Rhodoquinol-fumarate reductase is a key enzyme in the anaerobic respiratory chain of adult A. suum mitochondria. Its crystallization in the presence of a mixture of octaethyleneglycol monododecyl ether and n-dodecyl-β-d-maltopyranoside in a form suitable for X-ray structure analysis is reported.
Keywords: complex II, eukaryotic rhodoquinol-fumarate reductase, membrane proteins, flutolanil
Abstract
In adult Ascaris suum (roundworm) mitochondrial membrane-bound complex II acts as a rhodoquinol-fumarate reductase, which is the reverse reaction to that of mammalian complex II (succinate-ubiquinone reductase). The adult A. suum rhodoquinol-fumarate reductase was crystallized in the presence of octaethyleneglycol monododecyl ether and n-dodecyl-β-d-maltopyranoside in a 3:2 weight ratio. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 123.75, b = 129.08, c = 221.12 Å, and diffracted to 2.8 Å resolution using synchrotron radiation. The presence of two molecules in the asymmetric unit (120 kDa × 2) gives a crystal volume per protein mass (V M) of 3.6 Å3 Da−1.
1. Introduction
In parasites, fumarate plays an important role in redox homeostasis. The parasitic protozoan Trypanosoma cruzi utilizes bacterial-type dihydroorotate dehydrogenase (TcDHOD), which is the fourth enzyme in the pyrimidine-biosynthesis pathway and catalyzes the oxidation of dihydroorotate and the reduction of fumarate to succinate. We have elucidated the catalytic mechanisms of these sequential reactions by determination of the three-dimensional structures of TcDHOD in the ligand-free form and in complex with the substrates (dihydroorotate and fumarate), product (orotate and succinate) and inhibitor (oxonate) at atomic resolution (Inaoka et al., 2008 ▶).
In parasitic helminths, fumarate is the terminal electron acceptor of the anaerobic respiratory chain, which is essential for the survival of the parasites in the host (Kita & Takamiya, 2002 ▶). Complex II catalyzes fumarate reduction in anaerobic respiration and functions as a terminal oxidase. In eukaryotes, complex II (succinate-ubiquinone reductase in aerobic respiration; SQR) is located in the inner mitochondrial membrane and is generally composed of four polypeptides. The flavoprotein (Fp) subunit is the largest, with an approximate molecular mass of 70 kDa, and contains flavin adenine dinucleotide (FAD) as a prosthetic group. Complex II contains a relatively hydrophilic catalytic region formed by the Fp subunit and an iron–sulfur cluster (Ip) subunit, which has a molecular mass of approximately 30 kDa. The remaining subunits comprise cytochrome b, which contains a haem b. Cytochrome b is composed of two hydrophobic membrane-anchoring polypeptide subunits, namely a 15 kDa large subunit (CybL) and a 13 kDa small subunit (CybS). These cytochrome b subunits are necessary for the interaction of complex II with hydrophobic membrane-associated quinones such as ubiquinone (UQ) and rhodoquinone (RQ).
Our recent study on the respiratory chain of the parasitic nematode Ascaris suum has shown that the mitochondrial NADH-fumarate reductase system plays an important role in the anaerobic energy metabolism of adult parasites inhabiting hosts and that they undergo unique developmental changes during their life cycle (Kita & Takamiya, 2002 ▶; Iwata et al., 2008 ▶). In anaerobic metabolism by A. suum mitochondria, the transfer of a reducing equivalent from NADH to the low-potential RQ is conducted by the NADH–RQ reductase complex (complex I). This pathway ends with the production of succinate by the rhodoquinol-fumarate reductase (QFR) activity of complex II. Electron transfer from NADH to fumarate is coupled to site I phosphorylation of complex I via the generation of a proton gradient. The difference in redox potential between the NAD+/NADH couple (E′m = −320 mV) and the fumarate/succinate couple (E′m = +30 mV) is sufficient to drive ATP synthesis.
The anaerobic NADH-fumarate reductase system is found not only in nematodes but also in bacteria and many other parasites and is a promising target for chemotherapy (Omura et al., 2001 ▶; Tielens et al., 2002 ▶; Matsumoto et al., 2008 ▶). The most potent inhibitor of complex II, atpenin A5, was found during screening for inhibitors of A. suum complex II (Miyadera et al., 2003 ▶). However, the mammalian complex II is much more sensitive to atpenin A5 than the A. suum enzyme. By further screening, we have found that flutolanil, a commercially available fungicide (Ito et al., 2004 ▶), specifically inhibits the A. suum complex II. Therefore, we have taken flutolanil as a lead compound for structure-based drug design. In the current study, we have purified, crystallized and performed preliminary X-ray diffraction studies on the adult A. suum QFR.
2. Methods
2.1. Purification
Mitochondria were prepared from the muscle of adult A. suum as described by Takamiya et al. (1984 ▶), except that Chappell–Perry medium (100 mM KCl, 50 mM Tris–HCl pH 7.4, 1 mM ATP, 5 mM MgSO4 and 1 mM EDTA; Ernster & Nordenbrand, 1967 ▶) was used instead of MSE medium (210 mM mannitol, 70 mM sucrose and 0.1 mM EDTA). The QFR was solubilized from adult A. suum mitochondria in 1.0%(w/v) sucrose monolaurate (Dojindo) and purified in the presence of 0.1%(w/v) sucrose monolaurate. A. suum mitochondria (1 g protein) were homogenized in 500 ml buffer A (10 mM Tris–HCl pH 7.5, 1 mM sodium malonate) containing 1.0%(w/v) sucrose monolaurate. After incubating the mixture for 30 min at 277 K, it was centrifuged for 1 h at 200 000g. The clear reddish-brown supernatant containing the solubilized QFR was applied onto a GE Healthcare DEAE Sepharose FF column (2.6 × 24 cm) pre-equilibrated with buffer A containing 0.1%(w/v) sucrose monolaurate. After washing the column with the same buffer, the QFR was eluted with 2400 ml of the buffer containing a linear gradient of 0.0–0.3 M NaCl. Fractions containing the QFR, which started to elute at approximately 0.1 M NaCl, were pooled and adjusted to 0.15 g ml−1 polyethylene glycol 3350 (Hampton Research) to precipitate the QFR. The mixture was centrifuged for 20 min at 15 000g and the precipitate was dissolved in buffer A containing 0.1%(w/v) sucrose monolaurate. The QFR was then further loaded onto a GE Healthcare Source 15Q column (1.6 × 10 cm) and eluted with 400 ml of buffer containing a linear gradient of 0–0.3 M NaCl. Fractions containing pure QFR as judged by SDS–PAGE (Fig. 1 ▶) were pooled. The purified QFR was then precipitated by adding solid polyethylene glycol (PEG) 3350 to 0.15 g ml−1 and stored at 193 K.
Figure 1.
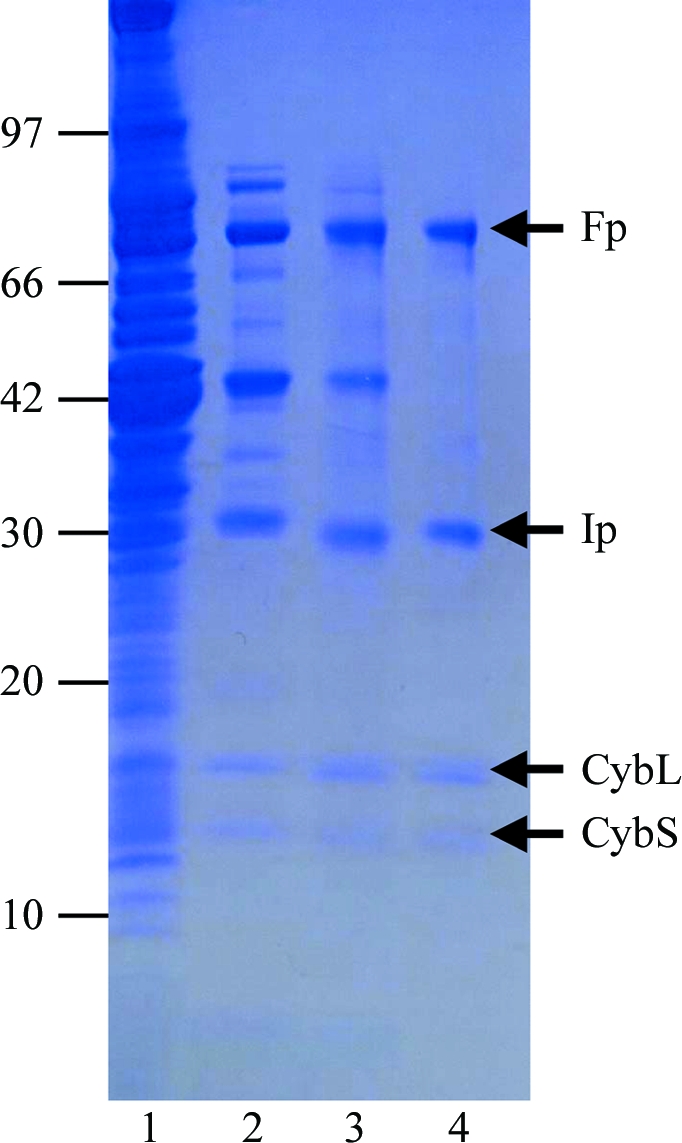
Purity of A. suum QFR at different stages of purification. Samples were separated by SDS–PAGE and the gel was stained with Coomassie Blue. The positions of molecular-weight markers are indicated on the left (in kDa) and the four subunits of A. suum QFR (Fp, flavoprotein subunit; Ip, iron–sulfur cluster subunit; CybL, cytochrome b large subunit; CybS, cytochrome b small subunit) are labelled on the right. Lane 1, whole mitochondria; lane 2, supernatant obtained after ultracentrifugation of the detergent extract; lane 3, pooled fractions from the DEAE Sepharose FF column; lane 4, pooled fractions from the Source 15Q column.
2.2. Crystallization
Conditions for crystallizing the A. suum QFR were screened using Crystal Screen (Jancarik & Kim, 1991 ▶) and Crystal Screen II (Hampton Research). Crystallization by hanging-drop vapour diffusion was carried out using 96-well CrystalClear Strips (Hampton Research). A droplet containing equal volumes of approximately 20 mg ml−1 QFR in buffer A containing detergent and reservoir solution was equilibrated against 100 µl reservoir solution.
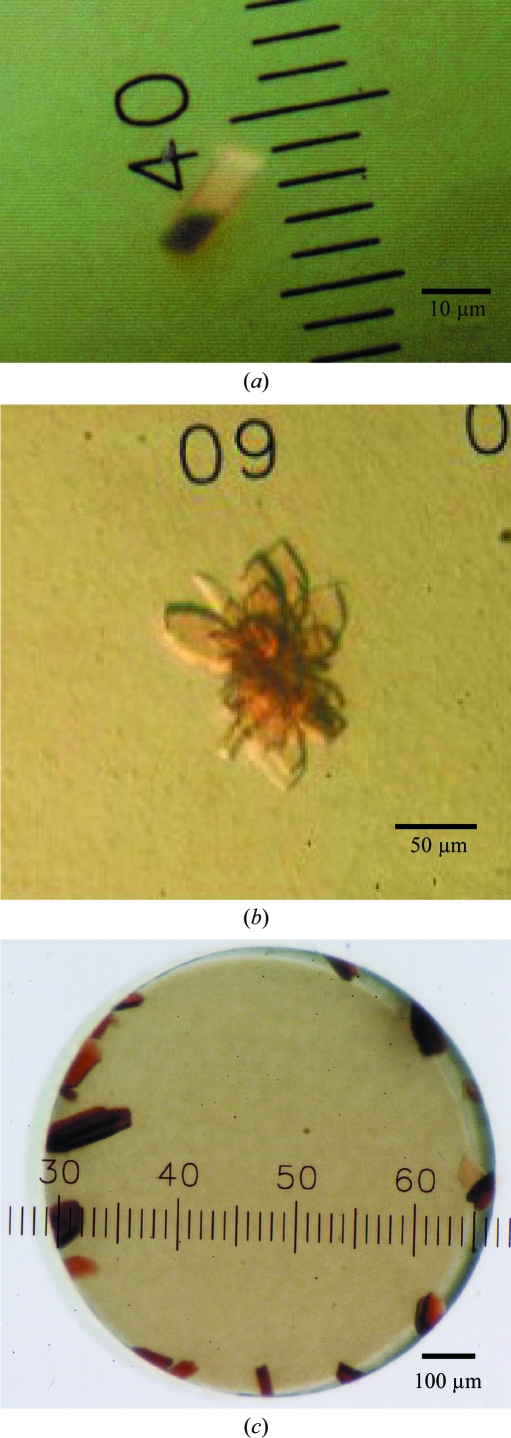
Aggregates of microcrystals were observed at 293 K from reservoir solutions containing polyethylene glycols with medium molecular weights and 200 mM salts when octaethyleneglycol monododecyl ether (C12E8) was used as a detergent. Attempts to optimize the conditions by altering the PEG type (3350, 4000 and 6000), PEG concentration and pH and by using Additive Screen kits (Hampton Research) did not succeed in improving the crystallization. We therefore examined the effect of adding another detergent as an additive. 0.1 volume of detergent solution was added to a droplet of approximately 20 mg ml−1 QFR in buffer A containing 0.5%(w/v) C12E8 and an equal volume of reservoir solution and crystallization by hanging-drop vapour diffusion was carried out. After several days, small crystals (∼10 µm; Fig. 2 ▶ a) appeared at 293 K in drops from reservoir solutions composed of 14–18%(w/v) PEG 3350, 100 mM Tris–HCl pH 7.5–8.6, 200 mM NaCl and 1 mM sodium malonate when the drops included 0.3–0.5%(w/v) n-dodecyl-β-d-maltopyranoside (C12M). To determine the optimal ratio of C12E8 to C12M, crystallization was carried out using QFR dissolved in buffer A containing different concentrations of C12E8 and C12M. The best condition for crystallization (Fig. 2 ▶ b) was achieved at a 3:2 C12E8:C12M weight ratio.
Figure 2.
Typical crystals of A. suum QFR. Crystals of A. suum QFR obtained (a) by the hanging-drop vapour-diffusion method using C12E8 as the main detergent and C12M as the additive detergent, (b) using a 3:2 weight ratio of C12E8 and C12M and (c) after detergent exchange to the 3:2 C12E8:C12M mixture by microdialysis.
Crystals larger than 100 µm in size were grown by the microdialysis method as follows. The precipitate of the purified QFR stored at 193 K was dissolved in buffer A (approximately 10 mg ml−1) containing 0.6%(w/v) C12E8, 0.4%(w/v) C12M and 200 mM NaCl. After incubation for 20 min on ice, the QFR was precipitated by adding an equal volume of 40%(w/v) PEG 3350. The precipitate obtained by centrifugation was dissolved in the same buffer, incubated for 20 min on ice and mixed with an equal volume of 40%(w/v) PEG 3350 to precipitate the QFR. This procedure was repeated several times in order to replace sucrose monolaurate with the added detergent. The precipitate was finally dissolved in buffer A containing 0.06%(w/v) C12E8, 0.04%(w/v) C12M and 0.2 M NaCl, giving an approximately 40 mg ml−1 QFR solution. After adding an equal volume of 23%(w/v) PEG 3350 to the QFR solution, undissolved materials were removed by centrifugation for 20 min at 20 000g. The supernatant was then sealed in a 5 µl microdialysis button and dialyzed against reservoir solution containing 15.0%(w/v) PEG 3350, 100 mM Tris–HCl pH 8.4, 200 mM NaCl, 1 mM sodium malonate, 0.06%(w/v) C12E8 and 0.04%(w/v) C12M. Dark red plate-shaped crystals appeared within 24 h and grew to 100–200 µm after 2–3 d at 293 K (Fig. 2 ▶ c).
X-ray diffraction data were collected on BL44XU at SPring-8 (Harima, Japan) and on BL-5A and NW12A at the Photon Factory (Tsukuba, Japan) by the rotation method. For X-ray diffraction experiments at 100 K, a QFR crystal mounted on a nylon loop was transferred successively to reservoir solution supplemented with 3.75, 7.5, 11.25 and 15% glycerol and was then frozen by rapid submergence in liquid nitrogen. Data were processed and scaled using HKL-2000 and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
To obtain sufficient A. suum mitochondria for purification of the QFR, we slightly modified the standard protocol. Specifically, we used Chappell–Perry medium (Ernster & Nordenbrand, 1967 ▶) in place of the standard medium. This resulted in 0.95 mg mitochondria per gram of muscle and 0.3 µmol min−1 mg−1 mitochondrial succinate dehydrogenase activity, which represents a threefold increase in recovery and a fourfold increase in specific activity compared with the previous method (Takamiya et al., 1984 ▶). Using this method, we obtained 7.5 mg pure enzyme (Fig. 1 ▶, lane 4) from 1 kg of adult A. suum muscle.
Because the success of membrane-protein crystallization strongly depends on which detergent is used, we tested a variety of commercially available nonionic detergents in the screening of crystallization conditions for the A. suum QFR. Aggregates of microcrystals were obtained under several crystallization conditions using the detergent C12E8, but we were unable to optimize the conditions. Instead, the optimal condition for producing crystals suitable for X-ray structure analysis was achieved when the sucrose monolaurate was exchanged for a 3:2 weight ratio of C12E8:C12M (Fig. 2 ▶ c). Crystals grew to larger than 100 µm in 2–3 d by dialyzing QFR, which was dissolved in buffer A containing 11.5%(w/v) PEG 3350, 0.06%(w/v) C12E8, 0.04%(w/v) C12M and 200 mM NaCl, against reservoir solution containing 15.0%(w/v) PEG 3350, 100 mM Tris–HCl pH 8.4, 200 mM NaCl, 1 mM sodium malonate, 0.06%(w/v) C12E8 and 0.04%(w/v) C12M. Adding 11.5%(w/v) PEG 3350 to the QFR solution in advance prevented serious bubble formation in the microdialysis button, which was unfavourable for crystallization.
X-ray diffraction patterns were recorded from a single crystal at 100 K with an oscillation angle of 1.0° using a Bruker DIP-6040 imaging-plate detector on the BL44XU beamline at SPring-8. Analysis of the symmetry and the systematic absences in the recorded diffraction pattern indicated that the crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 123.75, b = 129.08, c = 221.12 Å. Assuming the presence of two QFR molecules (120 kDa × 2) in the asymmetric unit, the calculated Matthews coefficient V M (Matthews, 1968 ▶) was 3.6 Å3 Da−1. A total of 587 189 observed reflections recorded on 180 images were merged to 75 372 unique reflections from 50.0 to 2.8 Å resolution.
During the screening of inhibitors, we found that flutolanil (Ito et al., 2004 ▶), a commercially available fungicide, specifically inhibits A. suum SQR. The IC50 of flutolanil against A. suum and bovine SQR was 0.081 and 16 µM, respectively, indicating that flutolanil is a promising lead compound for anthelminthics. To enable rational drug optimization, we prepared crystals of the A. suum QFR complexed with flutolanil by the soaking method. X-ray diffraction patterns were recorded at 100 K on 130 frames with an oscillation angle of 1° using an ADSC Quantum 315 CCD detector on NW12 at Photon Factory. A total of 54 964 unique reflections from 50.0 to 3.2 Å resolution were obtained. The data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Statistics of data collection and processing.
Values in parentheses are for the highest resolution shell.
| A. suum QFR | A. suum QFR with flutolanil | |
|---|---|---|
| X-ray source | BL44XU (SPring-8) | NW12A (Photon Factory) |
| Wavelength (Å) | 0.900 | 1.000 |
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 123.75 | 124.31 |
| b (Å) | 129.08 | 131.63 |
| c (Å) | 221.12 | 222.53 |
| Resolution range (Å) | 50.0–2.8 (2.9–2.8) | 50.0–3.20 (3.35–3.20) |
| No. of reflections | 587189 | 207156 |
| Unique reflections | 75372 | 54964 |
| Completeness (%) | 89.2 (58.8) | 93.9 (84.1) |
| Rmerge† (%) | 10.5 (36.6) | 11.5 (40.0) |
| I/σ(I) | 8.4 (3.5) | 17.4 (1.6) |
R
merge = 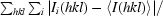
 .
.
We attempted to solve the structure of the A. suum QFR by the molecular-replacement method using the MOLREP program (Navaza, 1994 ▶) as implemented within CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) and the refined coordinates of SQR from pig heart mitochondria (Sun et al., 2005 ▶; PDB code 1zoy). The sequence identities between the pig and A. suum enzymes are 70.4, 68.3, 34.8 and 46.3% for the Fp, Ip, CybL and CybS subunits, respectively. Using X-ray diffraction data in the resolution range 15.0–2.8 Å collected from the flutolanil-free QFR crystal, a promising solution with two molecules per asymmetric unit was obtained and an R factor of 0.45 was achieved when the model was subsequently subjected to rigid-body refinement. Starting from the molecular-replacement solution, the structures of the flutolanil-free and flutolanil-bound forms of the A. suum QFR are currently being refined and electron density corresponding to bound flutolanil has been identified. The structures of the A. suum QFR together with those of the QFRs from Wolinella succinogenes (Lancaster et al., 1999 ▶) and Escherichia coli (Iverson et al., 1999 ▶) and the SQRs from E. coli (Yankovskaya et al., 2003 ▶), pig heart mitochondria (Sun et al., 2005 ▶) and avian heart mitochondria (Huang et al., 2006 ▶) should help to clarify the structure–function relationships in complex II. In addition, the structure of the A. suum QFR complexed with flutolanil should provide information for the structure-based design of anthelminthics.
Acknowledgments
We are grateful to the staff of BL44XU at SPring-8 and the staff of NW12 and BL-5A at Photon Factory for their help with the collection of X-ray diffraction data. This work was supported in part by a grant from the Japan Aerospace Exploration Agency and by Grants-in-Aid for Scientific Research on Priority Areas from the 21st Century COE Program (F-3), for Creative Scientific Research and Targeted Proteins Research Program from the Japanese Ministry of Education, Culture, Sports, Science and Technology (180 73004, 18GS0314 and 1903610), and for Scientific Research (B) from the Japan Society for the Promotion of Science (18370042). DKI was a research fellow supported by the Japan Society for the Promotion of Science.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Ernster, L. & Nordenbrand, K. (1967). Methods Enzymol.10, 86–94. [DOI] [PubMed]
- Huang, L. S., Sun, G., Cobessi, D., Wang, A. C., Shen, J. T., Tung, E. Y., Anderson, V. E. & Berry, E. A. (2006). J. Biol. Chem.281, 5965–5972. [DOI] [PMC free article] [PubMed]
- Inaoka, D. K., Sakamoto, K., Shimizu, H., Shiba, T., Kurisu, G., Nara, T., Aoki, T., Kita, K. & Harada, S. (2008). Biochemistry, 47, 10881–10891. [DOI] [PubMed]
- Ito, Y., Muraguchi, H., Seshime, Y., Oita, S. & Yanagi, S. O. (2004). Mol. Genet. Genomics, 272, 328–335. [DOI] [PubMed]
- Iverson, T. M., Luna-Chaves, C., Cecchini, G. & Rees, D. C. (1999). Science, 284, 1961–1966. [DOI] [PubMed]
- Iwata, F., Shinjyo, N., Amino, H., Sakamoto, K., Islam, M. K., Tsuji, N. & Kita, K. (2008). Parasitol. Int.57, 54–61. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Kita, K. & Takamiya, S. (2002). Adv. Parasitol.51, 95–131. [DOI] [PubMed]
- Lancaster, C. R. D., Kröger, A., Auer, M. & Michel, H. (1999). Nature (London), 402, 377–385. [DOI] [PubMed]
- Matsumoto, J., Sakamoto, K., Shinjyo, N., Kido, Y., Yamamoto, N., Yagi, K., Miyoshi, H., Nonaka, N., Katakura, K., Kita, K. & Oku, Y. (2008). Antimicrob. Agents. Chemother.52, 164–170. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Miyadera, H., Shiomi, K., Ui, H., Yamaguchi, Y., Masuma, R., Tomoda, H., Miyoshi, H., Osanai, A., Kita, K. & Omura, S. (2003). Proc. Natl Acad. Sci. USA, 100, 473–477. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Omura, S. et al. (2001). Proc. Natl Acad. Sci. USA, 98, 60–62.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sun, F., Huo, X., Zhai, Y., Wang, A., Xu, J., Su, D., Bartlam, M. & Rao, Z. (2005). Cell, 121, 1043–1057. [DOI] [PubMed]
- Takamiya, S., Furushima, R. & Oya, H. (1984). Mol. Biochem. Parasitol.13, 121–134. [DOI] [PubMed]
- Tielens, A. G. M., Rotte, C., van Hellemond, J. J. & Martin, W. (2002). Trends Biochem. Sci.27, 564–572. [DOI] [PubMed]
- Yankovskaya, V., Horsefield, R., Törnroth, S., Luna-Chaves, C., Miyoshi, H., Léger, C., Byrne, B., Cecchini, C. & Iwata, S. (2003). Science, 299, 700–704. [DOI] [PubMed]