl) and 2 µl precipitant solution [condition No. 20: 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6, 25% polyethylene glycol 4000 (PEG 4000)] equilibrated against 500 µl precipitant solution. Small bipyramidal crystals (approximately 0.05 × 0.03 × 0.03 mm) appeared after three weeks. Further investigation around the screening condition led to marginally larger crystals (0.07 × 0.05 × 0.05 mm; Fig. 1 ▶ b) using a drop consisting of 5 µl MG289 and 5 µl of a precipitant solution containing 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6 and 15% PEG 4000 equilibrated against 1 ml precipitant solution.
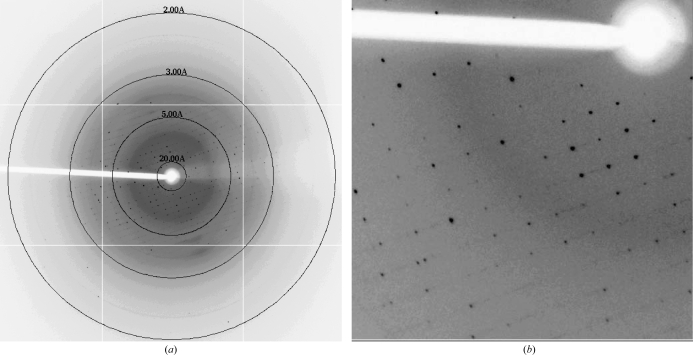
A single crystal was flash-cooled directly with no additional cryoprotectant. X-ray diffraction data were collected on the X29 beamline at Brookhaven National Laboratory (λ = 1.0809 Å) using an ADSC Quantum-315r nine-quadrant CCD detector. The crystal-to-detector distance was 200 mm. The oscillation width was 1°, with an exposure time of 1 s per image. 130 usable images were collected to a maximum resolution of 2.8 Å (Figs. 2 ▶ a and 2 ▶ b). The reflections were indexed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). Based on the unit-cell parameters, inspection of the intensities of the h00, 0k0 and 00l reflections and R merge statistics, the space group was assigned as P212121, with unit-cell parameters a = 49.7, b = 90.9, c = 176.1 Å. A total of 104 409 reflections were collected, 20 425 of which were unique. The data were 99.6% complete, with an average redundancy of 5.1 and an overall R merge of 0.087 (0.542 in the outer shell). Considering the presence of two molecules in the asymmetric unit, the Matthews coefficient was calculated in CNS to be 2.37 Å3 Da−1 with a solvent content of ∼46.0%, assuming a molecular density of 1.3 g cm−3 (Matthews, 1968 ▶; Brünger et al., 1998 ▶). A complete summary of the data statistics is given in Table 1 ▶. Initial attempts to solve the structure via molecular-replacement methods using the structure of the M. hyorhinis ortholog of MG289, Cypl (PDB code 3eki), as a model are under way (Sippel et al., 2009 ▶).
Figure 2.
X-ray diffraction pattern of MG289 collected on the X29 beamline (λ = 1.0809 Å) with an ADSC Quantum-315r CCD detector at Brookhaven National Laboratory. The data were collected using a crystal-to-detector distance of 200 mm, an exposure time of 1 s per image and an oscillation range of 1°. (a) A full diffraction image. Concentric rings show the location of the 20, 5, 3 and 2 Å resolution shells. (b) Enlarged view of a selected region of (a). This figure was produced using HKL-2000 (Otwinowski & Minor, 1997 ▶).
Table 1. Summary of data statistics.
Values in parentheses are for the outer resolution shell.
| Temperature (K) | 100 |
| Resolution range (Å) | 50–2.8 (2.9–2.8) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 49.7, b = 90.9, c = 176.1 |
| Wavelength (Å) | 1.0809 |
| Solvent content (%) | 46.1 |
| Crystal dimensions (mm) | 0.07 × 0.05 × 0.05 |
| VM (Å3 Da−1) | 2.37 |
| Total No. of reflections | 104409 |
| No. of unique reflections | 20425 |
| Rmerge† | 0.087 (0.542) |
| Completeness (%) | 99.6 (98.0) |
| Average I/σ(I) | 15.8 (2.2) |
R
merge = 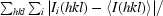
 .
.
Acknowledgments
The authors would like to thank the staff at Brookhaven National Laboratory beamline X29. They would also like to thank Dr Hyun-Joo Nam for help in data collection and Drs Arthur Robbins and Lakshmanan Govindasamy for helpful discussions.
References
- Adams, M. D. & Oxender, D. L. (1989). J. Biol. Chem.264, 15739–15742. [PubMed]
- Anagrius, C., Lore, B. & Jensen, J. S. (2005). Sex. Transm. Infect.81, 458–462. [DOI] [PMC free article] [PubMed]
- Baseman, J. B., Dallo, S. F., Tully, J. G. & Rose, D. L. (1988). J. Clin. Microbiol.26, 2266–2269. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Dudler, R., Schmidhauser, C., Parish, R. W., Wettenhall, R. E. & Schmidt, T. (1988). EMBO J.7, 3963–3970. [DOI] [PMC free article] [PubMed]
- Fraser, C. M. et al. (1995). Science, 270, 397–403. [DOI] [PubMed]
- Gaydos, C. A., Maldeis, N., Hardick, A., Hardick, J. & Quinn, T. C. (2009). Sex. Transm. Infect. doi:10.1136/sti.2008.035477. [DOI] [PMC free article] [PubMed]
- Gilson, E., Alloing, G., Schmidt, T., Claverys, J. P., Dudler, R. & Hofnung, M. (1988). EMBO J.7, 3971–3974. [DOI] [PMC free article] [PubMed]
- Glass, J. I., Assad-Garcia, N., Alperovich, N., Yooseph, S., Lewis, M. R., Maruf, M., Hutchison, C. A. III, Smith, H. O. & Venter, J. C. (2006). Proc. Natl Acad. Sci. USA, 103, 425–430. [DOI] [PMC free article] [PubMed]
- Ketcham, C. M., Anai, S., Reutzel, R., Sheng, S., Schuster, S. M., Brenes, R. B., Agbandje-McKenna, M., McKenna, R., Rosser, C. J. & Boehlein, S. K. (2005). Mol. Cancer Ther.4, 1031–1038. [DOI] [PubMed]
- Manhart, L. E., Holmes, K. K., Hughes, J. P., Houston, L. S. & Totten, P. A. (2007). Am. J. Public Health, 97, 1118–1125. [DOI] [PMC free article] [PubMed]
- Maniloff, J., McElhaney, R. M. & Baseman, J. B. (1992). Editors. Mycoplasmas: Molecular Biology and Pathogenesis. Washington DC: American Society for Microbiology.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: Wiley.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 306–326. [DOI] [PubMed]
- Sippel, K. H., Robbins, A. H., Reutzel, R., Boehlein, S. K., Namiki, K., Goodison, S., Agbandje-McKenna, M., Rosser, C. J. & McKenna, R. (2009). J. Bacteriol.191, 2585–2592. [DOI] [PMC free article] [PubMed]
- Sippel, K. H., Robbins, A. H., Reutzel, R., Domsic, J., Boehlein, S. K., Govindasamy, L., Agbandje-McKenna, M., Rosser, C. J. & McKenna, R. (2008). Acta Cryst. D64, 1172–1178. [DOI] [PubMed]
- Taylor-Robinson, D. (2008). Clin. Infect. Dis.47, 1554–1555. [DOI] [PubMed]
- Tully, J. G., Taylor-Robinson, D., Cole, R. M. & Rose, D. L. (1981). Lancet, 1, 1288–1291. [DOI] [PubMed]